Breaking the Chains: Advances in Substance Addiction Research through Single-Cell Sequencing, Epigenetics, and Epitranscriptomic
Abstract
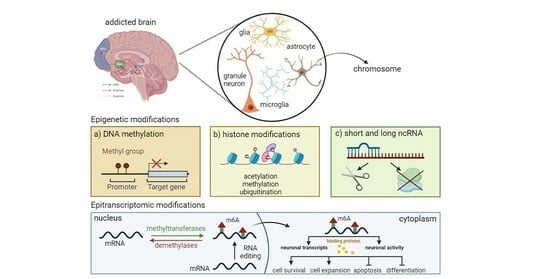
:1. Introduction
2. Single-Cell Sequencing Techniques
2.1. Application of Single-Cell Sequencing in Addiction Research
2.1.1. Identifying Cell Populations by Molecular Clustering
2.1.2. Identifying Transcriptome Mechanism
3. Epigenetic Mechanisms and Psychostimulant Addiction
3.1. Cocaine-induced Epigenetic Modifications
3.1.1. DNA Methylation
3.1.2. Histone Modifications
Histones Methylation
Histones Acetylation
Histones Ubiquitination
3.1.3. ncRNAs
| Type of Epigenetic Modifications | Localization of Modification ↓↑ * | Brain Region | Neuronal Cell Type, Pathways, and Receptors | Reference | |
|---|---|---|---|---|---|
| DNA methylation | DNMT ↑↓ 3A, 3B ↑↓, MeCP2 ↑↓ | NAc, DS, PFC | BDNF ↑ | [100,101,102,103,104] | |
| Histone modifications | methylation | H3R2me2a ↓ PRMT6 ↓ Histone Arg(R) ↑ | NAc, DS, PFC | BDNF ↑, D1-MSN ↓, D2-MSN ↑ | [100,101,102,103,104] |
| acetylation | H3K9/14Ac ↑,H4K5 ↑, HDAC3 ↑, HDAC4 ↑, HDAC5 ↓↑, SIRT1 ↑, SIRT2 ↑ | DS, NAc, mPFC, VTA | Bdnf↑ Creb1↑ Cbp↑, Cdk5↑ DA ↑ Glu/DA ↓↑ GABA ↑ | [116,117,118,120,128] | |
| ubiquitination | SMURF1 ↑↓ | NAc | AP1 ↑ RUNX2 ↑ SMAD1/5 ↑ | [123,124] | |
| ncRNAs | miRNAs | miRNA212 ↑ miRNA132 ↑ miRNA382 ↑ | DS, NAc-MSN | AP1 ↑ RUNX2 ↑ SMAD1/5 ↑ | [104,126,127] |
3.2. Methamphetamine-Induced Epigenetic Modifications
3.2.1. DNA Methylation
3.2.2. Histone Modifications
Histones Methylation
Histones Acetylation
Histones Ubiquitination
3.3. ncRNAs
| Type of Epigenetic Modifications | Localization of Modification ↓↑ * | Brain Region | Neuronal Cell Type, Pathways, and Receptors | Reference | |
|---|---|---|---|---|---|
| DNA methylation | DNMT ↑↓, MeCP2 ↑↓, LINE1 ↑ | DS, Hip, NAc, PFC, mPFC | BDNF ↑, OT ↓, K+ channel ↑↓, Syp ↑↓, Glu ↑, GluA1,2 ↑, LINE-1 ↑, NR4A1, GABA ↓ | [131,132,133,161,162] | |
| Histone modifications | methylation | KDM5C ↑, KMT2A ↑, HMT ↑, Mll1 ↑ | DS, Hip, NAc, PFC, mPFC | BDNF ↑, OT ↓, K+ channel ↑↓, Syp ↑↓, Glu ↑, GluA1,2 ↑, LINE-1 ↑, NR4A1, GABA ↓ | [131,132,133,161,162] |
| acetylation | H3 ↑, H4 ↑, H2Bac ↑, H3K9Ac ↑, H4K12Ac ↑ HTA (ATF-2 ↑, p300 ↑) HDAC1 ↑↓, 2 ↑↓, 3 ↓, 5 ↑, 6 ↑, 8 ↑↓, 9 ↓, 10 ↓, 11 ↓ | DS, Hip, mPFC, NAc | D1 ↑, D2 ↑, HCRTR1 ↓↑, HCRTR2 ↑, HRH1, 3 ↑, NMDA ↑ Glu ↑ α-adrenergic receptors ↑, BDNF ↑, NMDA ↑, D1 ↑, D2 ↓, CREB ↓ | [135,141,163] | |
| ubiquitination | Parkin ↑, SYVN1 ↓ | BLA, CeA, DS | D1, 2 ↑, NMDA ↑, AMPA ↑, GABAAα1 ↑ | [62,154,164] | |
| ncRNAs | miRNAs | miRNA128 ↑, 237 ↑, 296 ↑, 501 ↑ 31-3p ↑, 34a-5p ↑, 183–5p ↑, 9a-5p ↑, 369–3p ↑, 29a ↑, 181a/d ↑ | DS, Hip, NAc, VTA | PKG ↑, PI3K ↑, Wnt ↑, Ago2 ↑, BDNF ↑, GluN1 ↑ | [137,138,157] |
4. Epitranscriptomics and Psychostimulant Addiction
5. Challenges of the Rewarding Molecular Pathway
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Steketee, J.D.; Kalivas, P.W. Drug Wanting: Behavioral Sensitization and Relapse to Drug-Seeking Behavior. Pharmacol. Rev. 2011, 63, 348–365. [Google Scholar] [CrossRef]
- Robbins, T.W.; Everitt, B.J. Neurobehavioural mechanisms of reward and motivation. Curr. Opin. Neurobiol. 1996, 6, 228–236. [Google Scholar] [CrossRef]
- Robinson, T.E.; Berridge, K.C. The incentive sensitization theory of addiction: Some current issues. Philos. Trans. R. Soc. B Biol. Sci. 2008, 363, 3137. [Google Scholar] [CrossRef] [PubMed]
- Lewis, R.G.; Florio, E.; Punzo, D.; Borrelli, E. The Brain’s Reward System in Health and Disease. Adv. Exp. Med. Biol. 2021, 1344, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Rezayof, A.; Ghasemzadeh, Z.; Sahafi, O.H. Addictive drugs modify neurogenesis, synaptogenesis and synaptic plasticity to impair memory formation through neurotransmitter imbalances and signaling dysfunction. Neurochem. Int. 2023, 169, 105572. [Google Scholar] [CrossRef]
- Peters, K.Z.; Cheer, J.F.; Tonini, R. Modulating the Neuromodulators: Dopamine, Serotonin, and the Endocannabinoid System. Trends Neurosci. 2021, 44, 464–477. [Google Scholar] [CrossRef]
- Lüscher, C.; Malenka, R.C. Drug-evoked synaptic plasticity in addiction: From molecular changes to circuit remodeling. Neuron 2011, 69, 650–663. [Google Scholar] [CrossRef]
- Kutlu, M.G.; Gould, T.J. Effects of drugs of abuse on hippocampal plasticity and hippocampus-dependent learning and memory: Contributions to development and maintenance of addiction. Learn. Mem. 2016, 23, 515–533. [Google Scholar] [CrossRef] [PubMed]
- Anghel, D.-M.C.; Nițescu, G.V.; Tiron, A.-T.; Guțu, C.M.; Baconi, D.L. Understanding the Mechanisms of Action and Effects of Drugs of Abuse. Molecules 2023, 28, 4969. [Google Scholar] [CrossRef]
- Perry, A.N.; Westenbroek, C.; Becker, J.B. The development of a preference for cocaine over food identifies individual rats with addiction-like behaviors. PLoS ONE 2013, 8, e79465. [Google Scholar] [CrossRef]
- Chiu, V.M.; Schenk, J.O. Mechanism of action of methamphetamine within the catecholamine and serotonin areas of the central nervous system. Curr. Drug Abus. Rev. 2012, 5, 227–242. [Google Scholar] [CrossRef] [PubMed]
- Chomchai, C.; Chomchai, S. Global patterns of methamphetamine use. Curr. Opin. Psychiatry 2015, 28, 269–274. [Google Scholar] [CrossRef] [PubMed]
- Gangu, K.; Bobba, A.; Basida, S.D.; Avula, S.; Chela, H.; Singh, S. Trends of Cocaine Use and Manifestations in Hospitalized Patients: A Cross-Sectional Study. Cureus 2022, 14, e22090. [Google Scholar] [CrossRef]
- Chen, G.; Lai, S.; Bao, G.; Ke, J.; Meng, X.; Lu, S.; Wu, X.; Xu, H.; Wu, F.; Xu, Y.; et al. Distinct reward processing by subregions of the nucleus accumbens. Cell Rep. 2023, 42, 112069. [Google Scholar] [CrossRef] [PubMed]
