- 1Department of Laboratory Medicine, Shenzhen Key Laboratory of Medical Laboratory and Molecular Diagnostics, Shenzhen Institute of Translational Medicine, The First Affiliated Hospital of Shenzhen University, Shenzhen Second People's Hospital, Shenzhen, China
- 2Guangdong Key Laboratory for Biomedical Measurements and Ultrasound Imaging, National-Regional Key Technology Engineering Laboratory for Medical Ultrasound, School of Biomedical Engineering, Shenzhen University Medical School, Shenzhen, China
- 3College of Life Science, Northwest A&F University, Yangling, Shaanxi, China
This review constitutes a summary of current knowledge on GlnR, a global regulator, that assumes a critical function in the regulation of nitrogen metabolism of Actinomycetes. In cross-regulation with other regulators, GlnR was also shown to play a role in the regulation of carbon and phosphate metabolisms as well as of secondary metabolism. A description of the structure of the GlnR protein and of its binding sites in various genes promoters regions is also provided. This review thus provides a global understanding of the critical function played by GlnR in the regulation of primary and secondary metabolism in Actinomycetes.
1 Introduction
Both organic and inorganic forms of nitrogen sources are essential for bacterial growth and important components of cellular biomolecules, such as nucleotides, amino acids, and amino sugars (Merrick and Edwards, 1995). Actinomycetes are gram-positive bacteria with a high GC content (Berdy, 2005) that are able to produce, usually late in growth, a great variety of different bio-active molecules such as anti-bacterial, anti-fungi, herbicide, insecticide, immunosuppressants, immunomodulators, and anti-cancer drugs etc. (Berdy, 2005). The biosynthesis of these molecules is regulated by nitrogen, carbon, and phosphate sources availability (Hodgson, 2000; Reuther and Wohlleben, 2007). Among Actinomycetes, Streptomyces are major active metabolite producers, able to synthetize a great variety of bioactive metabolites of industrial interest with potential medical and agricultural applications (Ossai et al., 2022). Streptomyces are soil-dwelling bacteria with a unique and complicated life cycle, including vegetative mycelium formation, aerial mycelium differentiation, and sporulation (Chater, 2001). This complex morphological differentiation process, that is usually accompanied by the synthesis of bio-active specialized metabolites, involves temporally and spatially regulated regulatory networks (Hopwood et al., 1995). In soil habitats, Streptomyces compete with other bacteria for nutrients and especially for nitrogen sources (Reuther and Wohlleben, 2007), that are used to synthesize almost all important biological molecules (Arcondeguy et al., 2001) and also play a crucial role in the regulation specialized metabolism (Reuther and Wohlleben, 2007). Actinomycetes have thus developed complex transcriptional, translational and post-translationnal regulatory systems to regulate the expression of genes and activity of proteins involved in nitrogen metabolism in response to changes in environmental nitrogen levels (Leigh and Dodsworth, 2007). Diverse nitrogen resources, either mineral (nitrate, ammonium, etc.) or organic (urea, glutamine, etc.), are present in the environment. When the bacteria is facing nitrogen organic source shortage in its environment, it can assimilate inorganic nitrogen resources into organic ones (Sun et al., 2017). The two amino acids, glutamate and glutamine, that are the main intracellular nitrogen donors, can be synthesized from various mineral nitrogen sources and especially ammonium (Reitzer and Schneider, 2001; Sun et al., 2017). The nitrogen of glutamate is used for the biosynthesis of numerous amino acids (Fisher, 1989) whereas glutamine is used for the biosynthesis of both purines and pyrimidines, as well as other nitrogenous metabolites (Fisher, 1992). Depending on nitrogen availability, there are two key pathways of ammonium assimilation: the glutamate dehydrogenase (GDH) and the glutamine synthetase/glutamate synthase (GS/GOGAT) pathways (Fisher, 1989; Fisher, 1992) (Figure 1). In Streptomyces coelicolor, when the nitrogen supply is high, glutamine is synthesized from ammonium and 2-oxoglutarate by NADPH-dependent GDH whereas under low nitrogen concentrations, the GS of the GS/GOGAT system catalyzes the formation of glutamine from ammonium and glutamate, consuming ATP whereas GOGAT catalyzes the formation of two glutamate molecules from glutamine and 2-oxoglutarate (Magasanik, 1982; Tiffert et al., 2008). Most studies on microbial nitrogen metabolism and its regulatory network come from enterobacter (Magasanik, 1982; Merrick and Edwards, 1995; Oelze and Klein, 1996; Dubbs and Tabita, 2004; Zhang et al., 2014). In gram-negative Escherichia coli, ammonium assimilation is regulated by the two-component system NtrB/NtrC (Weiss et al., 2002) that requires σ54 to activate the transcription of target genes (Zimmer et al., 2000; Reitzer and Schneider, 2001). When bacteria are starved of nitrogen sources, the kinase NtrB phosphorylates NtrC that stimulates the transcriptional expression of downstream genes involved into nitrogen metabolism (Weiss et al., 2002). In the gram-positive bacteria, Bacillus subtilis, with a low GC content, the intracellular nitrogen availability is mainly regulated by two regulators of the MerR family, TnrA and GlnR (GlnRbsu) (Schreier et al., 1989; Wray et al., 1996). In excess of nitrogen, GlnRbsu represses the transcription of genes of nitrogen metabolism (Tiffert et al., 2008) whereas in condition of nitrogen limitation, TnrA activates the expression of the nrgAB, nasB, gabP, and ureA genes and represses that of glnRA (Wray et al., 1996). GlnR senses nitrogen excess indirectly by binding glutamine-feedback-inhibited-GS (FBI-GS). FBI-GS, as a GlnR chaperone, binds the GlnR C-terminal domain within its active-site cavity, promoting the formation of a DNA-binding-competent GlnR dimer and leading to GlnR DNA-binding activation (Travis et al., 2022).
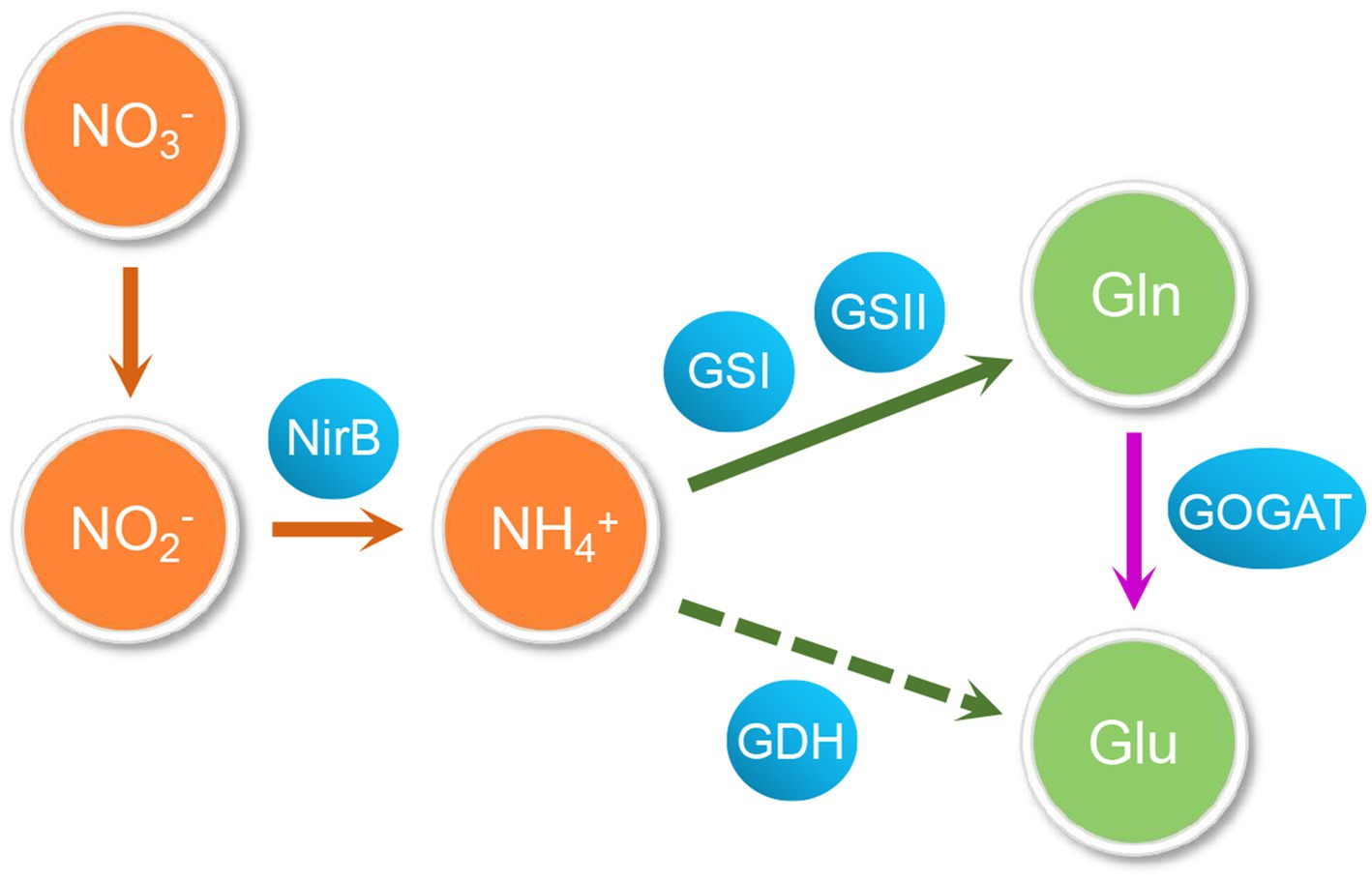
Figure 1. The nitrogen metabolism pathway in actinomycetes. Glutamate and glutamine are synthesized from various nitrogen sources through assimilation and they are the main intracellular nitrogen donors. The glutamate dehydrogenase (GDH) and the glutamine synthetase/glutamate synthetase (GS/GOGAT) systems are two key pathways of ammonium assimilation: When nitrogen supply is sufficient, glutamine is synthesized from ammonia and 2-oxoglutarate through GDH mediates with NADPH consumption; Under low nitrogen concentrations, the GS/GOGAT system consumes ATP to catalyze the formation of glutamine and glutamate. GS converts ammonium and glutamate to form glutamine, and GOGAT catalyzes glutamine and 2-oxoglutarate into two molecules of glutamate.
