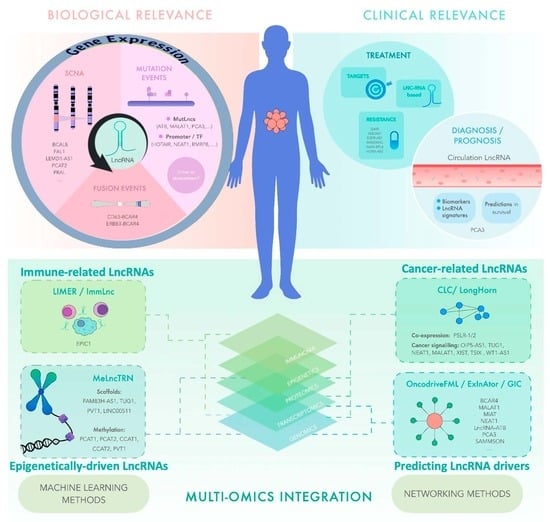
Multi-Omics Mining of lncRNAs with Biological and Clinical Relevance in Cancer
Abstract
:1. Introduction
2. lncRNAs Associated with Cancer Driver Somatic Mutations Profiles
3. Approaches for Prioritizing Cancer Driver lncRNAs Using Somatic Mutations Profiles
4. Copy-Number Alterations in Genomic Regions Encoding lncRNAs
5. The Revolution of Non-Coding Transcriptome in Cancer Studies
6. Multi-Omics Network Approaches Reveal lncRNA Biological Relevance on Cancer Biology
Describing the Novel lncRNAs Drivers in Cancer through Multi-Omics Integration
7. Challenges and Opportunities of Machine Learning to Deepen the Functional and Biological Roles of lncRNAs in Cancer
8. Perspective on the New Directions in lncRNA Research and Its Implication in Cancer through Multi-Omics Analysis
9. Perspective on the Future Use of lncRNAs for Therapeutic Purposes through Multi-Omics Oncology
10. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Francies, H.E.; McDermott, U.; Garnett, M.J. Genomics-Guided Pre-Clinical Development of Cancer Therapies. Nat. Cancer 2020, 1, 482–492. [Google Scholar] [CrossRef] [PubMed]
- Malone, E.R.; Oliva, M.; Sabatini, P.J.B.; Stockley, T.L.; Siu, L.L. Molecular Profiling for Precision Cancer Therapies. Genome Med. 2020, 12, 8. [Google Scholar] [CrossRef]
- Perakakis, N.; Yazdani, A.; Karniadakis, G.E.; Mantzoros, C. Omics, Big Data and Machine Learning as Tools to Propel Understanding of Biological Mechanisms and to Discover Novel Diagnostics and Therapeutics. Metabolism 2018, 87, A1–A9. [Google Scholar] [CrossRef] [PubMed]
- Vlachavas, E.I.; Bohn, J.; Ückert, F.; Nürnberg, S. A Detailed Catalogue of Multi-Omics Methodologies for Identification of Putative Biomarkers and Causal Molecular Networks in Translational Cancer Research. Int. J. Mol. Sci. 2021, 22, 2822. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A Revolutionary Tool for Transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Hong, M.; Tao, S.; Zhang, L.; Diao, L.-T.; Huang, X.; Huang, S.; Xie, S.-J.; Xiao, Z.-D.; Zhang, H. RNA Sequencing: New Technologies and Applications in Cancer Research. J. Hematol. Oncol. 2020, 13, 166. [Google Scholar] [CrossRef]
- Adelman, K.; Egan, E. Non-Coding RNA: More Uses for Genomic Junk. Nature 2017, 543, 183–185. [Google Scholar] [CrossRef]
- Moore, J.E.; Purcaro, M.J.; Pratt, H.E.; Epstein, C.B.; Shoresh, N.; Adrian, J.; Kawli, T.; Davis, C.A.; Dobin, A.; ENCODE Project Consortium; et al. Expanded Encyclopaedias of DNA Elements in the Human and Mouse Genomes. Nature 2020, 583, 699–710. [Google Scholar]
- Iyer, M.K.; Niknafs, Y.S.; Malik, R.; Singhal, U.; Sahu, A.; Hosono, Y.; Barrette, T.R.; Prensner, J.R.; Evans, J.R.; Zhao, S.; et al. The Landscape of Long Noncoding RNAs in the Human Transcriptome. Nat. Genet. 2015, 47, 199–208. [Google Scholar] [CrossRef]
- Zhang, X.; Meyerson, M. Illuminating the Noncoding Genome in Cancer. Nat. Cancer 2020, 1, 864–872. [Google Scholar] [CrossRef]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef] [PubMed]
- Matsui, M.; Corey, D.R. Non-Coding RNAs as Drug Targets. Nat. Rev. Drug Discov. 2017, 16, 167–179. [Google Scholar] [CrossRef]
- Amodio, N.; Stamato, M.A.; Juli, G.; Morelli, E.; Fulciniti, M.; Manzoni, M.; Taiana, E.; Agnelli, L.; Cantafio, M.E.G.; Romeo, E.; et al. Drugging the lncRNA MALAT1 via LNA gapmeR ASO Inhibits Gene Expression of Proteasome Subunits and Triggers Anti-Multiple Myeloma Activity. Leukemia 2018, 32, 1948–1957. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Zheng, F.; Liu, Z.; Wu, J.; Chai, X.; He, C.; Li, L.; Hann, S.S. Novel Reciprocal Interaction of lncRNA HOTAIR and miR-214-3p Contribute to the Solamargine-Inhibited PDPK1 Gene Expression in Human Lung Cancer. J. Cell. Mol. Med. 2019, 23, 7749–7761. [Google Scholar] [CrossRef]
- Cieślik, M.; Chinnaiyan, A.M. Cancer Transcriptome Profiling at the Juncture of Clinical Translation. Nat. Rev. Genet. 2018, 19, 93–109. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Hu, L.; Zhang, Z.; Wu, N.; Sun, J.; Su, J. Recurrence-Associated Long Non-Coding RNA Signature for Determining the Risk of Recurrence in Patients with Colon Cancer. Mol. Ther. Nucleic Acids 2018, 12, 518–529. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Zhao, J.; Sun, T.; Shen, Z. A Machine Learning Framework That Integrates Multi-Omics Data Predicts Cancer-Related lncRNAs. BMC Bioinform. 2021, 22, 332. [Google Scholar] [CrossRef] [PubMed]
- Heo, Y.J.; Hwa, C.; Lee, G.-H.; Park, J.-M.; An, J.-Y. Integrative Multi-Omics Approaches in Cancer Research: From Biological Networks to Clinical Subtypes. Mol. Cells 2021, 44, 433–443. [Google Scholar] [CrossRef]
- Qian, Y.; Shi, L.; Luo, Z. Long Non-Coding RNAs in Cancer: Implications for Diagnosis, Prognosis, and Therapy. Front. Med. 2020, 7, 612393. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Zhang, Z.; Bao, S.; Hou, P.; Zhou, M.; Xu, C.; Sun, J. Computational Methods and Applications for Identifying Disease-Associated lncRNAs as Potential Biomarkers and Therapeutic Targets. Mol. Ther. Nucleic Acids 2020, 21, 156–171. [Google Scholar] [CrossRef]
- Chen, G.; Wang, Z.; Wang, D.; Qiu, C.; Liu, M.; Chen, X.; Zhang, Q.; Yan, G.; Cui, Q. lncRNADisease: A Database for Long-Non-Coding RNA-Associated Diseases. Nucleic Acids Res. 2013, 41, D983–D986. [Google Scholar] [CrossRef]
- Aprile, M.; Katopodi, V.; Leucci, E.; Costa, V. lncRNAs in Cancer: From Garbage to Junk. Cancers 2020, 12, 3220. [Google Scholar] [CrossRef]
- Sheng, N.; Huang, L.; Gao, L.; Cao, Y.; Xie, X.; Wang, Y. A Survey of Computational Methods and Databases for lncRNA-MiRNA Interaction Prediction. IEEE/ACM Trans. Comput. Biol. Bioinform. 2023, 20, 2810–2826. [Google Scholar] [CrossRef]
- Raufaste-Cazavieille, V.; Santiago, R.; Droit, A. Multi-Omics Analysis: Paving the Path toward Achieving Precision Medicine in Cancer Treatment and Immuno-Oncology. Front. Mol. Biosci. 2022, 9, 962743. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Negrini, S.; Gorgoulis, V.G.; Halazonetis, T.D. Genomic Instability—An Evolving Hallmark of Cancer. Nat. Rev. Mol. Cell Biol. 2010, 11, 220–228. [Google Scholar] [CrossRef]
- Vogelstein, B.; Papadopoulos, N.; Velculescu, V.E.; Zhou, S.; Diaz, L.A., Jr.; Kinzler, K.W. Cancer Genome Landscapes. Science 2013, 339, 1546–1558. [Google Scholar] [CrossRef]
- Lawrence, M.S.; Stojanov, P.; Polak, P.; Kryukov, G.V.; Cibulskis, K.; Sivachenko, A.; Carter, S.L.; Stewart, C.; Mermel, C.H.; Roberts, S.A.; et al. Mutational Heterogeneity in Cancer and the Search for New Cancer-Associated Genes. Nature 2013, 499, 214–218. [Google Scholar] [CrossRef]
- Korf, B.R.; Rehm, H.L. New Approaches to Molecular Diagnosis. JAMA 2013, 309, 1511–1521. [Google Scholar] [CrossRef]
- Piraino, S.W.; Furney, S.J. Beyond the Exome: The Role of Non-Coding Somatic Mutations in Cancer. Ann. Oncol. 2016, 27, 240–248. [Google Scholar] [CrossRef]
- Rheinbay, E.; Nielsen, M.M.; Abascal, F.; Wala, J.A.; Shapira, O.; Tiao, G.; Hornshøj, H.; Hess, J.M.; Juul, R.I.; Lin, Z.; et al. Analyses of Non-Coding Somatic Drivers in 2658 Cancer Whole Genomes. Nature 2020, 578, 102–111. [Google Scholar] [CrossRef]