- Becker-Krail, D.D.; Walker, W.H.; Nelson, R.J. The Ventral Tegmental Area and Nucleus Accumbens as Circadian Oscillators: Implications for Drug Abuse and Substance Use Disorders. Front. Physiol. 2022, 13, 886704. Available online: https://www.frontiersin.org/articles/10.3389/fphys.2022.886704 (accessed on 11 December 2023). [CrossRef] [PubMed]
- Lee, A.M.; Messing, R.O. Protein Kinases and Addiction. Ann. N. Y. Acad. Sci. 2008, 1141, 22–57. [Google Scholar] [CrossRef] [PubMed]
- Amaral, I.M.; Scheffauer, L.; Hofer, A.; El Rawas, R. Protein kinases in natural versus drug reward. Pharmacol. Biochem. Behav. 2022, 221, 173472. [Google Scholar] [CrossRef]
- Shi, X.; McGinty, J.F. Repeated amphetamine treatment increases phosphorylation of extracellular signal-regulated kinase, protein kinase B, and cyclase response element-binding protein in the rat striatum. J. Neurochem. 2007, 103, 706–713. [Google Scholar] [CrossRef]
- Jia, W.; Kawahata, I.; Cheng, A.; Fukunaga, K. The Role of CaMKII and ERK Signaling in Addiction. Int. J. Mol. Sci. 2021, 22, 3189. [Google Scholar] [CrossRef]
- Wheeler, J.M.; Reed, C.; Burkhart-Kasch, S.; Li, N.; Cunningham, C.L.; Janowsky, A.; Franken, F.H.; Wiren, K.M.; Hashimoto, J.G.; Scibelli, A.C.; et al. Genetically correlated effects of selective breeding for high and low methamphetamine consumption. Genes Brain Behav. 2009, 8, 758–771. [Google Scholar] [CrossRef]
- Hitzemann, R.; Iancu, O.D.; Reed, C.; Baba, H.; Lockwood, D.R.; Phillips, T.J. Regional Analysis of the Brain Transcriptome in Mice Bred for High and Low Methamphetamine Consumption. Brain Sci. 2019, 9, 155. [Google Scholar] [CrossRef]
- Phillips, T.J.; Roy, T.; Aldrich, S.J.; Baba, H.; Erk, J.; Mootz, J.R.K.; Reed, C.; Chesler, E.J. Confirmation of a Causal Taar1 Allelic Variant in Addiction-Relevant Methamphetamine Behaviors. Front. Psychiatry 2021, 12, 725839. Available online: https://www.frontiersin.org/articles/10.3389/fpsyt.2021.725839 (accessed on 18 January 2024). [CrossRef]
- Shabani, S.; Houlton, S.; Ghimire, B.; Tonello, D.; Reed, C.; Baba, H.; Aldrich, S.; Phillips, T.J. Robust aversive effects of trace amine-associated receptor 1 activation in mice. Neuropsychopharmacology 2023, 48, 1–9. [Google Scholar] [CrossRef]
- Stafford, A.M.; Reed, C.; Baba, H.; Walter, N.A.; Mootz, J.R.; Williams, R.W.; Neve, K.A.; Fedorov, L.M.; Janowsky, A.J.; Phillips, T.J.; et al. Taar1 gene variants have a causal role in methamphetamine intake and response and interact with Oprm1. eLife 2019, 8, e46472. [Google Scholar] [CrossRef] [PubMed]
- Althobaiti, Y.S.; Almalki, A.H. Effects of environmental enrichment on reinstatement of methamphetamine-induced conditioned place preference. Behav. Brain Res. 2020, 379, 112372. [Google Scholar] [CrossRef] [PubMed]
- Stafford, A.M.; Reed, C.; Phillips, T.J. Non-genetic factors that influence methamphetamine intake in a genetic model of differential methamphetamine consumption. Psychopharmacology 2020, 237, 3315–3336. [Google Scholar] [CrossRef] [PubMed]
- Gibney, E.R.; Nolan, C.M. Epigenetics and gene expression. Heredity 2010, 105, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Nachtergaele, S.; He, C. The emerging biology of RNA post-transcriptional modifications. RNA Biol. 2017, 14, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017, 169, 1187–1200. [Google Scholar] [CrossRef] [PubMed]
- Lence, T.; Akhtar, J.; Bayer, M.; Schmid, K.; Spindler, L.; Ho, C.H.; Kreim, N.; Andrade-Navarro, M.A.; Poeck, B.; Helm, M.; et al. m6A modulates neuronal functions and sex determination in Drosophila. Nature 2016, 540, 242–247. [Google Scholar] [CrossRef]
- Zeng, H.; Sanes, J.R. Neuronal cell-type classification: Challenges, opportunities and the path forward. Nat. Rev. Neurosci. 2017, 18, 530–546. [Google Scholar] [CrossRef] [PubMed]
- Arendt, D. The evolution of cell types in animals: Emerging principles from molecular studies. Nat. Rev. Genet. 2008, 9, 868–882. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H. What is a cell type and how to define it? Cell 2022, 185, 2739–2755. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.A.; Gouwens, N.W.; Tasic, B.; Collman, F.; van Velthoven, C.T.; Bakken, T.E.; Hawrylycz, M.J.; Zeng, H.; Lein, E.S.; Bernard, A. Common cell type nomenclature for the mammalian brain. eLife 2020, 9, e59928. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.D.; Asan, L.; John, J.; Beretta, C.; Kuner, T.; Knabbe, J. Accurate classification of major brain cell types using in vivo imaging and neural network processing. PLoS Biol. 2023, 21, e3002357. [Google Scholar] [CrossRef]
- O’Donnell, E.A.; Ernst, D.N.; Hingorani, R. Multiparameter flow cytometry: Advances in high resolution analysis. Immune Netw. 2013, 13, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Baron, C.S.; Barve, A.; Muraro, M.J.; van der Linden, R.; Dharmadhikari, G.; Lyubimova, A.; de Koning, E.J.; van Oudenaarden, A. Cell Type Purification by Single-Cell Transcriptome-Trained Sorting. Cell 2019, 179, 527–542.e19. [Google Scholar] [CrossRef]
- Klein, A.M.; Mazutis, L.; Akartuna, I.; Tallapragada, N.; Veres, A.; Li, V.; Peshkin, L.; Weitz, D.A.; Kirschner, M.W. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 2015, 161, 1187–1201. [Google Scholar] [CrossRef]
- Macosko, E.Z.; Basu, A.; Satija, R.; Nemesh, J.; Shekhar, K.; Goldman, M.; Tirosh, I.; Bialas, A.R.; Kamitaki, N.; Martersteck, E.M.; et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 2015, 161, 1202–1214. [Google Scholar] [CrossRef]