In gram-positive Actinomycetes, with high GC content, the regulation mechanism of nitrogen metabolism evolved completely differently from that of E. coli and B. subtilis (Burkovski, 2003). The nitrogen metabolism of most Actinomycetes (such as S. coelicolor, Amycolatopsis mediterranei) is mediated by GlnR (Amon et al., 2009), a regulator of the OmpR family that is an orphan response regulator, significantly different from GlnRbsu of B. subtilis (Hutchings et al., 2004). In contrast, a small number of Actinomycetes (such as Mycobacterium smegmatis, Streptomyces avermitilis) bear two regulatory proteins, GlnR and AmtR, involved in the regulation of nitrogen metabolism whereas only AmtR regulates nitrogen metabolism in Corynebacterium (Burkovski, 2003; Beckers et al., 2005). In 1988, the glnA gene encoding glutamine synthase GSI was located in S. coelicolor, and GSI enzyme activity was determined (Wray and Fisher, 1988). In the 1990s, the research mainly focused on GlnR and GSI/GSII. In 1990, a second gene, glnII, encoding glutamine synthase GSII was identified in Frankia and Streptomyces viridochromogenes (Behrmann et al., 1990; Rochefort and Benson, 1990). In 1991, the isolation of six glutamine-deficient mutants of S. coelicolor led to the identification of the glnR encoding gene whose transcription start sites were determined (Wray et al., 1991). In 1993, the positive role that GlnR plays in the regulation of glnA expression was demonstrated (Wray and Fisher, 1993) as well as the post-translational modification of GSI (Fisher, 1992). With the completion of the genome sequence of S. coelicolor in 2002 and that of other Streptomyces subsequently, the conserved GlnR binding sites were identified (Tiffert et al., 2008) and used to identify GlnR target genes. Moreover, cross-regulation between GlnR and other transcription regulators was demonstrated (Rodriguez-Garcia et al., 2007; Yang et al., 2009; Wang R. et al., 2013). Since 2000, the functional identification of GlnR was achieved in other Actinomycetes (such as A. mediterranei U32, M. smegmatis, Saccharopolyspora erythraea, Streptomyces variabilis). In 2011, Tiffert et al. performed a proteomic analysis of the glnR deletion mutant of S. coelicolor and found that GlnR affects the expression of numerous genes belonging to different metabolic pathways (Tiffert et al., 2011).
This review focuses on the functional studies of GlnR mainly in S. coelicolor but also in other antibiotic producing Streptomyces species as well as in other Actinomycetes. This review reports what is known on the role played by GlnR in the regulation of nitrogen as well as of carbon and phosphate metabolisms and on the cross-regulation between GlnR and other regulators. The structure of the protein GlnR and of that of its target sites is also commented. This review thus provides a systematic understanding of the critical role played by the global regulator GlnR in the regulation of both primary and specialized metabolism in Actinomycetes.
2 Regulatory role and regulatory targets of GlnR in Streptomyces coelicolor and other Actinomycetes
Tiffert et al. performed a comparative proteome analysis of the wild type strain of S. coelicolor M145 and a glnR-deficient mutant and discovered that GlnR regulates the expression of numerous proteins (Tiffert et al., 2011) involved (1) amino acid metabolism, GlnR inhibits the biosynthesis and degradation of certain types of amino acids (methionine, tryptophan, serine), (2) carbon metabolism, GlnR enhances the synthesis of acetyl-CoA and inhibits the pentose phosphate pathway, (3) stress response, GlnR promotes the transcription of numerous stress-response genes (Tiffert et al., 2011), and (4) antibiotic biosynthesis. Whether GlnR directly or indirectly affects these processes requires further experimental demonstration, but it is clear that GlnR plays a pleiotropic role in the regulation of S. coelicolor M145 metabolism.
2.1 In nitrogen metabolism
2.1.1 In Streptomyces coelicolor
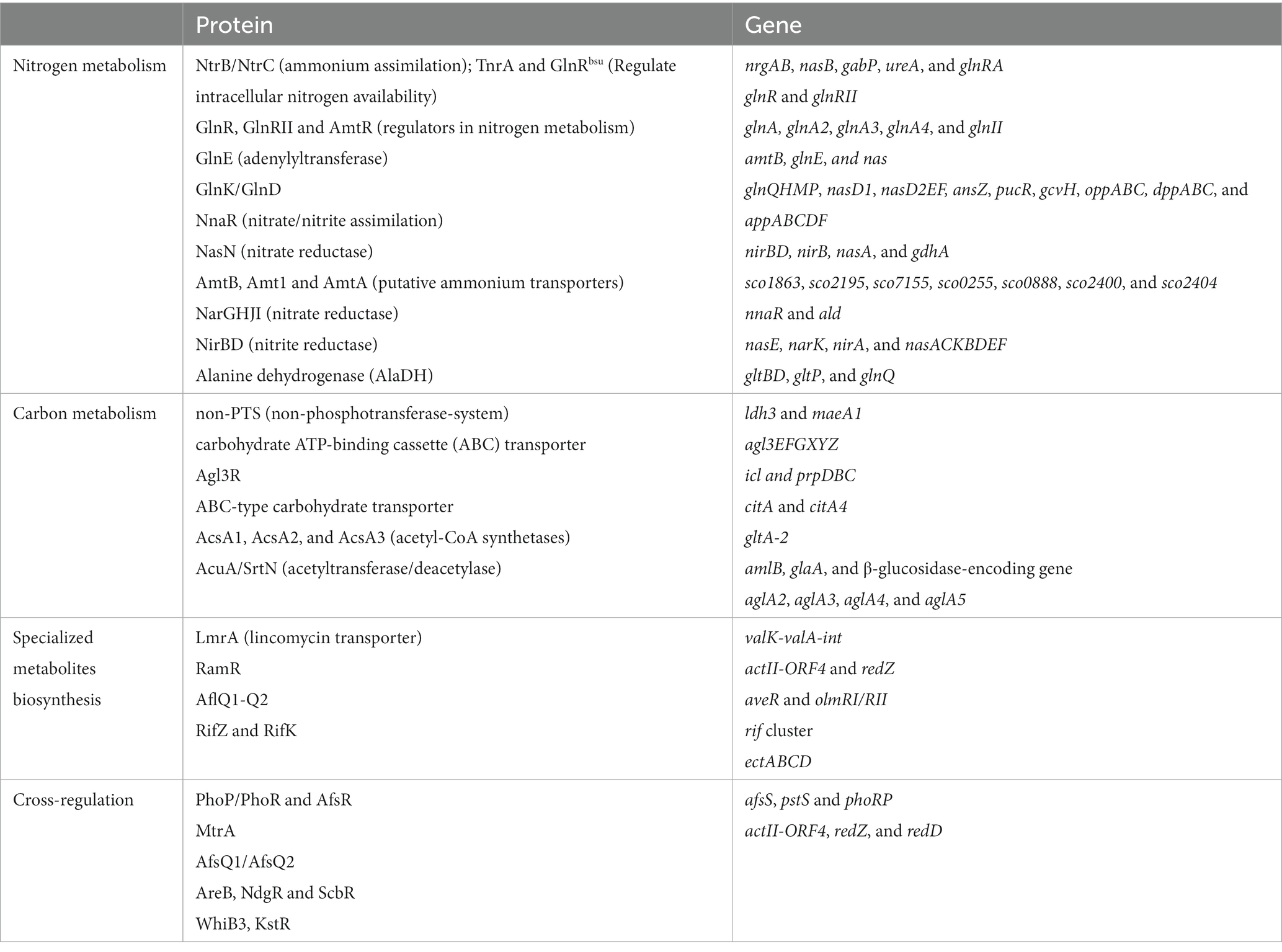
In S. coelicolor as well as in other Streptomyces nitrogen metabolism is mainly regulated by the regulator GlnR. However a second GlnR-like regulator was identified and called GlnRII. GlnR and GlnRII share 31% identity (Fink et al., 2002). GlnRII can interact with the promoter region of some genes of nitrogen metabolism, such as glnA, glnII, and amtB, but differ from GlnR in its physiological function since a mutant deleted for the glnRII gene does not behave as a glnR mutant (Reuther and Wohlleben, 2007). When nitrate is used as sole nitrogen source, the glnR mutant of S. coelicolor exhibits glutamine-auxotrophy but can survive with extremely low amounts of ammonium uptake and assimilation but that is not the case of the glnRII mutant (Tiffert et al., 2008). GlnR is thus the primary regulator of nitrogen metabolism and GlnRII may promote cell differentiation and antibiotic production under nitrogen limitation (Fink et al., 2002). GlnR regulates the expression of many essential genes involved into nitrogen metabolism, including nine genes directly involved into nitrogen metabolism (Table 1): amtB-glnK-glnD operon (encoding a putative ammonium transporter and nitrogen signaling proteins), nasA (encoding a putative nitrate reductase), nirB (encoding a putative nitrite reductase), ureA (encoding urease), glnA and glnII (encoding glutamine synthetase), gdhA (encoding glutamate dehydrogenase); and seven other genes indirectly involved in nitrogen assimilation: the putative transcriptional regulator SCO0255, the putative NADPH-dependent FMN reductase SCO0888, the putative membrane protein SCO2400, the putative sugar-binding protein SCO2404 of an ABC transporter, and three other hypothetical proteins of unknown function: SCO1863, SCO2195, SCO7155 (Tiffert et al., 2008). The nitrogen metabolism genes/operons (glnQHMP, nasD1, nasD2EF, ansZ, pucR, gcvH, oppABC, dppABC, and appABCDF) were also recently shown to belong to the GlnR regulon (Wang T. S. et al., 2023). In nitrogen-limited environments, GlnR positively regulates the expressions of glnA, glnII, amtB, nirB, and nasA (Tiffert et al., 2008; Wang and Zhao, 2009; Qiu et al., 2022) and negatively regulates the expressions of gdhA, ureA, sco0255, sco0888, sco2400, and sco2404 (Tiffert et al., 2008). Glutamine synthetase (GS) is crucial in ammonium assimilation, and many bacteria have GS isoenzymes (Murray et al., 2013). S. coelicolor possesses two types of GS, GSI encoded by glnA and GSII encoded by glnII (Wray and Fisher, 1988; Behrmann et al., 1990). In addition to the glnAsco (also named as glnA1sco), there are three glnA-type genes glnA2sco (encoding γ-glutamylpolyamine synthetase), glnA3sco (encoding γ-glutamylpolyamine synthetase), and glnA4sco (encoding γ-glutamylethanolamide synthetase), which are not directly involved in L-glutamine synthesis and nitrogen assimilation (Rexer et al., 2006; Krysenko et al., 2022). It has been demonstrated that the first step in polyamine catabolism is catalyzed by GlnA2sco (Krysenko et al., 2022). The GlnA3sco and GlnA4sco catalyze the first step in poly−/monoamine assimilation, respectively (Krysenko et al., 2021). GSI and GSII are homomultimeric proteins composed of 12 and 10 identical subunits, respectively (Weissschuh et al., 2000; Zeth, 2013). The expression of GSI is positively regulated by GlnR at the transcriptional level and the activity of the GSI enzyme is regulated post-translationally by adenylylation by the adenylyltransferase GlnE (Fisher and Wray, 1989). Under nitrogen limitation, GSI is activated, whereas under nitrogen excess, GSI is adenylylated by GlnE and GSI-AMP is inactive (Hesketh et al., 2002). In contrast, GSII is not modified by adenylylation (Brown et al., 1994). The GlnE-dependent modulation of GSI activity in S. coelicolor obviously does not require the GlnK/GlnD system (Hesketh et al., 2002). In S. coelicolor, GlnR also positively regulates the transcriptional expression of nnaR (nitrate/nitrite assimilation regulator) and NnaR activates directly the transcriptional expression of 4 target genes: narK (encoding a putative nitrate extrusion protein), nirB (encoding a putative nitrite reductase), nirA (encoding a putative nitrite/sulfite reductase), and nasA (encoding a putative nitrate reductase) (Amin et al., 2012). NnaR cooperatively binds the nirB promoter together with GlnR, indicating that NnaR may function as a co-activator of GlnR and participate in the transcriptional control of nitrate/nitrite assimilation genes (Amin et al., 2012). The regulatory role of NnaR in nitrate and nitrite assimilation had also been demonstrated in mycobacteria (Antczak et al., 2018).