- Rheinbay, E.; Parasuraman, P.; Grimsby, J.; Tiao, G.; Engreitz, J.M.; Kim, J.; Lawrence, M.S.; Taylor-Weiner, A.; Rodriguez-Cuevas, S.; Rosenberg, M.; et al. Recurrent and Functional Regulatory Mutations in Breast Cancer. Nature 2017, 547, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, A.; Furuta, M.; Totoki, Y.; Tsunoda, T.; Kato, M.; Shiraishi, Y.; Tanaka, H.; Taniguchi, H.; Kawakami, Y.; Ueno, M.; et al. Whole-Genome Mutational Landscape and Characterization of Noncoding and Structural Mutations in Liver Cancer. Nat. Genet. 2016, 48, 500–509. [Google Scholar] [CrossRef] [PubMed]
- Gasic, V.; Karan-Djurasevic, T.; Pavlovic, D.; Zukic, B.; Pavlovic, S.; Tosic, N. Diagnostic and Therapeutic Implications of Long Non-Coding RNAs in Leukemia. Life 2022, 12, 1770. [Google Scholar] [CrossRef]
- Puente, X.S.; Beà, S.; Valdés-Mas, R.; Villamor, N.; Gutiérrez-Abril, J.; Martín-Subero, J.I.; Munar, M.; Rubio-Pérez, C.; Jares, P.; Aymerich, M.; et al. Non-Coding Recurrent Mutations in Chronic Lymphocytic Leukaemia. Nature 2015, 526, 519–524. [Google Scholar] [CrossRef]
- Nik-Zainal, S.; Davies, H.; Staaf, J.; Ramakrishna, M.; Glodzik, D.; Zou, X.; Martincorena, I.; Alexandrov, L.B.; Martin, S.; Wedge, D.C.; et al. Author Correction: Landscape of Somatic Mutations in 560 Breast Cancer Whole-Genome Sequences. Nature 2019, 566, E1. [Google Scholar] [CrossRef]
- Rezaie, N.; Bayati, M.; Hamidi, M.; Tahaei, M.S.; Khorasani, S.; Lovell, N.H.; Breen, J.; Rabiee, H.R.; Alinejad-Rokny, H. Somatic Point Mutations Are Enriched in Non-Coding RNAs with Possible Regulatory Function in Breast Cancer. Commun. Biol. 2022, 5, 556. [Google Scholar] [CrossRef]
- Iraola-Guzmán, S.; Brunet-Vega, A.; Pegueroles, C.; Saus, E.; Hovhannisyan, H.; Casalots, A.; Pericay, C.; Gabaldón, T. Target Enrichment Enables the Discovery of lncRNAs with Somatic Mutations or Altered Expression in Paraffin-Embedded Colorectal Cancer Samples. Cancers 2020, 12, 2844. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Liu, X.; Lu, L.; Liu, G. Identification of a Somatic Mutation-Derived Long Non-Coding RNA Signatures of Genomic Instability in Renal Cell Carcinoma. Front. Oncol. 2021, 11, 728181. [Google Scholar] [CrossRef]
- Wang, Y.; Yan, K.; Wang, L.; Bi, J. Genome Instability-Related Long Non-Coding RNA in Clear Renal Cell Carcinoma Determined Using Computational Biology. BMC Cancer 2021, 21, 727. [Google Scholar] [CrossRef]
- Xu, Q.; Guo, T. Somatic Mutation-Associated Risk Index Based on lncRNA Expression for Predicting Prognosis in Acute Myeloid Leukemia. Hematology 2022, 27, 659–671. [Google Scholar] [CrossRef]
- Yun, D.; Yang, Z. Identification of a Four-lncRNA Prognostic Signature for Colon Cancer Based on Genome Instability. J. Oncol. 2021, 2021, 7408893. [Google Scholar] [CrossRef]
- Yin, T.; Zhao, D.; Yao, S. Identification of a Genome Instability-Associated lncRNA Signature for Prognosis Prediction in Colon Cancer. Front. Genet. 2021, 12, 679150. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ding, N.; He, Y.; Tao, C.; Liang, Z.; Xin, W.; Zhang, Q.; Wang, F. Bioinformatic Identification of Genomic Instability-Associated lncRNAs Signatures for Improving the Clinical Outcome of Cervical Cancer by a Prognostic Model. Sci. Rep. 2021, 11, 20929. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Zhao, J.-S.; Huang, X.-Q.; Yang, X.-Z.; Niu, F.-Y.; Lin, J.-R.; Ma, L.; Shi, Y.-X.; Li, X.-S.; Jiang, P.; et al. A Somatic Mutation-Derived lncRNA Signatures of Genomic Instability Predicts the Prognosis and Tumor Microenvironment Immune Characters in Hepatocellular Carcinoma. Hepatol. Int. 2022, 16, 1220–1233. [Google Scholar] [CrossRef]
- Wu, J.; Ren, X.; Wang, N.; Zhou, R.; Chen, M.; Cai, Y.; Lin, S.; Zhang, H.; Xie, X.; Dang, C.; et al. A Mutation-Related Long Noncoding RNA Signature of Genome Instability Predicts Immune Infiltration and Hepatocellular Carcinoma Prognosis. Front. Genet. 2021, 12, 779554. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Hu, Z.; Feng, Y.; Hu, X.; Yuan, J.; Zhao, S.D.; Zhang, Y.; Yang, L.; Shan, W.; He, Q.; et al. Comprehensive Genomic Characterization of Long Non-Coding RNAs across Human Cancers. Cancer Cell 2015, 28, 529–540. [Google Scholar] [CrossRef]
- Zhong, Q.; Lu, M.; Yuan, W.; Cui, Y.; Ouyang, H.; Fan, Y.; Wang, Z.; Wu, C.; Qiao, J.; Hang, J. Eight-lncRNA Signature of Cervical Cancer Were Identified by Integrating DNA Methylation, Copy Number Variation and Transcriptome Data. J. Transl. Med. 2021, 19, 58. [Google Scholar] [CrossRef]
- Zhou, C.-C.; Yang, F.; Yuan, S.-X.; Ma, J.-Z.; Liu, F.; Yuan, J.-H.; Bi, F.-R.; Lin, K.-Y.; Yin, J.-H.; Cao, G.-W.; et al. Systemic Genome Screening Identifies the Outcome Associated Focal Loss of Long Noncoding RNA PRAL in Hepatocellular Carcinoma. Hepatology 2016, 63, 850–863. [Google Scholar] [CrossRef]
- Liu, H.; Gu, X.; Wang, G.; Huang, Y.; Ju, S.; Huang, J.; Wang, X. Copy Number Variations Primed lncRNAs Deregulation Contribute to Poor Prognosis in Colorectal Cancer. Aging 2019, 11, 6089–6108. [Google Scholar] [CrossRef]
- Zhang, Y.; Liao, G.; Bai, J.; Zhang, X.; Xu, L.; Deng, C.; Yan, M.; Xie, A.; Luo, T.; Long, Z.; et al. Identifying Cancer Driver lncRNAs Bridged by Functional Effectors through Integrating Multi-Omics Data in Human Cancers. Mol. Ther. Nucleic Acids 2019, 17, 362–373. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Tan, Z.-Y.; Fang, X.-C. Identification of m6A-Related lncRNAs-Based Signature for Predicting the Prognosis of Patients with Skin Cutaneous Melanoma. SLAS Technol. 2023, in press. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Yu, R.; Peng, Y.; Miao, Y.; Jiang, K.; Li, Q. Identification of Genomic Instability Related lncRNA Signature with Prognostic Value and Its Role in Cancer Immunotherapy in Pancreatic Cancer. Front. Genet. 2022, 13, 990661. [Google Scholar] [CrossRef]
- Ning, J.; Wang, F.; Zhu, K.; Li, B.; Shu, Q.; Liu, W. Characterizing the Copy Number Variation of Non-Coding RNAs Reveals Potential Therapeutic Targets and Prognostic Markers of LUSC. Front. Genet. 2021, 12, 779155. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Xiao, L.; Li, J.; Dong, B.; Wang, C. Integrative Analysis Reveals Driver Long Non-Coding RNAs in Osteosarcoma. Medicine 2019, 98, e14302. [Google Scholar] [CrossRef]
- Athie, A.; Marchese, F.P.; González, J.; Lozano, T.; Raimondi, I.; Juvvuna, P.K.; Abad, A.; Marin-Bejar, O.; Serizay, J.; Martínez, D.; et al. Analysis of Copy Number Alterations Reveals the lncRNA ALAL-1 as a Regulator of Lung Cancer Immune Evasion. J. Cell Biol. 2020, 219, e201908078. [Google Scholar] [CrossRef]
- Cheng, Z.; Guo, Y.; Sun, J.; Zheng, L. Four-Copy Number Alteration (CNA)-Related lncRNA Prognostic Signature for Liver Cancer. Sci. Rep. 2022, 12, 14261. [Google Scholar] [CrossRef]
- Du, Z.; Fei, T.; Verhaak, R.G.W.; Su, Z.; Zhang, Y.; Brown, M.; Chen, Y.; Liu, X.S. Integrative Genomic Analyses Reveal Clinically Relevant Long Noncoding RNAs in Human Cancer. Nat. Struct. Mol. Biol. 2013, 20, 908–913. [Google Scholar] [CrossRef]
- Zheng, M.; Hu, Y.; Gou, R.; Nie, X.; Li, X.; Liu, J.; Lin, B. Identification Three lncRNA Prognostic Signature of Ovarian Cancer Based on Genome-Wide Copy Number Variation. Biomed. Pharmacother. 2020, 124, 109810. [Google Scholar] [CrossRef]