- Zheng, G.X.Y.; Terry, J.M.; Belgrader, P.; Ryvkin, P.; Bent, Z.W.; Wilson, R.; Ziraldo, S.B.; Wheeler, T.D.; McDermott, G.P.; Zhu, J.; et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017, 8, 14049. [Google Scholar] [CrossRef]
- Buenrostro, J.D.; Wu, B.; Chang, H.Y.; Greenleaf, W.J. ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr. Protoc. Mol. Biol. 2015, 109, 21.29.1–21.29.9. [Google Scholar] [CrossRef]
- Grandi, F.C.; Modi, H.; Kampman, L.; Corces, M.R. Chromatin accessibility profiling by ATAC-seq. Nat. Protoc. 2022, 17, 1518–1552. [Google Scholar] [CrossRef]
- Jansen, C.; Ramirez, R.N.; El-Ali, N.C.; Gomez-Cabrero, D.; Tegner, J.; Merkenschlager, M.; Conesa, A.; Mortazavi, A. Building gene regulatory networks from scATAC-seq and scRNA-seq using Linked Self Organizing Maps. PLoS Comput. Biol. 2019, 15, e1006555. [Google Scholar] [CrossRef]
- Zhang, Z.; Yang, C.; Zhang, X. scDART: Integrating unmatched scRNA-seq and scATAC-seq data and learning cross-modality relationship simultaneously. Genome Biol. 2022, 23, 139. [Google Scholar] [CrossRef]
- Lipovsek, M.; Bardy, C.; Cadwell, C.R.; Hadley, K.; Kobak, D.; Tripathy, S.J. Patch-seq: Past, Present, and Future. J. Neurosci. Off. J. Soc. Neurosci. 2021, 41, 937–946. [Google Scholar] [CrossRef]
- Lee, B.R.; Budzillo, A.; Hadley, K.; Miller, J.A.; Jarsky, T.; Baker, K.; Hill, D.; Kim, L.; Mann, R.; Ng, L.; et al. Scaled, high fidelity electrophysiological, morphological, and transcriptomic cell characterization. eLife 2021, 10, e65482. [Google Scholar] [CrossRef] [PubMed]
- van den Hurk, M.; Erwin, J.A.; Yeo, G.W.; Gage, F.H.; Bardy, C. Patch-Seq Protocol to Analyze the Electrophysiology, Morphology and Transcriptome of Whole Single Neurons Derived From Human Pluripotent Stem Cells. Front. Mol. Neurosci. 2018, 11, 261. Available online: https://www.frontiersin.org/articles/10.3389/fnmol.2018.00261 (accessed on 11 December 2023). [CrossRef] [PubMed]
- Koob, G.F.; Volkow, N.D. Neurobiology of addiction: A neurocircuitry analysis. Lancet Psychiatry 2016, 3, 760–773. [Google Scholar] [CrossRef] [PubMed]
- Uhl, G.R.; Koob, G.F.; Cable, J. The neurobiology of addiction. Ann. N. Y. Acad. Sci. 2019, 1451, 5–28. [Google Scholar] [CrossRef] [PubMed]
- Poisson, C.L.; Engel, L.; Saunders, B.T. Dopamine Circuit Mechanisms of Addiction-Like Behaviors. Front. Neural Circuits 2021, 15, 125. Available online: https://www.frontiersin.org/articles/10.3389/fncir.2021.752420 (accessed on 11 December 2023). [CrossRef] [PubMed]
- Srinivasan, C.; Phan, B.N.; Lawler, A.J.; Ramamurthy, E.; Kleyman, M.; Brown, A.R.; Kaplow, I.M.; Wirthlin, M.E.; Pfenning, A.R. Addiction-Associated Genetic Variants Implicate Brain Cell Type- and Region-Specific Cis-Regulatory Elements in Addiction Neurobiology. J. Neurosci. Off. J. Soc. Neurosci. 2021, 41, 9008–9030. [Google Scholar] [CrossRef]
- Tran, M.N.; Maynard, K.R.; Spangler, A.; Huuki, L.A.; Montgomery, K.D.; Sadashivaiah, V.; Tippani, M.; Barry, B.K.; Hancock, D.B.; Hicks, S.C.; et al. Single-nucleus transcriptome analysis reveals cell-type-specific molecular signatures across reward circuitry in the human brain. Neuron 2021, 109, 3088–3103.e5. [Google Scholar] [CrossRef]
- Navandar, M.; Martín-García, E.; Maldonado, R.; Lutz, B.; Gerber, S.; de Azua, I.R. Transcriptional signatures in prefrontal cortex confer vulnerability versus resilience to food and cocaine addiction-like behavior. Sci. Rep. 2021, 11, 9076. [Google Scholar] [CrossRef]
- Chen, R.; Blosser, T.R.; Djekidel, M.N.; Hao, J.; Bhattacherjee, A.; Chen, W.; Tuesta, L.M.; Zhuang, X.; Zhang, Y. Decoding molecular and cellular heterogeneity of mouse nucleus accumbens. Nat. Neurosci. 2021, 24, 1757–1771. [Google Scholar] [CrossRef]
- Bhatia, P.; Yang, L.; Luo, J.X.J.; Xu, M.; Renthal, W. Epigenomic profiling of mouse nucleus accumbens at single-cell resolution. Mol. Cell. Neurosci. 2023, 126, 103857. [Google Scholar] [CrossRef]
- Gangarossa, G.; Espallergues, J.; D’Exaerde, A.d.K.; El Mestikawy, S.; Gerfen, C.R.; Hervé, D.; Girault, J.-A.; Valjent, E. Distribution and compartmental organization of GABAergic medium-sized spiny neurons in the mouse nucleus accumbens. Front. Neural Circuits 2013, 7, 22. [Google Scholar] [CrossRef] [PubMed]
- Phillips, R.A.; Tuscher, J.J.; Black, S.L.; Andraka, E.; Fitzgerald, N.D.; Ianov, L.; Day, J.J. An atlas of transcriptionally defined cell populations in the rat ventral tegmental area. Cell Rep. 2022, 39, 110616. [Google Scholar] [CrossRef] [PubMed]
- Savell, K.E.; Tuscher, J.J.; Zipperly, M.E.; Duke, C.G.; Phillips, R.A.; Bauman, A.J.; Thukral, S.; Sultan, F.A.; Goska, N.A.; Ianov, L.; et al. A dopamine-induced gene expression signature regulates neuronal function and cocaine response. Sci. Adv. 2020, 6, eaba4221. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.; Browne, C.J.; Nöbauer, T.; Vaziri, A.; Friedman, J.M.; Nestler, E.J. Drugs of abuse hijack a mesolimbic pathway that processes homeostatic need. bioRxiv 2023, 2023.09.03.556059. [Google Scholar] [CrossRef]
- Zhou, J.L.; de Guglielmo, G.; Ho, A.J.; Kallupi, M.; Pokhrel, N.; Li, H.-R.; Chitre, A.S.; Munro, D.; Mohammadi, P.; Carrette, L.L.G.; et al. Single-nucleus genomics in outbred rats with divergent cocaine addiction-like behaviors reveals changes in amygdala GABAergic inhibition. Nat. Neurosci. 2023, 26, 1868–1879. [Google Scholar] [CrossRef] [PubMed]
- Čechová, B.; Šlamberová, R. Methamphetamine, neurotransmitters and neurodevelopment. Physiol. Res. 2021, 70, S301–S315. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Dong, X.; Awan, M.U.N.; Bai, J. Epigenetic mechanisms involved in methamphetamine addiction. Front. Pharmacol. 2022, 13, 984997. Available online: https://www.frontiersin.org/articles/10.3389/fphar.2022.984997 (accessed on 11 December 2023). [CrossRef] [PubMed]