2.1.2 In mycobacteria
M. smegmatis, Mycobacterium tuberculosis, Mycobacterium leprae, and Mycobacterium bovis are the most commonly studied Mycobacterium genus (Ergan et al., 2004; Amon et al., 2009; Schuenemann et al., 2013; Meehan et al., 2019; European Food Safety Authority and European Centre for Disease Prevention and Control, 2021). Different from S. coelicolor, glnA1myc, glnA2myc, glnA3myc, and glnA4myc are all encoding GS genes present in M. tuberculosis (Rakovitsky et al., 2018). In all mycobacteria, the GS genes glnA1myc and glnA2myc are present and have conserved genomic positions near glnE. The GS genes glnA3myc and glnA4myc are present in most mycobacteria but not in M. leprae (Amon et al., 2010). The model strain, M. smegmatis, bears at least three orthologues of GlnA4myc (Amon et al., 2010). The gene gdhA is only present in M. smegmatis whereas this gene is absent in other mycobacteria that assimilate ammonium through GS/GOGAT pathway (Amon et al., 2010). M. smegmatis bears three putative ammonium transporters: AmtB, Amt1, and AmtA (Amon et al., 2009). The amtB forms an operon with glnK and glnD, as in Streptomyces. The amt1 gene is present in a cluster of three genes encoding glutamine synthetase, glutamine amidotransferase, and glutamate synthase, whereas amtA is monocistronic (Amon et al., 2009). As glnA, the amtB-glnK-glnD and amt1 operons are positively controlled by GlnR (Amon et al., 2008). Genomics analysis of M. smegmatis, identified GlnR as the global nitrogen response regulator. It directly controls the transcription of over 100 genes involved in crucial nitrogen stress survival processes, such as nitrate and urea utilization and the use of cellular components as a source of ammonium (Jenkins et al., 2013). Moreover, GlnR controls redox sensing and lipid anabolism through WhiB3 (You et al., 2019) and controls cholesterol catabolism through KstR (Ma et al., 2022). M. smegmatis also has another global nitrogen regulator, AmtR, whose expression is controlled by a cis-encoded small RNA (Petridis et al., 2016) and competes with GlnR to regulate urea metabolism. Furthermore, the nitrate reductase NasN, is required for growth when nitrate is the only nitrogen source available, and GlnR regulates nasN expression (Tan et al., 2020).
In M. tuberculosis, NarGHJI (nitrate reductase) and NirBD (nitrite reductase) mediate the assimilatory reduction of nitrate and nitrite and GlnR activates nirBD transcription (Malm et al., 2009). Nitrate is present in infected tissues (Bogdan, 2001) and nitrate assimilation by M. tuberculosis is essential for its survival in vitro and in vivo (Hedgecock and Costello, 1962). Under nitrogen limitation, GlnR controls the expression of at least 33 genes (Malm et al., 2009). In contrast to non-pathogenic mycobacteria, in M. tuberculosis GlnR regulates the expression of genes associated with nitric oxide detoxification and intracellular survival (Williams et al., 2015). Although M. tuberculosis has four genes encoding glutamine synthetases (GS), mainly GlnA1 is highly expressed and is a critical enzyme for growth both in vitro and in vivo (Harth et al., 2005). In addition, GlnA1 is critical for M. tuberculosis virulence and may be a potential drug target for tuberculosis treatment (Tullius et al., 2003). In most mycobacteria, GlnR activates its own transcription in response to nitrogen-limitation (He et al., 2023). The auto-regulation of glnR transcription is ubiquitous in other mycobacterial species and most mycobacteria glnR promoter regions have potential GlnR binding sites (Shi et al., 2023). The study also demonstrated that the purified GlnR protein of M. smegmatis can precisely attach 16 promoter regions present in different mycobacteria species (He et al., 2023).
2.1.3 In Pseudonocardiaceae
Pseudonocardiaceae, belonging to Actinobacteria genera, are able to degrade cellulose and to produce antibiotics (Carr et al., 2012; Cuesta et al., 2013; Mast et al., 2020).
In S. erythraea, 25 genes related to nitrogen utilization regulated by GlnR were identified as associated with ammonium uptake and assimilation, urea utilization, nitrite/nitrate assimilation, glutamate transport, arginine biosynthesis, nitric oxide biosynthesis, etc. (Yao et al., 2014). The GlnR-mediated regulation of the GDH and GS/GOGAT pathways of nitrogen metabolism is similar to that of Streptomyces, ammonium is the preferred nitrogen source of S. erythraea (Yao et al., 2014). In ammonium-rich conditions, the inhibition of gdhA expression by GlnR is relieved, and the GDH pathway is primarily used for ammonium assimilation. In condition of low ammonium concentration, the expression of GS gene and GOGAT (glutamine oxoglutarate aminotransferase) gene controlled by GlnR are significantly upregulated. Additionally, two of the three putative urease-encoding operons are positively controlled by GlnR (Yao et al., 2014). As TnrA, the global regulator of nitrogen metabolism of B. subtilis (Yoshida et al., 2003), GlnR directly regulates the expression of gltBD operons (encoding GOGAT), gltP genes (encoding Na(+)/glutamate: H(+) symporters), and glnQ (encoding the ATP-binding protein of a glutamate ABC-type transporter). Unlike Streptomyces, S. erythraea has no homolog of glnD, so one ignores how GlnK activity is regulated in this strain (Yao et al., 2014). The expression of the acetyltransferase AcuA is positively regulated by GlnR and AcuA acetylates GlnA1 and GlnA4 in S. erythraea. This lysine acetylation inactivates GlnA4 but does not affect GlnA1 activity. The acetylated GlnA1 enhances GlnR-DNA binding, and this regulatory effect of acetylated GlnA1 is highly conserved in Actinomycetes (You et al., 2016).
In A. mediterranei U32, GlnR positively regulates glnA, amtB and the nas operon in conditions of nitrogen limitation (Yu et al., 2006; Wang Y. et al., 2013). However, under different nitrogen source conditions, there is no linear relationship between the amount of GlnR protein and glnA expression in U32 (Yu et al., 2007). In addition, the activity of GDH does not change with nitrogen sources, so its encoding gene gdhA might not be regulated by GlnR (Wang et al., 2014). Alanine dehydrogenase (AlaDH) has also been confirmed to be associated with ammonium assimilation in U32 under nitrogen-rich conditions (Wang et al., 2014). Notably, the AlaDH pathway is the major route alternative to the GDH pathway under high ammonium conditions (Ni et al., 1984). At the same time, GlnR represses the transcription of the AlaDH encoding gene, ald, under nitrogen limitation (Wang et al., 2014). Furthermore, NasE, a homolog of NnaR in U32, also affects nitrate/nitrite assimilation (Shao et al., 2011).