- Hou, X.; Ma, B.; Liu, M.; Zhao, Y.; Chai, B.; Pan, J.; Wang, P.; Li, D.; Liu, S.; Song, F. The Transcriptional Risk Scores for Kidney Renal Clear Cell Carcinoma Using XGBoost and Multiple Omics Data. Math. Biosci. Eng. 2023, 20, 11676–11687. [Google Scholar] [CrossRef]
- Peng, W.; Bai, S.; Zheng, M.; Chen, W.; Li, Y.; Yang, Y.; Zhao, Y.; Xiong, S.; Wang, R.; Cheng, B. An Exosome-Related lncRNA Signature Correlates with Prognosis, Immune Microenvironment, and Therapeutic Responses in Hepatocellular Carcinoma. Transl. Oncol. 2023, 31, 101651. [Google Scholar] [CrossRef]
- Yu, Z.; Zhu, Y.; Ji, J. Multi-Omics Analysis Revealing a Senescence-Relevant lncRNAs Signature for the Assessment of Response to Immunotherapy for Breast Cancer. Medicine 2023, 102, e34287. [Google Scholar] [CrossRef]
- Zhou, Q.-N.; Lei, R.-E.; Liang, Y.-X.; Li, S.-Q.; Guo, X.-W.; Hu, B.-L. Oxaliplatin Related lncRNAs Prognostic Models Predict the Prognosis of Patients given Oxaliplatin-Based Chemotherapy. Cancer Cell Int. 2023, 23, 103. [Google Scholar] [CrossRef]
- Li, G.; Huo, D.; Guo, N.; Li, Y.; Ma, H.; Liu, L.; Xie, H.; Zhang, D.; Qu, B.; Chen, X. Integrating Multiple Machine Learning Algorithms for Prognostic Prediction of Gastric Cancer Based on Immune-Related lncRNAs. Front. Genet. 2023, 14, 1106724. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Ren, W.; He, Z.; Chen, Y.; Tan, Y.; Mao, L.; Ouyang, W.; Lu, N.; Ouyang, J.; Chen, K.; et al. Machine Learning Radiomics of Magnetic Resonance Imaging Predicts Recurrence-Free Survival after Surgery and Correlation of lncRNAs in Patients with Breast Cancer: A Multicenter Cohort Study. Breast Cancer Res. 2023, 25, 132. [Google Scholar] [CrossRef] [PubMed]
- Maimaiti, A.; Aili, Y.; Turhon, M.; Kadeer, K.; Aikelamu, P.; Wang, Z.; Niu, W.; Aisha, M.; Kasimu, M.; Wang, Y.; et al. Modification Patterns of DNA Methylation-Related lncRNAs Regulating Genomic Instability for Improving the Clinical Outcomes and Tumour Microenvironment Characterisation of Lower-Grade Gliomas. Front. Mol. Biosci. 2022, 9, 844973. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wang, R.; Gao, F.; Huang, J.; Zhao, X.; Li, D. Pan-Cancer Analysis of the DNA Methylation Patterns of Long Non-Coding RNA. Genomics 2022, 114, 110377. [Google Scholar] [CrossRef]
- Song, P.; Li, Y.; Wang, F.; Pu, L.; Bao, L.; Gao, H.; Zhu, C.; Wang, M.; Tao, L. Genome-Wide Screening for Differentially Methylated Long Noncoding RNAs Identifies LIFR-AS1 as an Epigenetically Regulated lncRNA That Inhibits the Progression of Colorectal Cancer. Clin. Epigenetics 2022, 14, 138. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Li, X.; Zhi, H.; Zhang, Y.; Wang, P.; Wang, Y.; Shang, S.; Fang, Y.; Shen, W.; Ning, S.; et al. Comprehensive Characterization of Somatic Mutations Impacting lncRNA Expression for Pan-Cancer. Mol. Ther. Nucleic Acids 2019, 18, 66–79. [Google Scholar] [CrossRef]
- Esposito, R.; Lanzós, A.; Uroda, T.; Ramnarayanan, S.; Büchi, I.; Polidori, T.; Guillen-Ramirez, H.; Mihaljevic, A.; Merlin, B.M.; Mela, L.; et al. Tumour Mutations in Long Noncoding RNAs Enhance Cell Fitness. Nat. Commun. 2023, 14, 3342. [Google Scholar] [CrossRef]
- Zhang, Y.; Han, P.; Guo, Q.; Hao, Y.; Qi, Y.; Xin, M.; Zhang, Y.; Cui, B.; Wang, P. Oncogenic Landscape of Somatic Mutations Perturbing Pan-Cancer lncRNA-ceRNA Regulation. Front. Cell Dev. Biol. 2021, 9, 658346. [Google Scholar] [CrossRef]
- Brady, S.W.; Gout, A.M.; Zhang, J. Therapeutic and Prognostic Insights from the Analysis of Cancer Mutational Signatures. Trends Genet. 2022, 38, 194–208. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, A.; Ricci, A.D.; Di Federico, A.; Frega, G.; Palloni, A.; Tavolari, S.; Brandi, G. Predictive Biomarkers for Checkpoint Inhibitor-Based Immunotherapy in Hepatocellular Carcinoma: Where Do We Stand? Front. Oncol. 2021, 11, 803133. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.; Shan, Z.; Li, M.; Xia, Y.; Jin, Z. Exploration of the Associations of lncRNA Expression Patterns with Tumor Mutation Burden and Prognosis in Colon Cancer. OncoTargets Ther. 2021, 14, 2893–2909. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.J.-K.; Park, S.; Park, H.; Kim, S.; Lee, J.; Lee, J.; Youk, J.; Yi, K.; An, Y.; Park, I.K.; et al. Tracing Oncogene Rearrangements in the Mutational History of Lung Adenocarcinoma. Cell 2019, 177, 1842–1857.e21. [Google Scholar] [CrossRef] [PubMed]
- Bae, K.; Kim, J.H.; Jung, H.; Kong, S.-Y.; Kim, Y.-H.; Kim, S.; Lee, G.K.; Lee, J.S.; Lee, J.J.-K.; Ju, Y.S.; et al. A Fusion of CD63-BCAR4 Identified in Lung Adenocarcinoma Promotes Tumorigenicity and Metastasis. Br. J. Cancer 2021, 124, 290–298. [Google Scholar] [CrossRef] [PubMed]
- Jordan, E.J.; Kim, H.R.; Arcila, M.E.; Barron, D.; Chakravarty, D.; Gao, J.; Chang, M.T.; Ni, A.; Kundra, R.; Jonsson, P.; et al. Prospective Comprehensive Molecular Characterization of Lung Adenocarcinomas for Efficient Patient Matching to Approved and Emerging Therapies. Cancer Discov. 2017, 7, 596–609. [Google Scholar] [CrossRef] [PubMed]
- Koivunen, J.P.; Mermel, C.; Zejnullahu, K.; Murphy, C.; Lifshits, E.; Holmes, A.J.; Choi, H.G.; Kim, J.; Chiang, D.; Thomas, R.; et al. EML4-ALK Fusion Gene and Efficacy of an ALK Kinase Inhibitor in Lung Cancer. Clin. Cancer Res. 2008, 14, 4275–4283. [Google Scholar] [CrossRef]
- Wang, C.; Yin, R.; Dai, J.; Gu, Y.; Cui, S.; Ma, H.; Zhang, Z.; Huang, J.; Qin, N.; Jiang, T.; et al. Whole-Genome Sequencing Reveals Genomic Signatures Associated with the Inflammatory Microenvironments in Chinese NSCLC Patients. Nat. Commun. 2018, 9, 2054. [Google Scholar] [CrossRef]
- Godinho, M.F.E.; Sieuwerts, A.M.; Look, M.P.; Meijer, D.; Foekens, J.A.; Dorssers, L.C.J.; van Agthoven, T. Relevance of BCAR4 in Tamoxifen Resistance and Tumour Aggressiveness of Human Breast Cancer. Br. J. Cancer 2010, 103, 1284–1291. [Google Scholar] [CrossRef]
- Guichard, C.; Amaddeo, G.; Imbeaud, S.; Ladeiro, Y.; Pelletier, L.; Maad, I.B.; Calderaro, J.; Bioulac-Sage, P.; Letexier, M.; Degos, F.; et al. Integrated Analysis of Somatic Mutations and Focal Copy-Number Changes Identifies Key Genes and Pathways in Hepatocellular Carcinoma. Nat. Genet. 2012, 44, 694–698. [Google Scholar] [CrossRef]
- Shao, X.; Lv, N.; Liao, J.; Long, J.; Xue, R.; Ai, N.; Xu, D.; Fan, X. Copy Number Variation Is Highly Correlated with Differential Gene Expression: A Pan-Cancer Study. BMC Med. Genet. 2019, 20, 175. [Google Scholar] [CrossRef] [PubMed]
- Zack, T.I.; Schumacher, S.E.; Carter, S.L.; Cherniack, A.D.; Saksena, G.; Tabak, B.; Lawrence, M.S.; Zhsng, C.-Z.; Wala, J.; Mermel, C.H.; et al. Pan-Cancer Patterns of Somatic Copy Number Alteration. Nat. Genet. 2013, 45, 1134–1140. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, L.; Kucherlapati, M.; Chen, F.; Hadjipanayis, A.; Pantazi, A.; Bristow, C.A.; Lee, E.A.; Mahadeshwar, H.S.; Tang, J.; et al. A Pan-Cancer Compendium of Genes Deregulated by Somatic Genomic Rearrangement across More Than 1400 Cases. Cell Rep. 2018, 24, 515–527. [Google Scholar] [CrossRef] [PubMed]
- Beroukhim, R.; Mermel, C.H.; Porter, D.; Wei, G.; Raychaudhuri, S.; Donovan, J.; Barretina, J.; Boehm, J.S.; Dobson, J.; Urashima, M.; et al. The Landscape of Somatic Copy-Number Alteration across Human Cancers. Nature 2010, 463, 899–905. [Google Scholar] [CrossRef]