- Li, K.; Ling, H.; Wang, X.; Xie, Q.; Gu, C.; Luo, W.; Qiu, P. The role of NF-κB signaling pathway in reactive astrocytes among neurodegeneration after methamphetamine exposure by integrated bioinformatics. Prog. Neuropsychopharmacol. Biol. Psychiatry 2024, 129, 110909. [Google Scholar] [CrossRef]
- Re, G.-F.; Jia, J.; Xu, Y.; Zhang, Z.; Xie, Z.-R.; Kong, D.; Lu, D.; Li, Y.; Peng, Q.-Y.; Yu, J.; et al. Dynamics and correlations in multiplex immune profiling reveal persistent immune inflammation in male drug users after withdrawal. Int. Immunopharmacol. 2022, 107, 108696. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Jin, X.; Li, T.; Ye, Z. Brain organoids: Establishment and application. Front. Cell Dev. Biol. 2022, 10, 1029873. Available online: https://www.frontiersin.org/articles/10.3389/fcell.2022.1029873 (accessed on 12 December 2023). [CrossRef]
- Dang, J.; Tiwari, S.K.; Agrawal, K.; Hui, H.; Qin, Y.; Rana, T.M. Glial cell diversity and methamphetamine-induced neuroinflammation in human cerebral organoids. Mol. Psychiatry 2021, 26, 1194–1207. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wang, L.; Bu, Q.; Wei, Q.; Jiang, L.; Dai, Y.; Zhang, N.; Kuang, W.; Zhao, Y.; Cen, X. Methamphetamine exposure drives cell cycle exit and aberrant differentiation in rat hippocampal-derived neurospheres. Front. Pharmacol. 2023, 14, 1242109. Available online: https://www.frontiersin.org/articles/10.3389/fphar.2023.1242109 (accessed on 13 December 2023). [CrossRef]
- Bhattacherjee, A.; Djekidel, M.N.; Chen, R.; Chen, W.; Tuesta, L.M.; Zhang, Y. Cell type-specific transcriptional programs in mouse prefrontal cortex during adolescence and addiction. Nat. Commun. 2019, 10, 4169. [Google Scholar] [CrossRef]
- Perrotti, L.I.; Bolaños, C.A.; Choi, K.; Russo, S.J.; Edwards, S.; Ulery, P.G.; Wallace, D.L.; Self, D.W.; Nestler, E.J.; Barrot, M. DeltaFosB accumulates in a GABAergic cell population in the posterior tail of the ventral tegmental area after psychostimulant treatment. Eur. J. Neurosci. 2005, 21, 2817–2824. [Google Scholar] [CrossRef]
- Vaillancourt, K.; Ernst, C.; Mash, D.; Turecki, G. DNA Methylation Dynamics and Cocaine in the Brain: Progress and Prospects. Genes 2017, 8, 138. [Google Scholar] [CrossRef]
- Eisch, A.J.; Bolaños, C.A.; de Wit, J.; Simonak, R.D.; Pudiak, C.M.; Barrot, M.; Verhaagen, J.; Nestler, E.J. Brain-derived neurotrophic factor in the ventral midbrain-nucleus accumbens pathway: A role in depression. Biol. Psychiatry 2003, 54, 994–1005. [Google Scholar] [CrossRef] [PubMed]
- Mews, P.; Cunningham, A.M.; Scarpa, J.; Ramakrishnan, A.; Hicks, E.M.; Bolnick, S.; Garamszegi, S.; Shen, L.; Mash, D.C.; Nestler, E.J. Convergent abnormalities in striatal gene networks in human cocaine use disorder and mouse cocaine administration models. Sci. Adv. 2023, 9, eadd8946. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.R.; Chun, J.-W.; Kwak, S.M.; Bang, S.H.; Jin, Y.-B.; Lee, Y.; Kim, H.-N.; Chang, K.-T.; Chai, Y.G.; Lee, S.-R.; et al. Effects of acute and chronic methamphetamine administration on cynomolgus monkey hippocampus structure and cellular transcriptome. Toxicol. Appl. Pharmacol. 2018, 355, 68–79. [Google Scholar] [CrossRef] [PubMed]
- Ujike, H.; Onoue, T.; Akiyama, K.; Hamamura, T.; Otsuki, S. Effects of selective D-1 and D-2 dopamine antagonists on development of methamphetamine-induced behavioral sensitization. Psychopharmacology 1989, 98, 89–92. [Google Scholar] [CrossRef] [PubMed]
- Gerfen, C.R.; Miyachi, S.; Paletzki, R.; Brown, P. D1 dopamine receptor supersensitivity in the dopamine-depleted striatum results from a switch in the regulation of ERK1/2/MAP kinase. J. Neurosci. Off. J. Soc. Neurosci. 2002, 22, 5042–5054. [Google Scholar] [CrossRef] [PubMed]
- Mizoguchi, H.; Yamada, K.; Mizuno, M.; Mizuno, T.; Nitta, A.; Noda, Y.; Nabeshima, T. Regulations of methamphetamine reward by extracellular signal-regulated kinase 1/2/ets-like gene-1 signaling pathway via the activation of dopamine receptors. Mol. Pharmacol. 2004, 65, 1293–1301. [Google Scholar] [CrossRef] [PubMed]
- Valjent, E.; Pascoli, V.; Svenningsson, P.; Paul, S.; Enslen, H.; Corvol, J.-C.; Stipanovich, A.; Caboche, J.; Lombroso, P.J.; Nairn, A.C.; et al. Regulation of a protein phosphatase cascade allows convergent dopamine and glutamate signals to activate ERK in the striatum. Proc. Natl. Acad. Sci. USA 2005, 102, 491–496. [Google Scholar] [CrossRef]
- Limanaqi, F.; Gambardella, S.; Biagioni, F.; Busceti, C.L.; Fornai, F. Epigenetic Effects Induced by Methamphetamine and Methamphetamine-Dependent Oxidative Stress. Oxid. Med. Cell. Longev. 2018, 2018, e4982453. [Google Scholar] [CrossRef]
- Mews, P.; Walker, D.M.; Nestler, E.J. Epigenetic Priming in Drug Addiction. Cold Spring Harb. Symp. Quant. Biol. 2018, 83, 131–139. [Google Scholar] [CrossRef]
- Stewart, A.F.; Fulton, S.L.; Maze, I. Epigenetics of Drug Addiction. Cold Spring Harb. Perspect. Med. 2021, 11, a040253. [Google Scholar] [CrossRef]
- Robison, A.J.; Nestler, E.J. Transcriptional and epigenetic mechanisms of addiction. Nat. Rev. Neurosci. 2011, 12, 623–637. [Google Scholar] [CrossRef] [PubMed]
- Liu, E.; Pang, K.; Liu, M.; Tan, X.; Hang, Z.; Mu, S.; Han, W.; Yue, Q.; Comai, S.; Sun, J. KCNQ3 Normalizes Hyperactivity of VTA-NAcLat Circuit and Attenuates Methamphetamine Addiction in Mic. 2022. Available online: https://www.researchsquare.com/article/rs-1977142/v1 (accessed on 11 December 2023).