2.2 In carbon metabolism
Consistently with the proteomic data from Tiffert’s study, GlnR was shown to regulate carbon metabolism (Liao et al., 2014, 2015; Cen et al., 2016; You et al., 2017; Wang T. S. et al., 2023). In Actinomycetes, GlnR was shown to directly regulate carbon source up-take of non-phosphotransferase-system (non-PTS) via its binding to promoter region of 13 of the 20 carbohydrate ATP-binding cassette (ABC) transporters encoded genes present in the genome of these bacteria (Liao et al., 2015). GlnR-mediated regulation of non-PTS carbon source utilization is highly conserved in S. coelicolor, M. smegmatis, and S. avermitilis, indicating the importance of GlnR for the regulation carbohydrate metabolism in Actinomycetes (Liao et al., 2015). In the latest research, carbon metabolism genes ldh3 (encoding lactate dehydrogenase) and maeA1 (encoding malic enzyme) are newly identified targets of GlnR (Wang T. S. et al., 2023).
2.2.1 In Streptomyces
In S. coelicolor, the expression of the agl3EFGXYZ operon encoding a putative ABC-type carbohydrate transporter, noted as a putative multiple-sugar transporter (Sprusansky et al., 2003), is repressed by both Agl3R, a regulator of the GntR-family but also by GlnR (Cen et al., 2016). It is only in condition of low carbon and high nitrogen availability that the transcription of the agl3EFGXYZ operon is derepressed, when both GlnR and Agl3R are inactivated (Cen et al., 2016). Since GlnR regulated both carbon and nitrogen metabolisms, overexpressing GlnR can improve the utilization of less preferred carbon sources (Liao et al., 2015). Furthermore GlnR inhibits the expression of the ect biosynthesis cluster (ectABCD) (Shao et al., 2015), that is widespread in microorganisms and directs the synthesis of ectoine and hydroxyectoine that plays a role is the resistance to osmotic stress and high temperature (Bursy et al., 2008; Kol et al., 2010). Since ectoine and hydroxyectoine production requires the consumption of glutamate, bacteria maintain intracellular glutamate concentration by inhibiting the transcription of ect biosynthesis cluster by GlnR when nitrogen availability is low (Shao et al., 2015). This function of GlnR has also been confirmed in A. mediterranei U32 and S. avermitilis (Shao et al., 2015), suggesting the possible of general function of GlnR to regulates osmotic pressure in Actinomycetes.
2.2.2 In mycobacteria
In M. smegmatis GlnR controls negatively the glyoxylate and methylcitrate cycles by directly inhibiting the transcription of the icl gene (encoding isocitrate lyase) (Qi et al., 2021) and the prpDBC operon (prpD encoding methylcitrate dehydratase, prpB encoding methylisocitrate lyase, and prpC encoding methylcitrate synthase) (Liu et al., 2019). In Mycobacterium neoaurum, GlnR also represses the expression of the prp operon in condition of low nitrogen availability so in a ΔglnR mutant strain the prp operon is overexpresses and this strain efficiently produces androstenedione (Zhang et al., 2020).
2.2.3 In Saccharopolyspora
GlnR also shown to regulate the expression of genes involved in carbon metabolism in S. erythraea. GlnR enhances the synthesis of acetyl-CoA via the direct activation of the transcription of genes encoding acetyl-CoA synthetases (acsA1, acsA2, and acsA3). Interestingly, the activities of the three acetyl-CoA synthetases are regulated post-translationally via acetylation of a lysine residue by the couple “protein acetyltransferase AcuA/deacetylase SrtN” (You et al., 2017). In response to unknown signals of nitrogen starvation, GlnR regulates acetate metabolism, thereby coordinating nitrogen and carbon metabolism (You et al., 2017). In S. erythraea GlnR also negatively controls the expression of the citrate synthase encoding genes citA and citA4 whereas its binding to the promoter region of another citrate synthase encoding gene gltA-2 has no impact on the level of expression of this gene (Liao et al., 2014).
2.3 In specialized metabolites biosynthesis
The regulatory effect of GlnR on antibiotic synthesis was analyzed in different Streptomyces species. In S. coelicolor, GlnR was directly associated with the biosynthesis of two antibiotics, actinorhodin and undecylprodigiosin, via the pathway-specific activator genes actII-ORF4 and redZ, respectively (He et al., 2016). In Streptomyces hygroscopicus, GlnR regulates the expression of the validamycin A (a C7N-aminocyclitol antibiotic) biosynthesis gene clusters via its direct binding to the valK-valA-int promoter region (Qu et al., 2015). In S. avermitilis, GlnR could stimulate avermectin production directly through the binding to the respective pathway-specific activator encoding genes, aveR and olmRI/RII, and the binding motif of GlnR was determined (Table 2) (He et al., 2016).
In Streptomyces lincolnensis, GlnR directly regulates the biosynthesis of lincomycin by enhancing the expression of nitrate-specific ABC transporter genes, nitrate assimilation genes, and lincomycin transporter LmrA (Meng et al., 2017). GlnR is upregulated when nitrate is provided as nitrogen source (Wang et al., 2022) and positively regulates the transcription of the lincomycin transporter LmrA (Meng et al., 2017) whereas RamR as well as AflQ1-Q2 negatively regulate lincomycin biosynthesis and promote morphological development (Wang et al., 2022). Therefore, the disruption of aflQ1-Q2 is likely to lead to an increase of lincomycin production (Wang R. D. et al., 2023) but not that of RamR that was shown to directly positively regulates GlnR expression (Wang et al., 2022).
In A. mediterranei U32, GlnR also regulates positively rifamycin production GlnR via the direct activation of the expression of the regulators RifZ and RifK that activate the transcription of genes of the rif cluster (Liu et al., 2020). Interestingly, the over-expression of GlnR of U32 in S. coelicolor led to a decrease of actinorhodin production and an increase of undecylprodigiosin production (Yu et al., 2007).
In conclusion, GlnR is a global regulator in actinomycetes that can affect not only nitrogen but also phosphate and carbon metabolisms, the adaptation to high osmotic pressure as well as antibiotic biosynthesis (Figure 2).
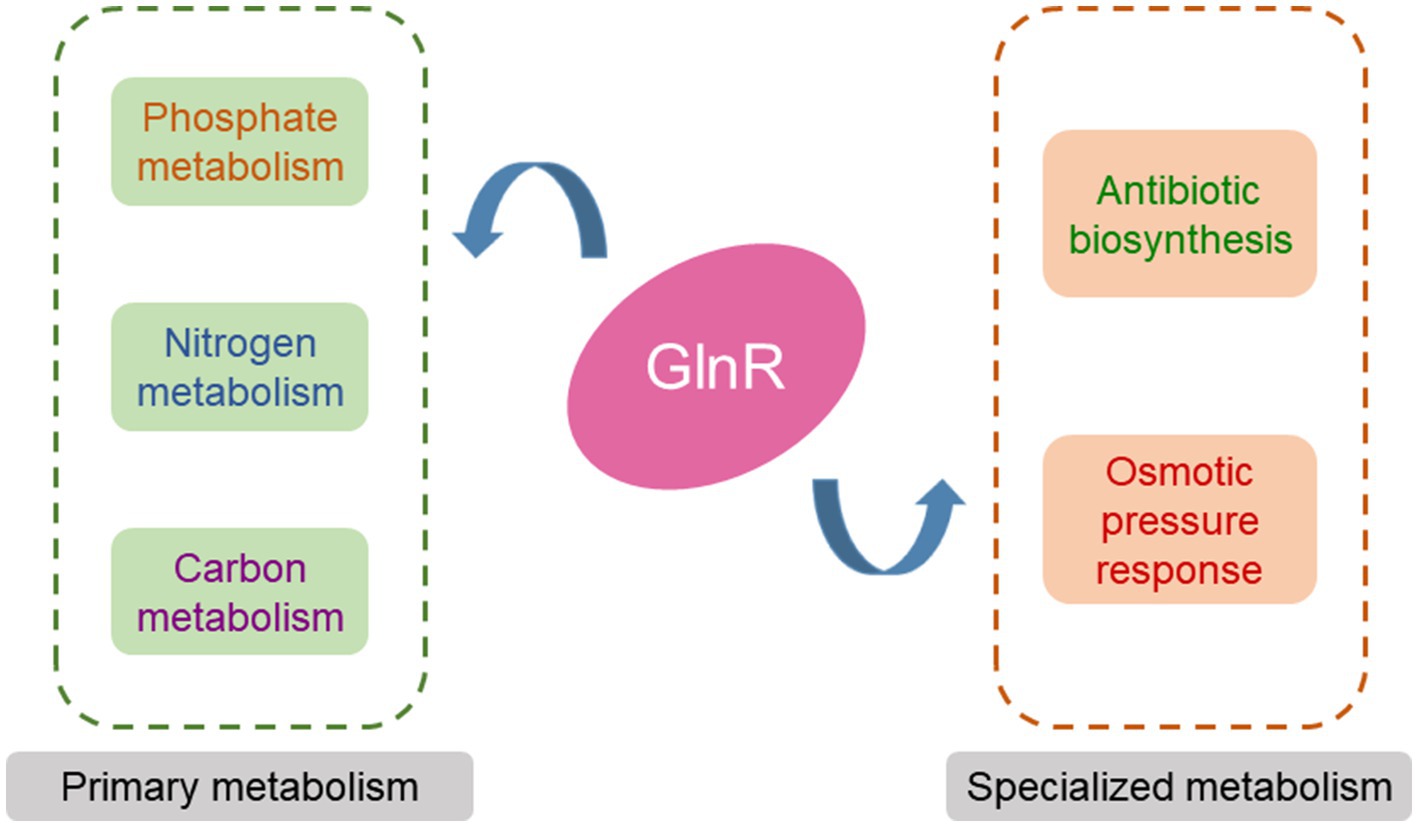
Figure 2. The GlnR regulatory network in actinomycetes. GlnR regulates nitrogen metabolism, phosphate metabolism, carbon metabolism, antibiotic biosynthesis, and the response of bacteria to osmotic pressure in actinomycetes.