- Ping, Y.; Zhou, Y.; Hu, J.; Pang, L.; Xu, C.; Xiao, Y. Dissecting the Functional Mechanisms of Somatic Copy-Number Alterations Based on Dysregulated ceRNA Networks across Cancers. Mol. Ther. Nucleic Acids 2020, 21, 464–479. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Feng, Y.; Zhang, D.; Zhao, S.D.; Hu, Z.; Greshock, J.; Zhang, Y.; Yang, L.; Zhong, X.; Wang, L.-P.; et al. A Functional Genomic Approach Identifies FAL1 as an Oncogenic Long Noncoding RNA That Associates with BMI1 and Represses p21 Expression in Cancer. Cancer Cell 2014, 26, 344–357. [Google Scholar] [CrossRef]
- Wang, Z.-L.; Li, B.; Piccolo, S.R.; Zhang, X.-Q.; Li, J.-H.; Zhou, H.; Yang, J.-H.; Qu, L.-H. Integrative Analysis Reveals Clinical Phenotypes and Oncogenic Potentials of Long Non-Coding RNAs across 15 Cancer Types. Oncotarget 2016, 7, 35044–35055. [Google Scholar] [CrossRef]
- Zheng, X.; Li, F.; Zhao, H.; Tang, Y.; Xue, K.; Zhang, X.; Liang, W.; Zhao, R.; Lv, X.; Song, X.; et al. A Novel Method to Identify and Characterize Personalized Functional Driver lncRNAs in Cancer Samples. Comput. Struct. Biotechnol. J. 2023, 21, 2471–2482. [Google Scholar] [CrossRef]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of Transcription in Human Cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef]
- Wei, R.; Zhao, M.; Zheng, C.-H.; Zhao, M.; Xia, J. Concordance between Somatic Copy Number Loss and down-Regulated Expression: A Pan-Cancer Study of Cancer Predisposition Genes. Sci. Rep. 2016, 6, 37358. [Google Scholar] [CrossRef]
- Dagogo-Jack, I.; Shaw, A.T. Tumour Heterogeneity and Resistance to Cancer Therapies. Nat. Rev. Clin. Oncol. 2018, 15, 81–94. [Google Scholar] [CrossRef]
- Fisher, R.; Pusztai, L.; Swanton, C. Cancer Heterogeneity: Implications for Targeted Therapeutics. Br. J. Cancer 2013, 108, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Xu, J.; Hong, J.; Tang, R.; Zhang, X.; Fang, J.-Y. Long Noncoding RNA Profiles Identify Five Distinct Molecular Subtypes of Colorectal Cancer with Clinical Relevance. Mol. Oncol. 2014, 8, 1393–1403. [Google Scholar] [CrossRef] [PubMed]
- Akrami, R.; Jacobsen, A.; Hoell, J.; Schultz, N.; Sander, C.; Larsson, E. Comprehensive Analysis of Long Non-Coding RNAs in Ovarian Cancer Reveals Global Patterns and Targeted DNA Amplification. PLoS ONE 2013, 8, e80306. [Google Scholar] [CrossRef]
- Volders, P.-J.; Anckaert, J.; Verheggen, K.; Nuytens, J.; Martens, L.; Mestdagh, P.; Vandesompele, J. LNCipedia 5: Towards a Reference Set of Human Long Non-Coding RNAs. Nucleic Acids Res. 2019, 47, D135–D139. [Google Scholar] [CrossRef]
- Sun, Y.; Ma, L. New Insights into Long Non-Coding RNA in Cancer and Metastasis. Cancers 2019, 11, 216. [Google Scholar] [CrossRef] [PubMed]
- Bhan, A.; Mandal, S.S. lncRNA HOTAIR: A Master Regulator of Chromatin Dynamics and Cancer. Biochim. Biophys. Acta 2015, 1856, 151–164. [Google Scholar] [CrossRef]
- Ferreira, L.B.; Palumbo, A.; de Mello, K.D.; Sternberg, C.; Caetano, M.S.; de Oliveira, F.L.; Neves, A.F.; Nasciutti, L.E.; Goulart, L.R.; Gimba, E.R.P. PCA3 Noncoding RNA Is Involved in the Control of Prostate-Cancer Cell Survival and Modulates Androgen Receptor Signaling. BMC Cancer 2012, 12, 507. [Google Scholar] [CrossRef]
- Ji, P.; Diederichs, S.; Wang, W.; Böing, S.; Metzger, R.; Schneider, P.M.; Tidow, N.; Brandt, B.; Buerger, H.; Bulk, E.; et al. MALAT-1, a Novel Noncoding RNA, and Thymosin beta4 Predict Metastasis and Survival in Early-Stage Non-Small Cell Lung Cancer. Oncogene 2003, 22, 8031–8041. [Google Scholar] [CrossRef]
- Okugawa, Y.; Toiyama, Y.; Hur, K.; Toden, S.; Saigusa, S.; Tanaka, K.; Inoue, Y.; Mohri, Y.; Kusunoki, M.; Boland, C.R.; et al. Metastasis-Associated Long Non-Coding RNA Drives Gastric Cancer Development and Promotes Peritoneal Metastasis. Carcinogenesis 2014, 35, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.-T.; Shi, D.-B.; Wang, Y.-W.; Li, X.-X.; Xu, Y.; Tripathi, P.; Gu, W.-L.; Cai, G.-X.; Cai, S.-J. High Expression of lncRNA MALAT1 Suggests a Biomarker of Poor Prognosis in Colorectal Cancer. Int. J. Clin. Exp. Pathol. 2014, 7, 3174–3181. [Google Scholar] [PubMed]
- Lu, R.; Zhang, J.; Zhang, W.; Huang, Y.; Wang, N.; Zhang, Q.; Qu, S. Circulating HOTAIR Expression Predicts the Clinical Response to Neoadjuvant Chemotherapy in Patients with Breast Cancer. Cancer Biomark. 2018, 22, 249–256. [Google Scholar] [CrossRef]
- Yang, Z.; Zhou, L.; Wu, L.-M.; Lai, M.-C.; Xie, H.-Y.; Zhang, F.; Zheng, S.-S. Overexpression of Long Non-Coding RNA HOTAIR Predicts Tumor Recurrence in Hepatocellular Carcinoma Patients Following Liver Transplantation. Ann. Surg. Oncol. 2011, 18, 1243–1250. [Google Scholar] [CrossRef]
- Lemos, A.E.G.; da Rocha Matos, A.; Ferreira, L.B.; Gimba, E.R.P. The Long Non-Coding RNA: An Update of Its Functions and Clinical Applications as a Biomarker in Prostate Cancer. Oncotarget 2019, 10, 6589–6603. [Google Scholar] [CrossRef] [PubMed]
- Pal, R.P.; Maitra, N.U.; Mellon, J.K.; Khan, M.A. Defining Prostate Cancer Risk before Prostate Biopsy. Urol. Oncol. 2013, 31, 1408–1418. [Google Scholar] [CrossRef]
- Li, Z.; Liu, L.; Feng, C.; Qin, Y.; Xiao, J.; Zhang, Z.; Ma, L. LncBook 2.0: Integrating Human Long Non-Coding RNAs with Multi-Omics Annotations. Nucleic Acids Res. 2023, 51, D186–D191. [Google Scholar] [CrossRef]
- Carlevaro-Fita, J.; Lanzós, A.; Feuerbach, L.; Hong, C.; Mas-Ponte, D.; Pedersen, J.S.; Johnson, R.; PCAWG Drivers and Functional Interpretation Group. PCAWG Consortium Author Correction: Cancer lncRNA Census Reveals Evidence for Deep Functional Conservation of Long Noncoding RNAs in Tumorigenesis. Commun. Biol. 2022, 5, 1347. [Google Scholar] [CrossRef]
- Mularoni, L.; Sabarinathan, R.; Deu-Pons, J.; Gonzalez-Perez, A.; López-Bigas, N. OncodriveFML: A General Framework to Identify Coding and Non-Coding Regions with Cancer Driver Mutations. Genome Biol. 2016, 17, 128. [Google Scholar] [CrossRef]
- Lanzós, A.; Carlevaro-Fita, J.; Mularoni, L.; Reverter, F.; Palumbo, E.; Guigó, R.; Johnson, R. Discovery of Cancer Driver Long Noncoding RNAs across 1112 Tumour Genomes: New Candidates and Distinguishing Features. Sci. Rep. 2017, 7, 41544. [Google Scholar] [CrossRef]
- Chiu, H.-S.; Somvanshi, S.; Patel, E.; Chen, T.-W.; Singh, V.P.; Zorman, B.; Patil, S.L.; Pan, Y.; Chatterjee, S.S.; Cancer Genome Atlas Research Network; et al. Pan-Cancer Analysis of lncRNA Regulation Supports Their Targeting of Cancer Genes in Each Tumor Context. Cell Rep. 2018, 23, 297–312.e12. [Google Scholar] [CrossRef] [PubMed]
- Mitra, R.; Adams, C.M.; Eischen, C.M. Systematic lncRNA Mapping to Genome-Wide Co-Essential Modules Uncovers Cancer Dependency on Uncharacterized lncRNAs. eLife 2022, 11, e77357. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Xu, F.; Wang, H.; Teschendorff, A.E.; Xie, F.; He, Y. Pan-Cancer Characterization of Long Non-Coding RNA and DNA Methylation Mediated Transcriptional Dysregulation. eBioMedicine 2021, 68, 103399. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef]
- Arunkumar, G.; Baek, S.; Sturgill, D.; Bui, M.; Dalal, Y. Oncogenic lncRNAs Alter Epigenetic Memory at a Fragile Chromosomal Site in Human Cancer Cells. Sci. Adv. 2022, 8, eabl5621. [Google Scholar] [CrossRef]
- Galon, J.; Bruni, D. Tumor Immunology and Tumor Evolution: Intertwined Histories. Immunity 2020, 52, 55–81. [Google Scholar] [CrossRef]