- Liu, E.; Pang, K.; Liu, M.; Tan, X.; Hang, Z.; Mu, S.; Han, W.; Yue, Q.; Comai, S.; Sun, J. Activation of Kv7 channels normalizes hyperactivity of the VTA-NAcLat circuit and attenuates methamphetamine-induced conditioned place preference and sensitization in mice. Mol. Psychiatry 2023, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Lähnemann, D.; Köster, J.; Szczurek, E.; McCarthy, D.J.; Hicks, S.C.; Robinson, M.D.; Vallejos, C.A.; Campbell, K.R.; Beerenwinkel, N.; Mahfouz, A.; et al. Eleven grand challenges in single-cell data science. Genome Biol. 2020, 21, 31. [Google Scholar] [CrossRef]
- Weinhold, B. Epigenetics: The Science of Change. Environ. Health Perspect. 2006, 114, A160–A167. [Google Scholar] [CrossRef]
- Cavalli, G.; Heard, E. Advances in epigenetics link genetics to the environment and disease. Nature 2019, 571, 489–499. [Google Scholar] [CrossRef]
- Fitz-James, M.H.; Cavalli, G. Molecular mechanisms of transgenerational epigenetic inheritance. Nat. Rev. Genet. 2022, 23, 325–341. [Google Scholar] [CrossRef]
- Li, S.; Tollefsbol, T.O. DNA methylation methods: Global DNA methylation and methylomic analyses. Methods 2021, 187, 28–43. [Google Scholar] [CrossRef]
- Geng, H.; Chen, H.; Wang, H.; Wang, L. The Histone Modifications of Neuronal Plasticity. Neural Plast. 2021, 2021, 6690523. [Google Scholar] [CrossRef]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Haberland, M.; Montgomery, R.L.; Olson, E.N. The many roles of histone deacetylases in development and physiology: Implications for disease and therapy. Nat. Rev. Genet. 2009, 10, 32–42. [Google Scholar] [CrossRef]
- Shi, Y. Histone lysine demethylases: Emerging roles in development, physiology and disease. Nat. Rev. Genet. 2007, 8, 829–833. [Google Scholar] [CrossRef] [PubMed]
- Bure, I.V.; Nemtsova, M.V.; Kuznetsova, E.B. Histone Modifications and Non-Coding RNAs: Mutual Epigenetic Regulation and Role in Pathogenesis. Int. J. Mol. Sci. 2022, 23, 5801. [Google Scholar] [CrossRef] [PubMed]
- Farrelly, L.A.; Zheng, S.; Schrode, N.; Topol, A.; Bhanu, N.V.; Bastle, R.M.; Ramakrishnan, A.; Chan, J.C.; Cetin, B.; Flaherty, E.; et al. Chromatin profiling in human neurons reveals aberrant roles for histone acetylation and BET family proteins in schizophrenia. Nat. Commun. 2022, 13, 2195. [Google Scholar] [CrossRef] [PubMed]
- Farrelly, L.A.; Thompson, R.E.; Zhao, S.; Lepack, A.E.; Lyu, Y.; Bhanu, N.V.; Zhang, B.; Loh, Y.-H.E.; Ramakrishnan, A.; Vadodaria, K.C.; et al. Histone serotonylation is a permissive modification that enhances TFIID binding to H3K4me3. Nature 2019, 567, 535–539. [Google Scholar] [CrossRef]
- Zhao, Y.; Garcia, B.A. Comprehensive Catalog of Currently Documented Histone Modifications. Cold Spring Harb. Perspect. Biol. 2015, 7, a025064. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.C.; Maze, I. Nothing Is Yet Set in (Hi)stone: Novel Post-Translational Modifications Regulating Chromatin Function. Trends Biochem. Sci. 2020, 45, 829–844. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.-S.; Wu, F.; Jin, Y.-M.; Chang, W.-Q.; Xu, T.-M. HDAC11: A rising star in epigenetics. Biomed. Pharmacother. Biomedecine Pharmacother. 2020, 131, 110607. [Google Scholar] [CrossRef]
- Mattick, J.S.; Makunin, I.V. Non-coding RNA. Hum. Mol. Genet. 2006, 15, R17–R29. [Google Scholar] [CrossRef]
- Suzuki, M.M.; Bird, A. DNA methylation landscapes: Provocative insights from epigenomics. Nat. Rev. Genet. 2008, 9, 465–476. [Google Scholar] [CrossRef]
- Guerrero-Bosagna, C.; Settles, M.; Lucker, B.; Skinner, M.K. Epigenetic transgenerational actions of vinclozolin on promoter regions of the sperm epigenome. PLoS ONE 2010, 5, e13100. [Google Scholar] [CrossRef]
- Franklin, T.B.; Russig, H.; Weiss, I.C.; Gräff, J.; Linder, N.; Michalon, A.; Vizi, S.; Mansuy, I.M. Epigenetic transmission of the impact of early stress across generations. Biol. Psychiatry 2010, 68, 408–415. [Google Scholar] [CrossRef] [PubMed]
- Anier, K.; Malinovskaja, K.; Aonurm-Helm, A.; Zharkovsky, A.; Kalda, A. DNA methylation regulates cocaine-induced behavioral sensitization in mice. Neuropsychopharmacol. Off. Publ. Am. Coll. Neuropsychopharmacol. 2010, 35, 2450–2461. [Google Scholar] [CrossRef] [PubMed]
- Im, H.-I.; Hollander, J.A.; Bali, P.; Kenny, P.J. MeCP2 controls BDNF expression and cocaine intake through homeostatic interactions with microRNA-212. Nat. Neurosci. 2010, 13, 1120–1127. [Google Scholar] [CrossRef] [PubMed]
- Bodetto, S.P.; Romieu, P.; Sartori, M.; Tesone-Coelho, C.; Majchrzak, M.; Barbelivien, A.; Zwiller, J.; Anglard, P. Differential regulation of MeCP2 and PP1 in passive or voluntary administration of cocaine or food. Int. J. Neuropsychopharmacol. 2014, 17, 2031–2044. [Google Scholar] [CrossRef] [PubMed]
- Greer, E.L.; Shi, Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012, 13, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Wilkinson, M.; Liu, X.; Purushothaman, I.; Ferguson, D.; Vialou, V.; Maze, I.; Shao, N.; Kennedy, P.; Koo, J.; et al. Chronic cocaine-regulated epigenomic changes in mouse nucleus accumbens. Genome Biol. 2014, 15, R65. [Google Scholar] [CrossRef]
- Damez-Werno, D.M.; Sun, H.; Scobie, K.N.; Shao, N.; Rabkin, J.; Dias, C.; Calipari, E.S.; Maze, I.; Pena, C.J.; Walker, D.M.; et al. Histone arginine methylation in cocaine action in the nucleus accumbens. Proc. Natl. Acad. Sci. USA 2016, 113, 9623–9628. [Google Scholar] [CrossRef]
- Renthal, W.; Kumar, A.; Xiao, G.; Wilkinson, M.; Covington, H.E.; Maze, I.; Sikder, D.; Robison, A.J.; LaPlant, Q.; Dietz, D.M.; et al. Genome Wide Analysis of Chromatin Regulation by Cocaine Reveals a Novel Role for Sirtuins. Neuron 2009, 62, 335–348. [Google Scholar] [CrossRef]
- Chrivia, J.C.; Kwok, R.P.S.; Lamb, N.; Hagiwara, M.; Montminy, M.R.; Goodman, R.H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature 1993, 365, 855–859. [Google Scholar] [CrossRef]
- Schmidt, H.D.; Sangrey, G.R.; Darnell, S.B.; Schassburger, R.L.; Cha, J.J.; Pierce, R.C.; Sadri-Vakili, G. Increased brain-derived neurotrophic factor (BDNF) expression in the ventral tegmental area during cocaine abstinence is associated with increased histone acetylation at BDNF exon I-containing promoters. J. Neurochem. 2012, 120, 202–209. [Google Scholar] [CrossRef]
- Taniguchi, M.; Carreira, M.B.; Smith, L.N.; Zirlin, B.C.; Neve, R.L.; Cowan, C.W. Histone deacetylase 5 limits cocaine reward through cAMP-induced nuclear import. Neuron 2012, 73, 108–120. [Google Scholar] [CrossRef]