3 Cross-regulation between GlnR and other regulators
The essential nutrients, such as carbon, nitrogen, and phosphorus, usually limit the growth of soil-dwelling bacteria. The deficiency of a particular nutrient will trigger a series of interrelated reactions governed by specific regulators to maintain metabolic balance (Martin et al., 2011).
3.1 With PhoP and AfsR
In S. coelicolor, the relationship between the phosphate and nitrogen metabolic pathways is intricate since PhoP was shown to directly negatively regulates the expression of glnR, glnA, glnII, and amtB-glnK-glnD operon (Rodriguez-Garcia et al., 2007, 2009). PhoP and GlnR compete for the binding to the glnA, glnII and amtB promoter regions since their binding sites overlap in these regions (Wang et al., 2012; Sola-Landa et al., 2013). Additionally, PhoP binds to other nitrogen-regulated genes: ureA (urease gamma subunit), sco0255 (putative transcriptional regulator), and sco1863 (hypothetical protein) (Sola-Landa et al., 2013) but does not bind to the glnRII promoter region, indicating another PhoP-independent regulatory pathway exists to control nitrogen metabolism (Rodriguez-Garcia et al., 2009). In contrast, GlnR cannot bind to several important genes of the Pho regulon such as pstS and phoRP (Martin et al., 2011). This cross-regulation between phosphate and nitrogen metabolism in S. coelicolor is thus not reciprocal (Martin et al., 2011). However, interestingly PhoP also binds specifically to the promoter region of afsS, a target gene of the SARP-like regulator AfsR that controls antibiotic synthesis (Santos-Beneit et al., 2011). AfsR, can bind to the promoter regions of PhoP-regulated genes such as afsS, pstS, and phoRP (Lee et al., 2002; Santos-Beneit et al., 2009). Furthermore, both PhoP and AfsR can bind to the glnR promoter region with overlapping binding sites but have different regulatory effects. PhoP negatively regulates glnR expression in condition of phosphate limitation whereas the regulatory effect of AfsR depends on the stage of the culture. AfsR activates glnR expression at early growth stages and inhibits it at later growth stages (Santos-Beneit et al., 2012).
In S. erythraea, the regulation between GlnR and PhoP is reciprocal, and GlnR can bind to the phoRP promoter region to regulate negatively phoRP transcription (Yao and Ye, 2016). Moreover, in contrast to S. coelicolor, PhoP has a positive rather than a negative role in the regulation of GlnR expression (Yao and Ye, 2016). It was also confirmed that in S. erythraea, GlnR and PhoP could synergistically or competitively activate genes associated with starch-degrading enzymes and cellulose-degrading enzymes: amlB (encoding an α-amylase), glaA (encoding a glucoamylase) and a β-glucosidase-encoding gene (Xu and Ye, 2018), and four genes encoding possible α-glucosidases aglA2, aglA3, aglA4, and aglA5 (Xu et al., 2016). Therefore, in response to changes in nutrient supplies, bacteria can coordinate the balance between carbon, nitrogen, and phosphate metabolisms through the regulation of polysaccharide degradation mediated by GlnR and PhoP. In addition, Ye et al. demonstrated that PhoP modulates erythromycin biosynthesis in S. erythraea by integrating phosphate/nitrogen signals, governing the interaction between phosphate/nitrogen metabolism and specialized metabolism (Xu et al., 2019). Furthermore, GlnR and PhoP enhance the production of erythromycin by regulating primary metabolism and thus precursors availability (Pei et al., 2022).
3.2 With MtrA
The regulator MtrA regulates nitrogen and phosphate metabolisms in a GlnR-dependent manner in several Streptomyces species (Zhang et al., 2017; Zhu et al., 2019, 2020, 2021, 2022). In some instances, MtrA competes with GlnR to control the expression of genes of the nitrogen metabolism but GlnR plays a decisive role in the control of nitrogen metabolism genes (Zhu et al., 2022). MtrA also regulates morphological differentiation and antibiotic production and its impact on antibiotic production is generally stronger than that of GlnR (Zhu et al., 2020).
3.3 With AfsQ1
In S. coelicolor, the regulator AfsQ1 of the two-component system AfsQ1/AfsQ2 directly interacts with the promoter regions of seven nitrogen assimilatory genes: gdhA, nirB, nasA, amtB, glnA, ureA and glnII but regulates negatively the expression of amtB, glnA, ureA, and glnII but not that of gdhA, nirB and nasA in a glutamate-based minimum medium (Wang R. et al., 2013). This finding is consistent with previous report of Nieselt et al. (2010) that indicated that in a medium containing excess of glutamate, the genes related to nitrogen metabolism are negatively regulated by other regulators besides PhoP/PhoR (Nieselt et al., 2010). AsfQ1, together with GlnR, also directly regulate the production of the specialized metabolites actinorhodin and undecylprodigiosin through their pathway-specific activators, actII-ORF4 and redZ (He et al., 2016). Interestingly, in Streptomyces venezuelae, the conserved binding site of AfsQ1 does not overlap the previously reported conserved binding site of GlnR (Pullan et al., 2011).
3.4 With NdgR and ScbR
In S. coelicolor, NdgR regulates nitrogen metabolism, morphological differentiation and antibiotic production. In minimal media containing Leu or Gln, ndgR deletion mutant of S. coelicolor showed a slow growth rate, a defective morphological differentiation and an altered antibiotic production (Yang et al., 2009). NdgR is unable to directly interact with the promoter regions of the nitrogen metabolism genes glnR, glnA, and glnII, as well as with the promoter region of the antibiotic pathway-specific activator genes actII-ORF4 and redD but it can directly activate the transcription of scbR (Yang et al., 2009) that regulates the expression of numerous genes, including antibiotic biosynthetic genes. Furthermore, ScbR can specifically interact with the glnR promoter region. Therefore, NdgR may indirectly regulate nitrogen metabolism and antibiotic synthesis through ScbR (Yang et al., 2009). NdgR is an IclR-like regulator, and these regulators have typical ligand binding domains at their C-terminal end (Molina-Henares et al., 2006). The interaction of NdgR with these ligands, that remain to be characterized, may affect its binding ability to its target genes.
In Streptomcyes peucetius NdgR is a homolog of AreB of Streptomyces clavuligerus that modulates leucine biosynthesis and antibiotic production (Santamarta et al., 2007). In S. peucetius, NdgR can directly specifically interact with the promoter regions of doxorubicin biosynthetic genes and regulate antibiotic production (Yang et al., 2009).
In conclusion, the ndgR gene that is highly conserved in Streptomcyes species and other genetically related bacteria, such as mycobacteria and Corynebacteria (Yang et al., 2009), connects primary metabolism to morphological differentiation and antibiotic production. The cross-regulations mentioned above between GlnR, PhoP, MtrA, AfsR, AfsQ1, NdgR and ScbR are regulating primary and secondary metabolisms to maintain the intracellular balance between nitrogen, phosphate and carbon to adjust the bacterial metabolism to the changes in the surrounding environment.
4 Protein structure of GlnR
In Actinomycetes, GlnR belongs to the OmpR family of proteins (Amon et al., 2010). OmpR-like proteins are response regulators of the bacterial two-component system. They are primarily transcriptional activators with typical DNA domain at the C-terminal and a signal-receiving domain at the N-terminal end of the protein (MartinezHackert and Stock, 1997). The comparison of GlnR amino acid sequences of 10 Actinomycetes, with the exception of that of Corynebacterium glutamicum, revealed that the GlnR proteins share 62 to 82% similarity. The DNA binding domain (α2-loop-α3) was relatively conserved and the α3 sequence, was identical (Tiffert et al., 2008). OmpR-like proteins typically contain a conserved Asp residue as their specific phosphorylation site (Delgado et al., 1993; Lee et al., 2002) whereas two residues, serine/threonine and tyrosine, are involved in phosphate transfer (Hengge, 2001). The OmpR crystal structure of E. coli shows that the α2-loop-α3 of the DNA binding domain forms the helix-turn-helix (HTH), where the α3 helix recognizes the specific DNA sequence through direct interaction with the major groove of DNA (Gross et al., 1989). The secondary structure of OmpR of E. coli showed that the conserved site of phosphorylation is an Asp residue (D-55) (Delgado et al., 1993), the serine/threonine residue is a Thr residue (T-83) and a tyrosine residue is present as Y-102 (Birck et al., 1999).
In S. coelicolor, GlnR is encoded by the glnR gene, which has three promoters: P1, P2, and P3. P2 has an identified −10 region, but P1 has no clearly identified −10 region (Wray and Fisher, 1993). The P2 promoter of glnR is preferentially recognized by a σ31-containing RNA polymerase during the stationary growth phase (Wray and Fisher, 1993). In bacteria, it is common that genes have several promoters and different σ factors of RNA polymerase selectively recognize these promoters, to adjust the transcription pattern to environmental changes (Storz and Toledano, 1994). GlnR is considered to be a typical orphan response regulator since glnR has no co-localized gene encoding a putative sensory component (Fink et al., 2002). GlnR of S. coelicolor contains a conserved Asp residue (D-50) that could constitute an alternative potential phosphorylation site. In GlnR, the other two conserved sites are replaced by a Tyr residue corresponding to the OmpR T-83 and a Val residue (V-95) corresponding to the OmpR Y-102 (Stock et al., 1989). Recently, Lin et al. characterized the structure of the GlnR-dependent transcription activation complex (GlnR-TAC). They reported the cryo-EM structure of GlnR-TAC and a co-crystal structure of the C-terminal DNA-binding domain of GlnR (GlnR_DBD) (Shi et al., 2023). The GlnR-TAC includes RNA polymerase, GlnR, and a promoter containing four conserved GlnR binding sites. These structures elucidate that four GlnR_DBDs bind around the promoter DNA in a head-to-tail manner, and four N-terminal receiver domain of GlnRs (GlnR-RECs) are formed in a tetramer state. The tetramerization of GlnR-RECs coordinately bridge domains of RNAP core enzyme with GlnR_DBDs, promoting stabilization of GlnR-TACs (Shi et al., 2023).