- Li, Y.; Jiang, T.; Zhou, W.; Li, J.; Li, X.; Wang, Q.; Jin, X.; Yin, J.; Chen, L.; Zhang, Y.; et al. Pan-Cancer Characterization of Immune-Related lncRNAs Identifies Potential Oncogenic Biomarkers. Nat. Commun. 2020, 11, 1000. [Google Scholar] [CrossRef]
- Hur, K.; Kim, S.-H.; Kim, J.-M. Potential Implications of Long Noncoding RNAs in Autoimmune Diseases. Immune Netw. 2019, 19, e4. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Wang, Y.; Yang, M.; Wang, Z.; Wang, Y.; Chaurasia, S.; Wu, Z.; Zhang, M.; Yadav, G.S.; Rathod, S.; et al. LincRNA-Immunity Landscape Analysis Identifies EPIC1 as a Regulator of Tumor Immune Evasion and Immunotherapy Resistance. Sci. Adv. 2021, 7, eabb3555. [Google Scholar] [CrossRef]
- Li, J.; Zhang, X.; Liu, C. The Computational Approaches of lncRNA Identification Based on Coding Potential: And Challenges. Comput. Struct. Biotechnol. J. 2020, 18, 3666–3677. [Google Scholar] [CrossRef]
- Chi, Y.; Wang, D.; Wang, J.; Yu, W.; Yang, J. Long Non-Coding RNA in the Pathogenesis of Cancers. Cells 2019, 8, 1015. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Zhen, M.; Sun, J.; Zhao, Q. Recent Advances on the Machine Learning Methods in Predicting ncRNA-Protein Interactions. Mol. Genet. Genom. 2021, 296, 243–258. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Luo, J.; Liang, C.; Xiao, Q.; Ding, P.; Zhang, Y. Prediction of lncRNA-Disease Associations Based on Network Consistency Projection. IEEE Access 2019, 7, 58849–58856. [Google Scholar] [CrossRef]
- Fan, X.-N.; Zhang, S.-W.; Zhang, S.-Y.; Zhu, K.; Lu, S. Prediction of lncRNA-Disease Associations by Integrating Diverse Heterogeneous Information Sources with RWR Algorithm and Positive Pointwise Mutual Information. BMC Bioinform. 2019, 20, 87. [Google Scholar] [CrossRef]
- Zhou, J.-R.; You, Z.-H.; Cheng, L.; Ji, B.-Y. Prediction of lncRNA-Disease Associations via an Embedding Learning HOPE in Heterogeneous Information Networks. Mol. Ther. Nucleic Acids 2021, 23, 277–285. [Google Scholar] [CrossRef]
- Wang, Y.; Juan, L.; Peng, J.; Zang, T.; Wang, Y. LncDisAP: A Computation Model for lncRNA-Disease Association Prediction Based on Multiple Biological Datasets. BMC Bioinform. 2019, 20, 582. [Google Scholar] [CrossRef]
- Yu, J.; Ping, P.; Wang, L.; Kuang, L.; Li, X.; Wu, Z. A Novel Probability Model for lncRNA–Disease Association Prediction Based on the Naïve Bayesian Classifier. Genes 2018, 9, 345. [Google Scholar] [CrossRef]
- Zhou, M.; Zhang, H.; Bai, Z.; Mann-Krzisnik, D.; Wang, F.; Li, Y. Single-Cell Multi-Omics Topic Embedding Reveals Cell-Type-Specific and COVID-19 Severity-Related Immune Signatures. Cell Rep. Methods 2023, 3, 100563. [Google Scholar] [CrossRef]
- Treppner, M.; Binder, H.; Hess, M. Interpretable Generative Deep Learning: An Illustration with Single Cell Gene Expression Data. Hum. Genet. 2022, 141, 1481–1498. [Google Scholar] [CrossRef]
- Nath, A.; Lau, E.Y.T.; Lee, A.M.; Geeleher, P.; Cho, W.C.S.; Huang, R.S. Discovering Long Noncoding RNA Predictors of Anticancer Drug Sensitivity beyond Protein-Coding Genes. Proc. Natl. Acad. Sci. USA 2019, 116, 22020–22029. [Google Scholar] [CrossRef]
- Ahmad, A.; Poltronieri, P.; Uddin, S. lncRNAs in Cancer Metastasis and Therapy Resistance; Frontiers Media SA: Lausanne, Switzerland, 2023; ISBN 9782832526125. [Google Scholar]
- Ding, Y.; Lei, X.; Liao, B.; Wu, F.-X. Machine Learning Approaches for Predicting Biomolecule-Disease Associations. Brief. Funct. Genom. 2021, 20, 273–287. [Google Scholar]
- Liu, Z.; Mi, M.; Li, X.; Zheng, X.; Wu, G.; Zhang, L. A lncRNA Prognostic Signature Associated with Immune Infiltration and Tumour Mutation Burden in Breast Cancer. J. Cell. Mol. Med. 2020, 24, 12444–12456. [Google Scholar] [CrossRef]
- Zaheed, O.; Kiniry, S.J.; Baranov, P.V.; Dean, K. Exploring Evidence of Non-Coding RNA Translation with Trips-Viz and GWIPS-Viz Browsers. Front. Cell Dev. Biol. 2021, 9, 703374. [Google Scholar]
- Leong, A.Z.-X.; Lee, P.Y.; Mohtar, M.A.; Syafruddin, S.E.; Pung, Y.-F.; Low, T.Y. Short Open Reading Frames (sORFs) and Microproteins: An Update on Their Identification and Validation Measures. J. Biomed. Sci. 2022, 29, 19. [Google Scholar]
- Zheng, C.; Wei, Y.; Zhang, P.; Lin, K.; He, D.; Teng, H.; Manyam, G.; Zhang, Z.; Liu, W.; Lee, H.R.L.; et al. CRISPR-Cas9-Based Functional Interrogation of Unconventional Translatome Reveals Human Cancer Dependency on Cryptic Non-Canonical Open Reading Frames. Nat. Struct. Mol. Biol. 2023. [Google Scholar] [CrossRef]
- Schlesinger, D.; Elsässer, S.J. Revisiting sORFs: Overcoming Challenges to Identify and Characterize Functional Microproteins. FEBS J. 2022, 289, 53–74. [Google Scholar]
- Patraquim, P.; Mumtaz, M.A.S.; Pueyo, J.I.; Aspden, J.L.; Couso, J.-P. Developmental Regulation of Canonical and Small ORF Translation from mRNAs. Genome Biol. 2020, 21, 128. [Google Scholar]
- Ingolia, N.T.; Ghaemmaghami, S.; Newman, J.R.S.; Weissman, J.S. Genome-Wide Analysis in Vivo of Translation with Nucleotide Resolution Using Ribosome Profiling. Science 2009, 324, 218–223. [Google Scholar]
- Patraquim, P.; Magny, E.G.; Pueyo, J.I.; Platero, A.I.; Couso, J.P. Translation and Natural Selection of Micropeptides from Long Non-Canonical RNAs. Nat. Commun. 2022, 13, 6515. [Google Scholar]
- Li, M.; Liu, G.; Jin, X.; Guo, H.; Setrerrahmane, S.; Xu, X.; Li, T.; Lin, Y.; Xu, H. Micropeptide MIAC Inhibits the Tumor Progression by Interacting with AQP2 and Inhibiting EREG/EGFR Signaling in Renal Cell Carcinoma. Mol. Cancer 2022, 21, 181. [Google Scholar]
- Zhang, M.; Zhao, K.; Xu, X.; Yang, Y.; Yan, S.; Wei, P.; Liu, H.; Xu, J.; Xiao, F.; Zhou, H.; et al. A Peptide Encoded by Circular Form of LINC-PINT Suppresses Oncogenic Transcriptional Elongation in Glioblastoma. Nat. Commun. 2018, 9, 4475. [Google Scholar] [PubMed]
- Jiang, L.; Yang, J.; Xu, Q.; Lv, K.; Cao, Y. Machine Learning for the Micropeptide Encoded by LINC02381 Regulates Ferroptosis through the Glucose Transporter SLC2A10 in Glioblastoma. BMC Cancer 2022, 22, 882. [Google Scholar] [CrossRef]
- Polycarpou-Schwarz, M.; Groß, M.; Mestdagh, P.; Schott, J.; Grund, S.E.; Hildenbrand, C.; Rom, J.; Aulmann, S.; Sinn, H.-P.; Vandesompele, J.; et al. The Cancer-Associated Microprotein CASIMO1 Controls Cell Proliferation and Interacts with Squalene Epoxidase Modulating Lipid Droplet Formation. Oncogene 2018, 37, 4750–4768. [Google Scholar] [CrossRef]
- Zhang, C.; Zhou, B.; Gu, F.; Liu, H.; Wu, H.; Yao, F.; Zheng, H.; Fu, H.; Chong, W.; Cai, S.; et al. Micropeptide PACMP Inhibition Elicits Synthetic Lethal Effects by Decreasing CtIP and poly(ADP-Ribosyl)ation. Mol. Cell 2022, 82, 1297–1312.e8. [Google Scholar] [PubMed]
- Huang, J.-Z.; Chen, M.; Chen, D.; Gao, X.-C.; Zhu, S.; Huang, H.; Hu, M.; Zhu, H.; Yan, G.-R. A Peptide Encoded by a Putative lncRNA HOXB-AS3 Suppresses Colon Cancer Growth. Mol. Cell 2017, 68, 171–184.e6. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Wu, S.; Zhu, X.; Zhang, L.; Deng, J.; Li, F.; Wang, Y.; Zhang, S.; Wu, R.; Lu, J.; et al. Micropeptide CIP2A-BP Encoded by LINC00665 Inhibits Triple-Negative Breast Cancer Progression. EMBO J. 2020, 39, e102190. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.-C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long Noncoding RNA as Modular Scaffold of Histone Modification Complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef]