- Host, L.; Dietrich, J.-B.; Carouge, D.; Aunis, D.; Zwiller, J. Cocaine self-administration alters the expression of chromatin-remodelling proteins; modulation by histone deacetylase inhibition. J. Psychopharmacol. Oxf. Engl. 2011, 25, 222–229. [Google Scholar] [CrossRef]
- Smith, A.C.W.; Kenny, P.J. HDAC5 Regulates the Formation of Drug Memories. Trends Mol. Med. 2018, 24, 106–108. [Google Scholar] [CrossRef]
- Taniguchi, M.; Carreira, M.B.; Cooper, Y.A.; Bobadilla, A.-C.; Heinsbroek, J.A.; Koike, N.; Larson, E.B.; Balmuth, E.A.; Hughes, B.W.; Penrod, R.D.; et al. HDAC5 and its target gene, NPAS4, function in the nucleus accumbens to regulate cocaine conditioned behaviors. Neuron 2017, 96, 130–144.e6. [Google Scholar] [CrossRef]
- Brami-Cherrier, K.; Valjent, E.; Hervé, D.; Darragh, J.; Corvol, J.-C.; Pages, C.; Simon, A.J.; Girault, J.-A.; Caboche, J. Parsing Molecular and Behavioral Effects of Cocaine in Mitogen- and Stress-Activated Protein Kinase-1-Deficient Mice. J. Neurosci. 2005, 25, 11444–11454. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Choi, K.-H.; Renthal, W.; Tsankova, N.M.; Theobald, D.E.; Truong, H.-T.; Russo, S.J.; LaPlant, Q.; Sasaki, T.S.; Whistler, K.N.; et al. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron 2005, 48, 303–314. [Google Scholar] [CrossRef] [PubMed]
- Renthal, W.; Maze, I.; Krishnan, V.; Covington, H.E.; Xiao, G.; Kumar, A.; Russo, S.J.; Graham, A.; Tsankova, N.; Kippin, T.E.; et al. Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron 2007, 56, 517–529. [Google Scholar] [CrossRef]
- Ferguson, D.; Koo, J.W.; Feng, J.; Heller, E.; Rabkin, J.; Heshmati, M.; Renthal, W.; Neve, R.; Liu, X.; Shao, N.; et al. Essential role of SIRT1 signaling in the nucleus accumbens in cocaine and morphine action. J. Neurosci. Off. J. Soc. Neurosci. 2013, 33, 16088–16098. [Google Scholar] [CrossRef]
- Zell, V.; Steinkellner, T.; Hollon, N.G.; Warlow, S.M.; Souter, E.; Faget, L.; Hunker, A.C.; Jin, X.; Zweifel, L.S.; Hnasko, T.S. VTA Glutamate Neuron Activity Drives Positive Reinforcement Absent Dopamine Co-release. Neuron 2020, 107, 864–873.e4. [Google Scholar] [CrossRef] [PubMed]
- Bach, S.V.; Hegde, A.N. The proteasome and epigenetics: Zooming in on histone modifications. Biomol. Concepts 2016, 7, 215–227. [Google Scholar] [CrossRef]
- Jarome, T.J.; Helmstetter, F.J. The ubiquitin–proteasome system as a critical regulator of synaptic plasticity and long-term memory formation. Neurobiol. Learn. Mem. 2013, 105, 107–116. [Google Scholar] [CrossRef]
- Werner, C.T.; Viswanathan, R.; Martin, J.A.; Gobira, P.H.; Mitra, S.; Thomas, S.A.; Wang, Z.-J.; Liu, J.-F.; Stewart, A.F.; Neve, R.L.; et al. E3 Ubiquitin-Protein Ligase SMURF1 in the Nucleus Accumbens Mediates Cocaine Seeking. Biol. Psychiatry 2018, 84, 881–892. [Google Scholar] [CrossRef]
- Zhang, Y. Transcriptional regulation by histone ubiquitination and deubiquitination. Genes Dev. 2003, 17, 2733–2740. [Google Scholar] [CrossRef]
- Lepack, A.E.; Werner, C.T.; Stewart, A.F.; Fulton, S.L.; Zhong, P.; Farrelly, L.A.; Smith, A.C.W.; Ramakrishnan, A.; Lyu, Y.; Bastle, R.M.; et al. Dopaminylation of histone H3 in ventral tegmental area regulates cocaine seeking. Science 2020, 368, 197–201. [Google Scholar] [CrossRef]
- Tapocik, J.D.; Barbier, E.; Flanigan, M.; Solomon, M.; Pincus, A.; Pilling, A.; Sun, H.; Schank, J.R.; King, C.; Heilig, M. microRNA-206 in rat medial prefrontal cortex regulates BDNF expression and alcohol drinking. J. Neurosci. Off. J. Soc. Neurosci. 2014, 34, 4581–4588. [Google Scholar] [CrossRef]
- Li, J.; Li, J.; Liu, X.; Qin, S.; Guan, Y.; Liu, Y.; Cheng, Y.; Chen, X.; Li, W.; Wang, S.; et al. MicroRNA expression profile and functional analysis reveal that miR-382 is a critical novel gene of alcohol addiction. EMBO Mol. Med. 2013, 5, 1402–1414. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.-J.; Seto, E. HATs and HDACs: From structure, function and regulation to novel strategies for therapy and prevention. Oncogene 2007, 26, 5310–5318. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Luo, T.; Dong, H.; Zhang, C.; Liu, T.; Zhang, X.; Hao, W. Genome-Wide DNA Methylation Analysis in Male Methamphetamine Users With Different Addiction Qualities. Front. Psychiatry 2020, 11, 588229. [Google Scholar] [CrossRef] [PubMed]
- Asaoka, Y.; Won, M.; Morita, T.; Ishikawa, E.; Lee, Y.-A.; Goto, Y. Monoamine and genome-wide DNA methylation investigation in behavioral addiction. Sci. Rep. 2020, 10, 11760. [Google Scholar] [CrossRef] [PubMed]
- Iamjan, S.-A.; Thanoi, S.; Watiktinkorn, P.; Fachim, H.; Dalton, C.F.; Nudmamud-Thanoi, S.; Reynolds, G.P. Changes of BDNF exon IV DNA methylation are associated with methamphetamine dependence. Epigenomics 2021, 13, 953–965. [Google Scholar] [CrossRef]
- Salehzadeh, S.A.; Mohammadian, A.; Salimi, F. Effect of chronic methamphetamine injection on levels of BDNF mRNA and its CpG island methylation in prefrontal cortex of rats. Asian J. Psychiatry 2020, 48, 101884. [Google Scholar] [CrossRef]
- Moszczynska, A.; Flack, A.; Qiu, P.; Muotri, A.R.; Killinger, B.A. Neurotoxic Methamphetamine Doses Increase LINE-1 Expression in the Neurogenic Zones of the Adult Rat Brain. Sci. Rep. 2015, 5, 14356. [Google Scholar] [CrossRef]
- Walker, D.M.; Nestler, E.J. Neuroepigenetics and addiction. Handb. Clin. Neurol. 2018, 148, 747–765. [Google Scholar] [CrossRef]
- Li, H.; Chen, J.-A.; Ding, Q.-Z.; Lu, G.-Y.; Wu, N.; Su, R.-B.; Li, F.; Li, J. Behavioral sensitization induced by methamphetamine causes differential alterations in gene expression and histone acetylation of the prefrontal cortex in rats. BMC Neurosci. 2021, 22, 24. [Google Scholar] [CrossRef]
- Wang, Y.; Wei, T.; Zhao, W.; Ren, Z.; Wang, Y.; Zhou, Y.; Song, X.; Zhou, R.; Zhang, X.; Jiao, D. MicroRNA-181a Is Involved in Methamphetamine Addiction through the ERAD Pathway. Front. Mol. Neurosci. 2021, 14, 667725. Available online: https://www.frontiersin.org/articles/10.3389/fnmol.2021.667725 (accessed on 12 December 2023). [CrossRef] [PubMed]
- Qian, H.; Shang, Q.; Liang, M.; Gao, B.; Xiao, J.; Wang, J.; Li, A.; Yang, C.; Yin, J.; Chen, G.; et al. MicroRNA-31-3p/RhoA signaling in the dorsal hippocampus modulates methamphetamine-induced conditioned place preference in mice. Psychopharmacology 2021, 238, 3207–3219. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Liang, M.; Zhu, L.; Zhou, T.-T.; Wang, Y.; Wang, R.; Wu, F.-F.; Goh, E.L.K.; Chen, T. Potential Ago2/miR-3068-5p Cascades in the Nucleus Accumbens Contribute to Methamphetamine-Induced Locomotor Sensitization of Mice. Front. Pharmacol. 2021, 12, 708034. Available online: https://www.frontiersin.org/articles/10.3389/fphar.2021.708034 (accessed on 12 December 2023). [CrossRef] [PubMed]