A crystal structural analysis of the N-terminal region of GlnR of A. mediterranei U32 was performed by Wang et al. and this study revealed that the previously referred to as “phosphorylation pocket” was not conserved but replaced by an Arg-52 present in the β3-α3 loop (Lin et al., 2014). The potential phosphorylation residue Asp-50 is not phosphorylated but is involved in the homodimerization of GlnR. Additionally, Asp-50 is involved in charge interactions between the receiver domain and the highly conserved residues Arg-52 and Thr-9. These interactions are likely to play a crucial role to maintain the correct conformation for homodimerization (Lin et al., 2014) that affects GlnR DNA-binding capacity (Wang and Zhao, 2009). The receiver domain of the N-terminal end of GlnR forms a homodimer through the α4-β5-α5 dimer interface, while the aspartate residue at the N-terminal does not need to be phosphorylated (West and Stock, 2001). However, this site is necessary to maintain the stability of homodimerization (Lin et al., 2014).
In M. smegmatis, the aspartate residue D-48 is essential for GlnR activity under nitrogen limitation. When D-48 is changed to alanine, its phenotype is similar to that of a glnR mutant and showed blocked growth (Jenkins et al., 2012).
GlnRII, another regulator of nitrogen metabolism, is also an OmpR family protein whose C-terminal DNA domain is highly similar to that of GlnR (Fink et al., 2002). The α3-DNA recognition helix of GlnRII is almost identical to that of GlnR except for two amino acids Alanine (Ala-163 in GlnRII and Arg-195 in GlnR) and Arginine (Arg-167 in GlnRII and Ala-199 in GlnR) that may be related to the DNA binding specificity (Fink et al., 2002). This may explain why GlnR and GlnRII bind some common target genes. Interestingly, CheY, another regulator of the OmpR family, has α4 and β5 helixes (Fink et al., 2002) that are involved in the contact with the kinase CheA (Welch et al., 1998) whereas the N-terminal part of GlnRII does not contain such helixes (Fink et al., 2002). Moreover, GlnRII does not have the three conserved phosphorylation sites of OmpR family proteins, and it is not co-localized with a kinase (Fink et al., 2002). Therefore, the phosphorylation mode of GlnRII may differ from that of OmpR family proteins. As regulators of nitrogen metabolism, GlnR and GlnRII may receive different signals and have different target genes due to their structural differences. Since GlnR cannot be phosphorylated in vitro and its not co-localized with an histidine kinase, the post-translational modification of GlnR has been challenging to study. Amin et al. found that the transcriptional expression level of GlnR in S. coelicolor did not change significantly with nitrogen availability (Amin et al., 2016) whereas the transcriptional level of downstream target genes regulated by GlnR changes significantly according nitrogen source nature and availability. This indicated that GlnR binding activity might be modulated by post-translational modification. Indeed, it was demonstrated that GlnR is mainly phosphorylated on serine/threonine and acetylated on lysine (Amin et al., 2016). When nitrogen sources are abundant, GlnR is primarily modified by phosphorylation and cannot interact with the promoter region of its target genes whereas under nitrogen limitation, the acetylation of GlnR takes place but does not alter its DNA-binding capacity (Amin et al., 2016).
5 The conserved binding sites of GlnR
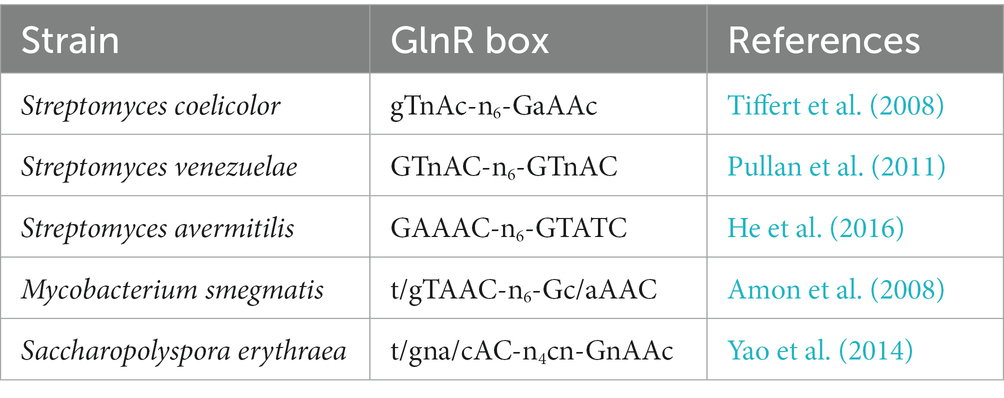
GlnR of S. coelicolor can bind to the glnA promoter regions of different Actinomycetes (Tiffert et al., 2008). Sequence comparisons of glnA promoter regions from various Actinomycetes including S. coelicolor, S. avermitilis, Bifidobacterium longum, Frankia sp. EAN1, M. bovis, M. tuberculosis, C. glutamicum, Propionibacterium acnes, Rhodococcus sp. RHA1, Nocardia farcinica, Nocardioides sp. JS614 showed that all Actinomycetes except C. glutamicum, whose nitrogen metabolism is regulated by AmtR, contain conserved GlnR binding sites (Tiffert et al., 2008). In S. coelicolor, the promoter sequences of the 13 GlnR target genes were compared, and the conserved GlnR binding sequence was determined to be gTnAc-n6-GaAAc-n6-GtnAC-n6-GAAAc-n6 (a-b-a-b), with each GlnR binding box having both a-site and b-site (Tiffert et al., 2008). Additionally, DNase I footprinting analysis accurately verifies the binding sequences on glnA and gdhA promoter regions. As predicted, two binding boxes were needed for GlnR binding in the glnA promoter region, while only one was necessary for GlnR binding in the gdhA promoter region (Tiffert et al., 2008). The GlnR binding box is frequently found on the coding strand of the target gene (Wang et al., 2012). However, there are exceptions. For example, SCO0888’s GlnR binding box is on the non-coding strand (Tiffert et al., 2008). Generally, when two GlnR binding boxes are present, they are continuous on the DNA sequence (Shi et al., 2023) but Wang et al. found that the two binding boxes of GlnR in the nasA promoter region were separated by 1 bp (Wang and Zhao, 2009). There are also three GlnR binding boxes on the promoter of the amtB gene, which is responsible for ammonium transport (Wang et al., 2012). A PhoP protected sequence also exists between the first and the second GlnR binding box (Wang et al., 2012). Wang et al. confirmed that the three binding boxes are necessary for amtB regulation by GlnR and revealed the molecular mechanism by which GlnR and PhoP cross-regulate amtB expression (Wang et al., 2012). Subsequently, Sola-landa et al. comprehensively analyzed the promoter regions of 14 GlnR target genes in S. coelicolor. They confirmed these targets by footprinting analysis, and proposed a novel 22-nt long consensus for GlnR DNA binding motifs (Sola-Landa et al., 2013). In this 22-nt long consensus, the second 11-nt long repeat is more highly conserved than the first one (Sola-Landa et al., 2013). In GlnR of S. venezuelae, a 16-bp GlnR box was proposed, which contained 5-bp a-site, 6-bp spacer and 5-bp highly conserved b-site (5 + 6 + 5-bp): GTnAC-n6-GTnAC (Pullan et al., 2011) but this consensus is fairly similar to the GlnR/22 nt long consensus. In addition, GlnR conserved binding sites were also identified in M. smegmatis (t/gTAAC-n6-Gc/aAAC) (Amon et al., 2008) and S. erythraea (t/gna/cAC-n4cn-GnAAc) (Yao et al., 2014) (Table 2). In A. mediterranei U32, the nasACKBDEF operon involved in nitrate assimilation (Shao et al., 2011), is directly regulated by GlnR (Wang Y. et al., 2013). Four GlnR binding sites were predicted by bioinformatics analysis in the promoter region of this operon (Wang Y. et al., 2013) but the studies revealed that only three sites (a1-b1-b2 sites) were necessary for GlnR binding: while a2-site was not required. EMSA (Electrophoretic Mobility Shift Assay) results demonstrated that GlnR yielded two binding bands with the nas promoter region forming two complexes (complexes I and II), where the complex with a lower mobility rate was designated complex II (Wang Y. et al., 2013). In this binding site pattern, a1-b1 sites were “5 + 6 + 5-bp,” just enough for the GlnR homodimer to combine and generate complex I. The generation of complex II required the b2-site. A 16-bp spacer between the b1-site and b2-site may cause a bend between the b1-site and b2-site upon GlnR binding as observed in other prokaryotic systems (PerezMartin and deLorenzo, 1997). Wang et al. determined the GlnR conserved DNA binding motif by changing the nucleotides of GlnR binding site on the glnA promoter in U32 one by one. This study revealed that the adenine at the fourth position was highly conserved and irreplaceable and proposed a 5-nt GlnR box as the fundamental motif for GlnR binding (Wang et al., 2015). Altogether these studies demonstrated that the GlnR binding mode is more adaptable than anticipated.