- Li, Q.; Shao, Y.; Zhang, X.; Zheng, T.; Miao, M.; Qin, L.; Wang, B.; Ye, G.; Xiao, B.; Guo, J. Plasma Long Noncoding RNA Protected by Exosomes as a Potential Stable Biomarker for Gastric Cancer. Tumour Biol. 2015, 36, 2007–2012. [Google Scholar] [CrossRef]
- Badowski, C.; He, B.; Garmire, L.X. Blood-Derived lncRNAs as Biomarkers for Cancer Diagnosis: The Good, the Bad and the Beauty. npj Precis. Oncol. 2022, 6, 40. [Google Scholar]
- Profumo, V.; Forte, B.; Percio, S.; Rotundo, F.; Doldi, V.; Ferrari, E.; Fenderico, N.; Dugo, M.; Romagnoli, D.; Benelli, M.; et al. LEADeR Role of miR-205 Host Gene as Long Noncoding RNA in Prostate Basal Cell Differentiation. Nat. Commun. 2019, 10, 307. [Google Scholar] [CrossRef]
- Garofoli, M.; Volpicella, M.; Guida, M.; Porcelli, L.; Azzariti, A. The Role of Non-Coding RNAs as Prognostic Factor, Predictor of Drug Response or Resistance and Pharmacological Targets, in the Cutaneous Squamous Cell Carcinoma. Cancers 2020, 12, 2552. [Google Scholar] [CrossRef] [PubMed]
- Smallegan, M.J.; Rinn, J.L. Linking Long Noncoding RNA to Drug Resistance. Proc. Natl. Acad. Sci. USA 2019, 116, 21963–21965. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Kong, H.; Peng, F.; Wang, C.; Zhang, D.; Tian, J.; Zhang, L. Inferences of Individual Drug Response-Related Long Non-Coding RNAs Based on Integrating Multi-Omics Data in Breast Cancer. Mol. Ther. Nucleic Acids 2020, 20, 128–139. [Google Scholar] [CrossRef]
- Liu, H.; Wang, S.; Zhou, S.; Meng, Q.; Ma, X.; Song, X.; Wang, L.; Jiang, W. Drug Resistance-Related Competing Interactions of lncRNA and mRNA across 19 Cancer Types. Mol. Ther. Nucleic Acids 2019, 16, 442–451. [Google Scholar] [CrossRef] [PubMed]
- Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA Therapeutics—Challenges and Potential Solutions. Nat. Rev. Drug Discov. 2021, 20, 629–651. [Google Scholar]
- Arun, G.; Diermeier, S.D.; Spector, D.L. Therapeutic Targeting of Long Non-Coding RNAs in Cancer. Trends Mol. Med. 2018, 24, 257–277. [Google Scholar] [CrossRef]




| lncRNA of Relevance from the Study | Potential Association Based on Biological or Clinical Data | Goal | Main Insight Regarding the lncRNA | Model Construction | Kaplan–Meier and Log-Rank Test | Receiver Operator Curve AUC | Reference |
|---|---|---|---|---|---|---|---|
| ESR1, TRPS1, ERG, RUNX1, SNHG16, and HOTAIR | Various cancer types | Molecular and biological characterization of the regulatory mutations on functional lncRNAs in cancer | Somatic mutations on the lncRNA TF binding site are associated with their expression and activity in cancer | NA | NA | NA | [32] |
| CASC8 | Breast cancer | Prioritize and predict lncRNAs with a functional impact through mutation analysis | Somatic mutations on the lncRNA loci site are associated with expression alterations that have a functional impact | NA | NA | NA | [37] |
| ENSG0000021403, ENSG00000261650, ENSG00000281406, and G001643 | Colorectal carcinoma | Molecular and biological characterization of the regulatory mutations on functional lncRNAs in cancer | Somatic mutations on the lncRNA loci site are associated with expression alterations that have a functional impact | NA | NA | NA | [38] |
| LINC00460, AC156455.1, AC015977.2, ‘PRDM16-dt’, AL139351.1, AL035661.1, and LINC01606 | Renal cell carcinoma | Identify the genome instability-related lncRNAs and their clinical significance | Discovery of 11 lncRNAs related to mutational burden and associated with patient poor overall survival | Univariable and multi-variable COX prognostic regression model | High-risk vs. low-risk groups; p < 0.001 | Train AUC = 0.743 Test = 0.770 | [39] |
| LINC00460, LINC01234 | Clear cell renal carcinoma | Identify the genome instability-related lncRNAs and their clinical significance | Identification of a lncRNAs signature related to the mutational burden and associated with patient poor overall survival | Univariable and multi-variable COX prognostic regression model | High-risk vs. low-risk groups; p < 0.001 | AUC = 0.681 | [40] |
| FAM30A, CACNA1C-AS1, LINC02595, LINC00926, AL589863.1, and AP000919.3 | AML | Construct a somatic mutation-associated risk index | lncRNAs related to the mutational burden and associated with patient poor overall survival | Selection of candidate lncRNAs using LASSO followed by the univariable and multi-variable COX prognostic regression model | High-risk vs. low-risk groups; p < 0.001 | AUC = 0.804 | [41] |
| AC007996.1, AC009237.14, AP003555.1, and AL590483.1 | Colorectal carcinoma | Evaluate the performance of a genome stability-related lncRNA signature as a risk predictor | Identification of a lncRNAs signature related to the mutational burden and associated with patient poor overall survival | Univariable and multi-variable COX prognostic regression model | High-risk vs. low-risk groups at 3 years; p < 0.001 | AUC = 0.713 | [42] |
| ZNF503-AS1, AL353747.2, AC129492.1, AP003555.1, and AC009237.14 | Colorectal carcinoma | Evaluate the performance of a genome stability-related lncRNA signature as a risk predictor | Identification of a lncRNAs signature related to the mutational burden and immune infiltration that are associated with patient poor overall survival | Univariable and multi-variable COX prognostic regression model | High-risk vs. low-risk groups; p < 0.001; in the validation set | AUC (1 year) = 0.750; AUC (3 years) = 0.757, AUC (5 years) = 0.711; in the validation set | [43] |
| AC107464.2, MIR100HG, and AP001527.2 | Cervical carcinoma | Evaluate the performance of a genome stability-related lncRNA signature as a risk predictor | Identification of a lncRNAs signature related to the mutational burden and associated with patient poor overall survival | Univariable and multi-variable COX prognostic regression model | High-risk (1.7 years) vs. low-risk (1.5 years) groups; p < 0.001; in the validation set | AUC (3 years) = 0.663; in the validation set | [44] |
| AC002511.2, LINC00501, LINC02055, LINC02714, LINC01508, LOC105371967, RP11_96A15.1, RP11_305F18.1, RP11_342M1.3, RP11_432J24.3, and U95743.1 | Hepatocellular carcinoma | Evaluate the performance of a genome stability-related lncRNA signature as a risk predictor | Identification of a lncRNAs signature related to the mutational burden and associated with patient poor overall survival | Selection of candidate lncRNAs using LASSO followed by the univariable and multi-variable COX prognostic regression model | High-risk vs. low-risk groups; p < 0.001 | NA | [45] |
| C116351.1, ZFPM2-AS1, AC145343.1, and MIR210HG | Hepatocellular carcinoma | Evaluate the performance of a genome stability-related lncRNA signature as a risk predictor | Identification of a lncRNAs signature related to the mutational burden and associated with patient poor overall survival | Univariable and multi-variable COX prognostic regression model | High-risk vs. low-risk groups at 3 and 5 years; p < 0.001 | AUC (3 years) = 0.710, AUC (5 years) = 0.707; in the validation set | [46] |
| BCAL8 | Breast cancer | Molecular and biological characterization of the functional regulatory mutations on cancer | Identification of a somatic SCNA-related lncRNAs signature associated with patient prognosis | Selection of candidate lncRNAs using filtering methods | NA | NA | [47] |