- Renthal, W.; Nestler, E.J. Epigenetic mechanisms in drug addiction. Trends Mol. Med. 2008, 14, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Ikegami, D.; Narita, M.; Imai, S.; Miyashita, K.; Tamura, R.; Narita, M.; Takagi, S.; Yokomizo, A.; Takeshima, H.; Ando, T.; et al. Epigenetic modulation at the CCR2 gene correlates with the maintenance of behavioral sensitization to methamphetamine. Addict. Biol. 2010, 15, 358–361. [Google Scholar] [CrossRef] [PubMed]
- Cadet, J.L.; Jayanthi, S. Epigenetic Landscape of Methamphetamine Use Disorder. Curr. Neuropharmacol. 2021, 19, 2060–2066. [Google Scholar] [CrossRef] [PubMed]
- Jayanthi, S.; McCoy, M.T.; Cadet, J.L. Epigenetic Regulatory Dynamics in Models of Methamphetamine-Use Disorder. Genes 2021, 12, 1614. [Google Scholar] [CrossRef] [PubMed]
- Martin, T.A.; Jayanthi, S.; McCoy, M.T.; Brannock, C.; Ladenheim, B.; Garrett, T.; Lehrmann, E.; Becker, K.G.; Cadet, J.L. Methamphetamine Causes Differential Alterations in Gene Expression and Patterns of Histone Acetylation/Hypoacetylation in the Rat Nucleus Accumbens. PLoS ONE 2012, 7, e34236. [Google Scholar] [CrossRef] [PubMed]
- Limanaqi, F.; Busceti, C.L.; Celli, R.; Biagioni, F.; Fornai, F. Autophagy as a gateway for the effects of methamphetamine: From neurotransmitter release and synaptic plasticity to psychiatric and neurodegenerative disorders. Prog. Neurobiol. 2021, 204, 102112. [Google Scholar] [CrossRef]
- González, B.; Torres, O.V.; Jayanthi, S.; Gomez, N.; Sosa, M.H.; Bernardi, A.; Urbano, F.J.; García-Rill, E.; Cadet, J.-L.; Bisagno, V. The effects of single-dose injections of modafinil and methamphetamine on epigenetic and functional markers in the mouse medial prefrontal cortex: Potential role of dopamine receptors. Prog. Neuropsychopharmacol. Biol. Psychiatry 2019, 88, 222–234. [Google Scholar] [CrossRef]
- González-Rojo, S.; Lombó, M.; Fernández-Díez, C.; Herráez, M.P. Male exposure to bisphenol a impairs spermatogenesis and triggers histone hyperacetylation in zebrafish testes. Environ. Pollut. 2019, 248, 368–379. [Google Scholar] [CrossRef]
- Torres, C.M.; Biran, A.; Burney, M.J.; Patel, H.; Henser-Brownhill, T.; Cohen, A.-H.S.; Li, Y.; Ben-Hamo, R.; Nye, E.; Spencer-Dene, B.; et al. The linker histone H1.0 generates epigenetic and functional intratumor heterogeneity. Science 2016, 353, aaf1644. [Google Scholar] [CrossRef]
- Jayanthi, S.; Ladenheim, B.; Sullivan, P.; McCoy, M.T.; Krasnova, I.N.; Goldstein, D.S.; Cadet, J.L. Biochemical Neuroadaptations in the Rat Striatal Dopaminergic System after Prolonged Exposure to Methamphetamine Self-Administration. Int. J. Mol. Sci. 2022, 23, 10092. [Google Scholar] [CrossRef]
- Doke, M.; Pendyala, G.; Samikkannu, T. Psychostimulants and opioids differentially influence the epigenetic modification of histone acetyltransferase and histone deacetylase in astrocytes. PLoS ONE 2021, 16, e0252895. [Google Scholar] [CrossRef]
- Limanaqi, F.; Biagioni, F.; Busceti, C.L.; Ryskalin, L.; Fornai, F. The effects of proteasome on baseline and methamphetamine-dependent dopamine transmission. Neurosci. Biobehav. Rev. 2019, 102, 308–317. [Google Scholar] [CrossRef] [PubMed]
- Briones-Lizardi, L.J.; Cortés, H.; Avalos-Fuentes, J.A.; Paz-Bermúdez, F.J.; Aceves, J.; Erlij, D.; Florán, B. Presynaptic control of [3H]-glutamate release by dopamine receptor subtypes in the rat substantia nigra. Central role of D1 and D3 receptors. Neuroscience 2019, 406, 563–579. [Google Scholar] [CrossRef]
- Dong, C.; Bach, S.V.; Haynes, K.A.; Hegde, A.N. Proteasome modulates positive and negative translational regulators in long-term synaptic plasticity. J. Neurosci. Off. J. Soc. Neurosci. 2014, 34, 3171–3182. [Google Scholar] [CrossRef] [PubMed]
- Hegde, A.N. Proteolysis, synaptic plasticity and memory. Neurobiol. Learn. Mem. 2017, 138, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Harutyunyan, A.; Schneider, B.L.; Moszczynska, A. Parkin regulates drug-taking behavior in rat model of methamphetamine use disorder. Transl. Psychiatry 2021, 11, 293. [Google Scholar] [CrossRef]
- Cates, H.M.; Li, X.; Purushothaman, I.; Kennedy, P.J.; Shen, L.; Shaham, Y.; Nestler, E.J. Genome-wide transcriptional profiling of central amygdala and orbitofrontal cortex during incubation of methamphetamine craving. Neuropsychopharmacol. Off. Publ. Am. Coll. Neuropsychopharmacol. 2018, 43, 2426–2434. [Google Scholar] [CrossRef] [PubMed]
- Jiao, D.-L.; Chen, Y.; Liu, Y.; Ju, Y.-Y.; Long, J.-D.; Du, J.; Yu, C.-X.; Wang, Y.-J.; Zhao, M.; Liu, J.-G. SYVN1, an ERAD E3 Ubiquitin Ligase, Is Involved in GABAAα1 Degradation Associated with Methamphetamine-Induced Conditioned Place Preference. Front. Mol. Neurosci. 2017, 10, 313. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, C.; Zhou, Y.; Luo, C.; Ou, J.; Li, J.; Mo, Z. Expression of microRNAs in the serum exosomes of methamphetamine-dependent rats vs. ketamine-dependent rats. Exp. Ther. Med. 2018, 15, 3369. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Li, L.; Hong, S.; Zhang, D.; Zhou, Y. Methamphetamine leads to the alterations of microRNA profiles in the nucleus accumbens of rats. Pharm. Biol. 2020, 58, 797–805. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhu, L.; Su, H.; Liu, D.; Yan, Z.; Ni, T.; Wei, H.; Goh, E.L.; Chen, T. Regulation of miR-128 in the nucleus accumbens affects methamphetamine-induced behavioral sensitization by modulating proteins involved in neuroplasticity. Addict. Biol. 2021, 26, e12881. [Google Scholar] [CrossRef]
- Chavoshi, H.; Boroujeni, M.E.; Abdollahifar, M.-A.; Amini, A.; Tehrani, A.M.; Moghaddam, M.H.; Norozian, M.; Farahani, R.M.; Aliaghaei, A. From dysregulated microRNAs to structural alterations in the striatal region of METH-injected rats. J. Chem. Neuroanat. 2020, 109, 101854. [Google Scholar] [CrossRef]
- Werner, C.T.; Altshuler, R.D.; Shaham, Y.; Li, X. Epigenetic Mechanisms in Drug Relapse. Biol. Psychiatry 2021, 89, 331–338. [Google Scholar] [CrossRef]
- Kaplan, G.; Xu, H.; Abreu, K.; Feng, J. DNA Epigenetics in Addiction Susceptibility. Front. Genet. 2022, 13, 806685. [Google Scholar] [CrossRef]
- Gonzalez, W.G.; Zhang, H.; Harutyunyan, A.; Lois, C. Persistence of neuronal representations through time and damage in the hippocampus. Science 2019, 365, 821–825. [Google Scholar] [CrossRef]