6 Summary and outlook
Under the stimulation of an unknown nitrogen signal(s), unknown sensory kinase(s) may transmit the signal(s) to GlnR, which then interacts with the promoter region of its target genes to regulate nitrogen metabolism. In addition, nitrogen metabolism is also cross-regulated by other regulators (Figure 3) such as PhoP, the regulatory protein of phosphate metabolism that can also control the expression of glnR and of its target genes belonging to nitrogen metabolism; MtrA and AfsQ1, which regulate antibiotic production but can also negatively regulate nitrogen metabolism, while AfsR, NdgR and ScbR positively regulate glnR expression. Genes and proteins in interaction with GlnR are summarized in Table 3. There are still many things to be revealed concerning GlnR regulatory role. Continuous research efforts should lead to the identification of the kinase(s) involved in the phosphorylation of GlnR as well as of the enzymes involved in its post-translational modification. The implementation of systems biology approaches including proteomics, transcriptomics, metabolomics, and bioinformatics will surely lead to the characterization of new direct and indirect GlnR target genes and novel cross-regulatory networks with other regulators.
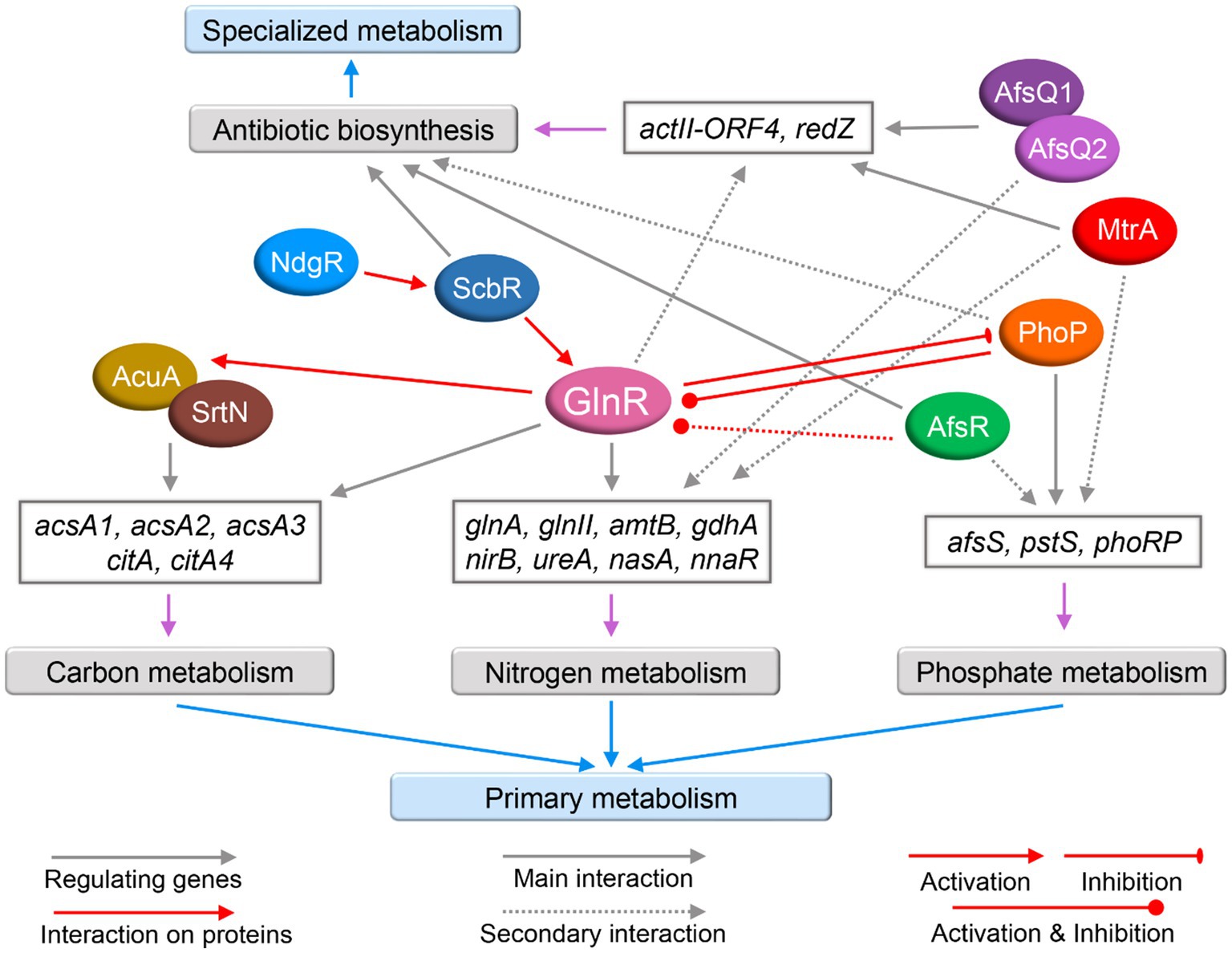
Figure 3. The cross-regulatory network of GlnR with other regulators in actinomycetes. The regulators, including GlnR, are represented by colored ellipses. Gray arrows represent certain proteins act as regulators for the expression of certain genes. Red arrows represent interactions at the protein level. Red triangle-headed arrows represent activation. Red bars represent inhibition. Red circular-headed arrows represent both activation and inhibition. Main interaction is indicated by solid lines and secondary interaction is indicated by dotted lines. The purple arrows indicate genes involved in the relevant nitrogen metabolism, phosphate metabolism, carbon metabolism, and antibiotic biosynthesis, and the corresponding genes are classified in gray boxes.
Author contributions
BG: Investigation, Validation, Writing – original draft, Writing – review & editing. GL: Investigation, Validation, Writing – review & editing. DG: Funding acquisition, Validation, Writing – review & editing. JW: Conceptualization, Funding acquisition, Validation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Key Research and Development Program of China [No. 2022YFC2302700]; Guangdong Science and Technology Foundation [No. 2021A1515220084, No. 2022B1111020001]; Shenzhen Science and Technology Foundation [ZDSYS20210623092001003, GJHZ20200731095604013, No. 201906133000069, JCYJ20190809160001751, JCYJ20200109120205924]; Medical-Engineering Interdisciplinary Research Foundation of ShenZhen University.
Acknowledgments
The authors thank AiMi Academic Services (www.aimieditor.com) for English language editing and review services.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Amin, R., Franz-Wachtel, M., Tiffert, Y., Heberer, M., Meky, M., Ahmed, Y., et al. (2016). Post-translational serine/threonine phosphorylation and lysine acetylation: a novel regulatory aspect of the global nitrogen response regulator GlnR in S. coelicolor M145. Front. Mol. Biosci. 3:38. doi: 10.3389/fmolb.2016.00038
Amin, R., Reuther, J., Bera, A., Wohlleben, W., and Mast, Y. (2012). A novel GInR target gene, nnaR, is involved in nitrate/nitrite assimilation in Streptomyces coelicolor. Microbiology 158, 1172–1182. doi: 10.1099/mic.0.054817-0
Amon, J., Brau, T., Grimrath, A., Hanbler, E., Hasselt, K., Holler, M., et al. (2008). Nitrogen control in Mycobacterium smegmatis: nitrogen-dependent expression of ammonium transport and assimilation proteins depends on the OmpR-type regulator GlnR. J. Bacteriol. 190, 7108–7116. doi: 10.1128/JB.00855-08
Amon, J., Titgemeyer, F., and Burkovski, A. (2009). A genomic view on nitrogen metabolism and nitrogen control in mycobacteria. J. Mol. Microbiol. Biotechnol. 17, 20–29. doi: 10.1159/000159195
Amon, J., Titgemeyer, F., and Burkovski, A. (2010). Common patterns - unique features: nitrogen metabolism and regulation in gram-positive bacteria. FEMS Microbiol. Rev. 34, 588–605. doi: 10.1111/j.1574-6976.2010.00216.x
Antczak, M., Płocińska, R., Płociński, P., Rumijowska-Galewicz, A., Żaczek, A., Strapagiel, D., et al. (2018). The NnaR orphan response regulator is essential for the utilization of nitrate and nitrite as sole nitrogen sources in mycobacteria. Sci. Rep. 8:17552. doi: 10.1038/s41598-018-35844-z
Arcondeguy, T., Jack, R., and Merrick, M. (2001). P-II signal transduction proteins, pivotal players in microbial nitrogen control. Microbiol. Mol. Biol. Rev. 65, 80–105. doi: 10.1128/MMBR.65.1.80-105.2001
Beckers, G., Strosser, J., Hildebrandt, U., Kalinowski, J., Farwick, M., Kramer, R., et al. (2005). Regulation of AmtR-controlled gene expression in Corynebacterium glutamicum: mechanism and characterization of the AmtR regulon. Mol. Microbiol. 58, 580–595. doi: 10.1111/j.1365-2958.2005.04855.x
Behrmann, I., Hillemann, D., Pühler, A., Strauch, E., and Wohlleben, W. (1990). Overexpression of a Streptomyces viridochromogenes gene (glnII) encoding a glutamine synthetase similar to those of eucaryotes confers resistance against the antibiotic phosphinothricyl-alanyl-alanine. J. Bacteriol. 172, 5326–5334. doi: 10.1128/jb.172.9.5326-5334.1990
Berdy, J. (2005). Bioactive microbial metabolites - a personal view. J. Antibiot. 58, 1–26. doi: 10.1038/ja.2005.1
Birck, C., Mourey, L., Gouet, P., Fabry, B., Schumacher, J., Rousseau, P., et al. (1999). Conformational changes induced by phosphorylation of the FixJ receiver domain. Structure 7, 1505–1515. doi: 10.1016/S0969-2126(00)88341-0
Bogdan, C. (2001). Nitric oxide and the immune response. Nat. Immunol. 2, 907–916. doi: 10.1038/ni1001-907
Brown, J. R., Masuchi, Y., Robb, F. T., and Doolittle, W. F. (1994). Evolutionary relationships of bacterial and archaeal glutamine-Synthetase genes. J. Mol. Evol. 38, 566–576. doi: 10.1007/BF00175876
Burkovski, A. (2003). Ammonium assimilation and nitrogen control in Corynebacterium glutamicum and its relatives: an example for new regulatory mechanisms in actinomycetes. FEMS Microbiol. Rev. 27, 617–628. doi: 10.1016/S0168-6445(03)00067-6
Bursy, J., Kuhlmann, A. U., Pittelkow, M., Hartmann, H., Jebbar, M., Pierik, A. J., et al. (2008). Synthesis and uptake of the compatible solutes Ectoine and 5-Hydroxyectoine by Streptomyces coelicolor A3(2) in response to salt and heat stresses. Appl. Environ. Microbiol. 74, 7286–7296. doi: 10.1128/AEM.00768-08
Carr, G., Derbyshire, E. R., Caldera, E., Currie, C. R., and Clardy, J. (2012). Antibiotic and antimalarial Quinones from fungus-growing ant-associated Pseudonocardia sp. J. Nat. Prod. 75, 1806–1809. doi: 10.1021/np300380t