| RUSC1-AS1, LINC01990, LINC01411, LINC02099, H19, LINC00452, ADPGK-AS1, and C1QTNF1-AS1 | Cervical carcinoma | Evaluate the performance of a SCNA-related lncRNA signature as a risk predictor | Identification of a somatic SCNA-related lncRNAs signature with a value as independent tumor-free survival | Univariable and multi-variable COX prognostic regression model | High-risk vs. low-risk groups at 1, 3, and 5 years; p < 0.001 | AUC (1 year), AUC (3 years), and AUC (5 years) > 0.750 | [48] |
| PRAL | Tumor suppressor | Molecular and biological characterization of SCNA-related lncRNAs in cancer | Identification of a somatic SCNA-related lncRNA with a value as an independent predictor for reduced tumor-free survival | Univariable and multi-variable COX prognostic regression model | High-risk vs. low-risk groups; p < 0.001 | NA | [49] |
| LOC101927604, LOC105377267, CASC15, LINC-PINT, CLDN10-AS1, C14orf132, LMF1, LINC00675, CCDC144NL-AS1, and LOC284454 | Colorectal carcinoma | Evaluate the performance of an SCNA-related lncRNA signature as a risk predictor for CRC | Identification of somatic SCNA-related lncRNA with a value as an independent predictor for reduced tumor-free survival | Clustering based on the gene expression, SCNA, and DNA methylation followed by the univariable and multi-variable COX prognostic regression model | High-risk vs. low-risk groups; p < 0.001 | NA | [50] |
| RP11-571M6.8 | Glioblastoma | Molecular and biological characterization of SCNA-related lncRNAs in cancer | Identification of a somatic SCNA-related lncRNA that is significantly predictive of disease-free survival | Selection of candidate lncRNAs using filtering methods followed by the univariable and multi-variable COX prognostic regression model | High-risk vs. low-risk groups; p < 0.001 | NA | [51] |
| RP11-1020A11.1 | Bladder carcinoma | Molecular and biological characterization of SCNA-related lncRNAs with risk prediction utility in cancer | Identification of a somatic SCNA-related lncRNA that is significantly predictive of disease-free survival | Selection of candidate lncRNAs using filtering methods followed by the univariable and multi-variable COX prognostic regression model | High-risk vs. low-risk groups; p < 0.001 | NA | [51] |
| LINC02528, SEMA6A-AS1, EBLN3P, MIR155HG, LYRM4-AS1, and HLA-DQB1-AS1 | Skin cutaneous melanoma | Identify a novel prognostic signature using m6A-related lncRNAs and evaluate the prognostic of survival performance | Identification of a lncRNAs signature-related somatic SCNA associated with patient overall survival | Selection of candidate lncRNAs using LASSO followed by the COX prognostic regression model | High-risk vs. low-risk groups at 1, 2, 3, and 5 years; p < 0.001 and construction of a nomogram for the clinical decision risk score | AUC (1, 2, 3, and 5 years) > 0.6 | [52] |
| AL121772.1, BX640514.2, LINC01133, and LYPLAL1-AS1 | Pancreatic cancer | Investigate the prognostic performance of a lncRNA signature and its relationship with the tumor immune microenvironment | Identification of a genomic instability-related lncRNAs signature associated with patient overall survival | Selection of candidate lncRNAs using filtering methods followed by the univariable and multi-variable COX prognostic regression model | High-risk vs. low-risk groups at 1 year; p < 0.001 | AUC (1 year) = 0.653 | [53] |
| CTD-2256P15.2 | Lung adenocarcinoma | Molecular and biological characterization of SCNA-related lncRNAs with risk prediction utility in cancer | Identification of a somatic SCNA-related lncRNAs signature associated with the prognosis of patients after methyl ethyl ketone (MEK) inhibitors treatment | Selection of candidate lncRNAs using filtering methods followed by the univariable and multi-variable COX prognostic regression model | High-risk vs. low-risk groups; p < 0.001 | NA | [54] |
| LINC00896, MCM8-AS1, LINC01251, LNX1-AS1, GPRC5D-AS1, CTD-2350J17.1, LINC01133, LINC01121, and AC073130.1 | Non-small cell lung cancer | Evaluate the performance of a SCNA-related lncRNA signature as a risk predictor | Identification of a lncRNAs signature related to somatic SCNA and associated with patient poor overall survival | Selection of candidate lncRNAs using LASSO followed by the COX prognostic regression model | High-risk vs. low-risk groups at 1 and 3 years; p < 0.001 | AUC (1 and 3 years) = 0.73 | [54] |
| RP11-241F15.10 | Osteosarcoma | Evaluate the performance of a SCNA-related lncRNA signature as a risk predictor | Identification of a lncRNAs signature-related somatic SCNA associated with patient disease-free and overall survival | Univariable and multi-variable COX prognostic regression model | High-risk vs. low-risk groups at 1 and 3 years; p < 0.001 | NA | [55] |
| ALAL-1 | Non–small cell lung cancer | Molecular and biological characterization of SCNA-related lncRNAs in cancer | Identification of a pro-oncogenic lncRNA that mediates cancer immune evasion, pointing to a new target for immune potentiation | NA | NA | NA | [56] |
| LOC339803, F11-AS1, and PCAT2 TMEM220-AS1 | Hepatocellular carcinoma | Evaluate the performance of a SCNA-related lncRNA signature as a risk predictor | Identification of CNA-related lncRNAs that can better evaluate the prognosis of patients with liver cancer | Selection of candidate lncRNAs using LASSO followed by the COX prognostic regression model | High-risk vs. low-risk groups at 1, 3, and 5 years; p < 0.001 | AUC (1, 3, and 5 year) > 0.7 | [57] |
| ENSG00000261582 | Lung adenocarcinoma and cervical carcinoma | Evaluate the performance of a SCNA-related lncRNA signature as a risk predictor | Identification of a somatic SCNA-related lncRNAs signature associated with patient overall survival | Univariable and multi-variable COX prognostic regression model | High-risk vs. low-risk groups at 5 years; p < 0.001 | NA | [58] |
| PCAN-R1 (Ensembl ID ENSG00000228288) and PCAN-R2 (Ensembl ID ENSG00000231806) | Prostate adenocarcinoma | Evaluate the performance of a SCNA-related lncRNA signature as a risk predictor | Identification of a somatic SCNA-related lncRNAs signature associated with patient overall survival | Univariable and multi-variable COX prognostic regression model | High-risk vs. low-risk groups at 5 years; p < 0.001 | NA | [58] |
| LOC101927151, LINC00861, and LEMD1-AS1 | Ovarian cancer | Evaluate the performance of a SCNA-related lncRNA signature as a risk predictor | Identification of a somatic SCNA-related lncRNAs signature associated with patient prognosis | Univariable and multi-variable COX prognostic regression model | High-risk vs. low-risk groups at 5 years; p < 0.001 | NA | [59] |
| TSPOAP1-AS1, CCNT2-AS1, LINC01094, AL033527.2, and LINC00460 | Gastric cancer | Evaluate the performance of a mutation-related lncRNA signature as a risk predictor and characterization of functional activity | Identification of an anoikis-related lncRNAs signature associated with patient poor overall survival and immunotherapy response | Selection of candidate lncRNAs using LASSO followed by the COX prognostic regression model | High-risk vs. low-risk groups; p < 0.001 | Train AUC = 0.707 Test = 0.646 | [53] |