- Peeler, J.C.; Schedin-Weiss, S.; Soula, M.; Kazmi, M.A.; Sakmar, T.P. Isopeptide and ester bond ubiquitination both regulate degradation of the human dopamine receptor 4. J. Biol. Chem. 2017, 292, 21623–21630. [Google Scholar] [CrossRef]
- Kan, R.L.; Chen, J.; Sallam, T. Crosstalk between epitranscriptomic and epigenetic mechanisms in gene regulation. Trends Genet. TIG 2022, 38, 182–193. [Google Scholar] [CrossRef]
- Flamand, M.N.; Meyer, K.D. The epitranscriptome and synaptic plasticity. Curr. Opin. Neurobiol. 2019, 59, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Angelova, M.T.; Dimitrova, D.G.; Dinges, N.; Lence, T.; Worpenberg, L.; Carré, C.; Roignant, J.-Y. The Emerging Field of Epitranscriptomics in Neurodevelopmental and Neuronal Disorders. Front. Bioeng. Biotechnol. 2018, 6, 46. [Google Scholar] [CrossRef] [PubMed]
- Chokkalla, A.K.; Mehta, S.L.; Vemuganti, R. Epitranscriptomic regulation by m6A RNA methylation in brain development and diseases. J. Cereb. Blood Flow Metab. Off. J. Int. Soc. Cereb. Blood Flow Metab. 2020, 40, 2331–2349. [Google Scholar] [CrossRef] [PubMed]
- Engel, M.; Chen, A. The emerging role of mRNA methylation in normal and pathological behavior. Genes Brain Behav. 2018, 17, e12428. [Google Scholar] [CrossRef]
- Weng, Y.-L.; Wang, X.; An, R.; Cassin, J.; Vissers, C.; Liu, Y.; Liu, Y.; Xu, T.; Wang, X.; Wong, S.Z.H.; et al. Epitranscriptomic m6A Regulation of Axon Regeneration in the Adult Mammalian Nervous System. Neuron 2018, 97, 313–325.e6. [Google Scholar] [CrossRef] [PubMed]
- Śledź, P.; Jinek, M. Structural insights into the molecular mechanism of the m6A writer complex. eLife 2016, 5, e18434. [Google Scholar] [CrossRef]
- Wang, P.; Doxtader, K.A.; Nam, Y. Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol. Cell 2016, 63, 306–317. [Google Scholar] [CrossRef]
- Schöller, E.; Weichmann, F.; Treiber, T.; Ringle, S.; Treiber, N.; Flatley, A.; Feederle, R.; Bruckmann, A.; Meister, G. Interactions, localization, and phosphorylation of the m6A generating METTL3–METTL14–WTAP complex. RNA 2018, 24, 499–512. [Google Scholar] [CrossRef]
- Ping, X.-L.; Sun, B.-F.; Wang, L.; Xiao, W.; Yang, X.; Wang, W.-J.; Adhikari, S.; Shi, Y.; Lv, Y.; Chen, Y.-S.; et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014, 24, 177–189. [Google Scholar] [CrossRef]
- Patil, D.P.; Chen, C.-K.; Pickering, B.F.; Chow, A.; Jackson, C.; Guttman, M.; Jaffrey, S.R. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 2016, 537, 369–373. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.-G.; et al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.-M.; Li, C.J.; Vågbø, C.B.; Shi, Y.; Wang, W.-L.; Song, S.-H.; et al. ALKBH5 Is a Mammalian RNA Demethylase that Impacts RNA Metabolism and Mouse Fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Vitali, P.; Basyuk, E.; Le Meur, E.; Bertrand, E.; Muscatelli, F.; Cavaillé, J.; Huttenhofer, A. ADAR2-mediated editing of RNA substrates in the nucleolus is inhibited by C/D small nucleolar RNAs. J. Cell Biol. 2005, 169, 745–753. [Google Scholar] [CrossRef] [PubMed]
- Purchal, M.K.; Eyler, D.E.; Tardu, M.; Franco, M.K.; Korn, M.M.; Khan, T.; McNassor, R.; Giles, R.; Lev, K.; Sharma, H.; et al. Pseudouridine synthase 7 is an opportunistic enzyme that binds and modifies substrates with diverse sequences and structures. Proc. Natl. Acad. Sci. USA 2022, 119, e2109708119. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, J.; Lugoboni, M.; Junion, G. m6A RNA modification in transcription regulation. Transcription 2021, 12, 266–276. [Google Scholar] [CrossRef] [PubMed]
- Livneh, I.; Moshitch-Moshkovitz, S.; Amariglio, N.; Rechavi, G.; Dominissini, D. The m6A epitranscriptome: Transcriptome plasticity in brain development and function. Nat. Rev. Neurosci. 2020, 21, 36–51. [Google Scholar] [CrossRef]
- Widagdo, J.; Wong, J.J.-L.; Anggono, V. The m6A-epitranscriptome in brain plasticity, learning and memory. Semin. Cell Dev. Biol. 2022, 125, 110–121. [Google Scholar] [CrossRef]
- Chen, X.; Yu, C.; Guo, M.; Zheng, X.; Ali, S.; Huang, H.; Zhang, L.; Wang, S.; Huang, Y.; Qie, S.; et al. Down-Regulation of m6A mRNA Methylation Is Involved in Dopaminergic Neuronal Death. ACS Chem. Neurosci. 2019, 10, 2355–2363. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Hales, C.M.; Garber, K.; Jin, P. Fat mass and obesity-associated (FTO) protein interacts with CaMKII and modulates the activity of CREB signaling pathway. Hum. Mol. Genet. 2014, 23, 3299–3306. [Google Scholar] [CrossRef]
- Sevgi, M.; Rigoux, L.; Kühn, A.B.; Mauer, J.; Schilbach, L.; Hess, M.E.; Gruendler, T.O.; Ullsperger, M.; Stephan, K.E.; Brüning, J.C.; et al. An Obesity-Predisposing Variant of the FTO Gene Regulates D2R-Dependent Reward Learning. J. Neurosci. 2015, 35, 12584–12592. [Google Scholar] [CrossRef] [PubMed]
- Spychala, A.; Rüther, U. FTO affects hippocampal function by regulation of BDNF processing. PLoS ONE 2019, 14, e0211937. [Google Scholar] [CrossRef] [PubMed]
- Xue, A.; Huang, Y.; Li, M.; Wei, Q.; Bu, Q. Comprehensive Analysis of Differential m6A RNA Methylomes in the Hippocampus of Cocaine-Conditioned Mice. Mol. Neurobiol. 2021, 58, 3759–3768. [Google Scholar] [CrossRef]
- Sun, L.; Ma, L.; Zhang, H.; Cao, Y.; Wang, C.; Hou, N.; Huang, N.; von Deneen, K.M.; Zhao, C.; Shi, Y.; et al. Fto Deficiency Reduces Anxiety- and Depression-Like Behaviors in Mice via Alterations in Gut Microbiota. Theranostics 2019, 9, 721–733. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Filošević Vujnović, A.; Stanković Matić, I.; Saftić Martinović, L.; Dević Pavlić, S. Breaking the Chains: Advances in Substance Addiction Research through Single-Cell Sequencing, Epigenetics, and Epitranscriptomic. Future Pharmacol. 2024, 4, 115-138. https://doi.org/10.3390/futurepharmacol4010009
Filošević Vujnović A, Stanković Matić I, Saftić Martinović L, Dević Pavlić S. Breaking the Chains: Advances in Substance Addiction Research through Single-Cell Sequencing, Epigenetics, and Epitranscriptomic. Future Pharmacology. 2024; 4(1):115-138. https://doi.org/10.3390/futurepharmacol4010009
Chicago/Turabian StyleFilošević Vujnović, Ana, Ivana Stanković Matić, Lara Saftić Martinović, and Sanja Dević Pavlić. 2024. "Breaking the Chains: Advances in Substance Addiction Research through Single-Cell Sequencing, Epigenetics, and Epitranscriptomic" Future Pharmacology 4, no. 1: 115-138. https://doi.org/10.3390/futurepharmacol4010009