| NA | Kidney renal clear cell carcinoma | Evaluate the performance of a multi-omics-derived lncRNA signature as a risk predictor | Identification of a lncRNAs signature derived from the analysis of transcriptomics and DNA methylation associated with patient poor overall survival | Selection of candidate lncRNAs using a novel TRS method utilizing multiple omics data and a XGBoost model followed by the COX prognostic regression model | High-risk vs. low-risk groups; p < 0.001 | AUC = 0.95; using the best predictor model | [60] |
| Twenty-six lncRNAs | Hepatocellular carcinoma | Evaluate the performance of a multi-omics-derived lncRNA signature as a risk predictor and characterization of functional activity | Identification of an exosome-related lncRNAs signature associated with patient poor overall survival and a response to transarterial chemoembolization (TACE) therapy and sorafenib therapy | Selection of candidate lncRNAs using a weighted correlation network analysis followed by the COX prognostic regression model | High-risk vs. low-risk groups; p < 0.001 | AUC > 0.7 | [61] |
| Ten lncRNAs, including: LINC00582, MIR205HG and TRG-S1, | Breast cancer | Evaluate the performance of a mutation-related lncRNA signature as a risk predictor and characterization of functional activity | Identification of a TMB-related lncRNAs signature associated with patient poor overall survival and a response to immunotherapy | Clustering based on the gene expression and selection of the gene predictors using LASSO followed by the COX prognostic regression model | High-risk vs. low-risk groups at 1, 3, and 5 years; p < 0.001 | AUC (1 year) = 0.722, AUC (3 years) = 0.745, AUC (5 years) = 0.811 | [62] |
| C20orf197, UCA1, MIR17HG, and MIR22HG | Various cancer types | Evaluate the performance of a lncRNA signature as a patient prognosis predictor | Oxiplatin sensitivity-related lncRNAs signature associated with the prognosis of patients given oxaliplatin-based chemotherapy | Selection of candidate lncRNAs using LASSO, decision tree, random forest, and a support vector machine followed by the COX prognostic regression model | High-risk vs. low-risk groups at 1, 3, and 5 years; p < 0.001 | AUC (1 year) = 0.76, AUC (3 years) = 0.79, AUC (5 years) = 0.88; using the best predictor model | [63] |
| 18 lncRNAs | Gastric cancer | Evaluate the performance of a lncRNA signature as a patient prognosis predictor | Identification of an immune-related lncRNAs signature that helps predict the prognosis of patients suffering from gastric cancer | Selection of candidate lncRNAs using the integration of multiple machine learning algorithms followed by the COX prognostic regression model | High-risk vs. low-risk groups at 1, 3, and 5 years; p < 0.001 | AUC (1 year) = 0.715, AUC (3 years) = 0.80, AUC (5 years) = 0.809; using the best predictor model | [64] |
| Various lncRNAs | Breast cancer | Develop an interpretable deep-learning-based network for classifying the recurrence risk and revealing the potential biological mechanisms | Construction of a lncRNa-based model associated with the radiomics of magnetic resonance features for predicting individual recurrence risk after surgery | Construction of a lncRNa-based model using the Cox proportional hazards deep neural network | High-risk vs. low-risk groups at 1, 2, and 3 years; p < 0.001 | AUC (1 year) = 0.98, AUC (2 years) = 0.94, AUC (3 years) = 0.92; using the best predictor model | [65] |
| CRNDE, AC010273.2, MPPED2-AS1, SNHG18, and CYTOR | Lower-grade gliomas | Evaluate the impact of DNA methylation-related lncRNAs with an effect on genome stability and the immune microenvironment on disease progression | Identification of a five DNA methylation-related signature with an independent prognostic value | Selection of lncRNAs using filtering methods followed by the COX prognostic regression model | High-risk vs. low-risk groups at 1, 2, and 3 years; p < 0.001 | AUC (1 year) = 0.893, AUC (2 years) = 0.919, AUC (3 years) = 0.866; using the best predictor model | [66] |
| CRNDE, AC010273.2, MPPED2-AS1, SNHG18, and CYTOR | Various cancer types | Explore the functional effects of lncRNAs related to DNA methylation and evaluate their predictive performance on patient survival | Identification of a DNA methylation and genome stability-related signature with an independent prognostic value | Construction of a lncRNa-based co-expression network of pADM–lncRNA | High-risk vs. low-risk groups; p < 0.001 | AUC = 0.6839 | [67] |
| LIFR-AS1 | Colorectal carcinoma | Evaluate the impact of DNA methylation-related lncRNAs with an effect on disease progression | Identification of LIFR-AS1 as a tumor suppressor RNA with an independent prognostic value | Selection using filtering methods followed by the COX prognostic regression model | High-risk vs. low-risk groups; p < 0.001 | AUC = 0.872 | [68] |
| lncRNA | Micropeptide | Biological Effect | Method | Citation |
|---|---|---|---|---|
| lncRNA AC025154.2 | MIAC | Inhibits the actin cytoskeleton by interacting with Aquaporin 2 to suppress tumor growth and metastasis | RNA-seq data from kidney renal clear cell carcinoma patients | [141] |
| LINC-PINT | PINT87aa | Suppresses glioblastoma cell proliferation by interacting with PAF1c and preventing the transcriptional elongation of cancer-related genes | RNA-seq of circRNAs (transcriptome sequencing) and RNC-RNAs (translatome sequencing) | [142] |
| LINC02381 | LINC02381-aa | LINC02381-aa contained in exosomes enhances ferroptosis by promoting the glucose transporter SLC2A10 in glioblastoma | Machine learning to integrate the multi-omics data to identify microproteins | [143] |
| NR_029453 | CASIMO1 | Interacts with the enzyme squalene epoxidase and increases the phosphorylation of ERK, a relevant actor of the MAPK pathway affecting cell proliferation | Transcriptome analysis and DNA conservation evaluation | [144] |
| lncRNA CTD-2256P15.2 | PACMP | PACMP acts as an activator of the PARP1-dependent pathways (DNA repair process), enhancing tumor growth and limiting the cell response to PARP inhibitors | RNA-seq | [145] |
| HOXB-AS3 | HOXB-AS3 peptide | Suppresses cancer growth by limiting PKM splicing and, subsequently, the metabolic reprogramming | Transcriptome analysis and RNA affinity purification analysis | [146] |
| LINC00665 | CIP2A-BP | Decreases cell invasion in vivo and correlates with better patient survival. Competes with PP2A for CIP2A binding, suppressing the oncogenic P13K/AKT/NFkB pathway | Ribosome profiling and RNA sequencing data analysis | [147] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salido-Guadarrama, I.; Romero-Cordoba, S.L.; Rueda-Zarazua, B. Multi-Omics Mining of lncRNAs with Biological and Clinical Relevance in Cancer. Int. J. Mol. Sci. 2023, 24, 16600. https://doi.org/10.3390/ijms242316600
Salido-Guadarrama I, Romero-Cordoba SL, Rueda-Zarazua B. Multi-Omics Mining of lncRNAs with Biological and Clinical Relevance in Cancer. International Journal of Molecular Sciences. 2023; 24(23):16600. https://doi.org/10.3390/ijms242316600
Chicago/Turabian StyleSalido-Guadarrama, Ivan, Sandra L. Romero-Cordoba, and Bertha Rueda-Zarazua. 2023. "Multi-Omics Mining of lncRNAs with Biological and Clinical Relevance in Cancer" International Journal of Molecular Sciences 24, no. 23: 16600. https://doi.org/10.3390/ijms242316600