Cen, X. F., Wang, J. Z., Zhao, G. P., Wang, Y., and Wang, J. (2016). Molecular evidence for the coordination of nitrogen and carbon metabolisms, revealed by a study on the transcriptional regulation of the agl3EFG operon that encodes a putative carbohydrate transporter in Streptomyces coelicolor. Biochem. Biophys. Res. Commun. 471, 510–514. doi: 10.1016/j.bbrc.2016.02.044
Chater, K. F. (2001). Regulation of sporulation in Streptomyces coelicolor A3(2): a checkpoint multiplex? Curr. Opin. Microbiol. 4, 667–673. doi: 10.1016/S1369-5274(01)00267-3
Cuesta, G., Soler, A., Alonso, J. L., Ruvira, M. A., Lucena, T., Arahal, D. R., et al. (2013). Pseudonocardia hispaniensis sp nov., a novel actinomycete isolated from industrial wastewater activated sludge. Anton Leeuw. Int. J. G. 103, 135–142. doi: 10.1007/s10482-012-9792-1
Delgado, J., Forst, S., Harlocker, S., and Inouye, M. (1993). Identification of a phosphorylation site and functional-analysis of conserved aspartic-acid residues of Ompr, a transcriptional activator for Ompf and Ompc in Escherichia-Coli. Mol. Microbiol. 10, 1037–1047. doi: 10.1111/j.1365-2958.1993.tb00974.x
Dubbs, J. M., and Tabita, F. R. (2004). Regulators of nonsulfur purple phototrophic bacteria and the interactive control of CO2 assimilation, nitrogen fixation, hydrogen metabolism and energy generation. FEMS Microbiol. Rev. 28, 353–376. doi: 10.1016/j.femsre.2004.01.002
Ergan, B., Coplu, L., Alp, A., and Artvinli, M. (2004). Mycobacterium smegmatis pneumonia. Respirology 9, 283–285. doi: 10.1111/j.1440-1843.2004.00570.x
European Food Safety Authority and European Centre for Disease Prevention and Control (2021). The European Union one health 2020 Zoonoses report. EFSA J. 19:6971. doi: 10.2903/j.efsa.2021.6971
Fink, D., Weissschuh, N., Reuther, J., Wohlleben, W., and Engels, A. (2002). Two transcriptional regulators GlnR and GlnRII are involved in regulation of nitrogen metabolism in Streptomyces coelicolor A3(2). Mol. Microbiol. 46, 331–347. doi: 10.1046/j.1365-2958.2002.03150.x
Fisher, S. H. (1989). Glutamate synthesis in Streptomyces coelicolor. J. Bacteriol. 171, 2372–2377. doi: 10.1128/jb.171.5.2372-2377.1989
Fisher, S. H. (1992). Glutamine synthesis in Streptomyces - a review. Gene 115, 13–17. doi: 10.1016/0378-1119(92)90534-V
Fisher, S. H., and Wray, L. V. (1989). Regulation of glutamine synthetase in Streptomyces coelicolor. J. Bacteriol. 171, 2378–2383. doi: 10.1128/jb.171.5.2378-2383.1989
Gross, R., Arico, B., and Rappuoli, R. (1989). Families of bacterial signal-transducing proteins. Mol. Microbiol. 3, 1661–1667. doi: 10.1111/j.1365-2958.1989.tb00152.x
Harth, G., Maslesa-Galic, S., Tullius, M. V., and Horwitz, M. A. (2005). All four Mycobacterium tuberculosis glnA genes encode glutamine synthetase activities but only GlnA1 is abundantly expressed and essential for bacterial homeostasis. Mol. Microbiol. 58, 1157–1172. doi: 10.1111/j.1365-2958.2005.04899.x
He, J. M., Kang, X. M., Wu, J. C., Shao, Z. H., Zhang, Z. H., Wu, Y. Q., et al. (2023). Transcriptional self-regulation of the master nitrogen regulator GlnR in mycobacteria. J. Bacteriol. 205:e0047922. doi: 10.1128/jb.00479-22
He, J. M., Zhu, H., Zheng, G. S., Liu, P. P., Wang, J., Zhao, G. P., et al. (2016). Direct involvement of the master nitrogen metabolism regulator GlnR in antibiotic biosynthesis in Streptomyces. J. Biol. Chem. 291, 26443–26454. doi: 10.1074/jbc.M116.762476
Hedgecock, L. W., and Costello, R. L. (1962). Utilization of nitrate by pathogenic and saprophytic mycobacteria. J. Bacteriol. 84, 195–205. doi: 10.1128/jb.84.2.195-205.1962
Hengge, A. C. (2001). Isotope effects in the study of enzymatic phosphoryl transfer reactions. FEBS Lett. 501, 99–102. doi: 10.1016/S0014-5793(01)02638-2
Hesketh, A., Fink, D., Gust, B., Rexer, H. U., Scheel, B., Chater, K., et al. (2002). The GlnD and GlnK homologues of Streptomyces coelicolor A3(2) are functionally dissimilar to their nitrogen regulatory system counterparts from enteric bacteria. Mol. Microbiol. 46, 319–330. doi: 10.1046/j.1365-2958.2002.03149.x
Hodgson, D. A. (2000). Primary metabolism and its control in streptomycetes: a most unusual group of bacteria. Adv. Microb. Physiol. 42, 47–238. doi: 10.1016/S0065-2911(00)42003-5
Hopwood, D. A., Chater, K. F., and Bibb, M. J. (1995). Genetics of antibiotic production in Streptomyces coelicolor A3(2), a model streptomycete. Biotechnology 28, 65–102.
Hutchings, M. I., Hoskisson, P. A., Chandra, G., and Buttner, M. J. (2004). Sensing and responding to diverse extracellular signals? Analysis of the sensor kinases and response regulators of Streptomyces coelicolor A3(2). Microbiology 150, 2795–2806. doi: 10.1099/mic.0.27181-0
Jenkins, V. A., Barton, G. R., Robertson, B. D., and Williams, K. J. (2013). Genome wide analysis of the complete GlnR nitrogen-response regulon in Mycobacterium smegmatis. BMC Genomics 14:14. doi: 10.1186/1471-2164-14-301
Jenkins, V. A., Robertson, B. D., and Williams, K. J. (2012). Aspartate D48 is essential for the GlnR-mediated transcriptional response to nitrogen limitation in Mycobacterium smegmatis. FEMS Microbiol. Lett. 330, 38–45. doi: 10.1111/j.1574-6968.2012.02530.x
Kol, S., Merlo, M. E., Scheltema, R. A., de Vries, M., Vonk, R. J., Kikkert, N. A., et al. (2010). Metabolomic characterization of the salt stress response in Streptomyces coelicolor. Appl. Environ. Microbiol. 76, 2574–2581. doi: 10.1128/AEM.01992-09
Krysenko, S., Matthews, A., Busche, T., Bera, A., and Wohlleben, W. (2021). Poly- and monoamine metabolism in Streptomyces coelicolor: the new role of glutamine Synthetase-like enzymes in the survival under environmental stress. Microbiol. Physiol. 31, 233–247. doi: 10.1159/000516644
Krysenko, S., Okoniewski, N., Nentwich, M., Matthews, A., Bauerle, M., Zinser, A., et al. (2022). A second gamma-Glutamylpolyamine Synthetase, GlnA2, is involved in polyamine catabolism in Streptomyces coelicolor. Int. J. Mol. Sci. 23:3752. doi: 10.3390/ijms23073752
Lee, P. C., Umeyama, T., and Horinouchi, S. (2002). afsS is a target of AfsR, a transcriptional factor with ATPase activity that globally controls secondary metabolism in Streptomyces coelicolor A3(2). Mol. Microbiol. 43, 1413–1430. doi: 10.1046/j.1365-2958.2002.02840.x
Leigh, J. A., and Dodsworth, J. A. (2007). Nitrogen regulation in bacteria and archaea. Annu. Rev. Microbiol. 61, 349–377. doi: 10.1146/annurev.micro.61.080706.093409
Liao, C. H., Yao, L. L., Xu, Y., Liu, W. B., Zhou, Y., and Ye, B. C. (2015). Nitrogen regulator GInR controls uptake and utilization of non-phosphotransferase-system carbon sources in actinomycetes. Proc. Natl. Acad. Sci. USA 112, 15630–15635. doi: 10.1073/pnas.1508465112
Liao, C. H., Yao, L. L., and Ye, B. C. (2014). Three genes encoding citrate synthases in Saccharopolyspora erythraea are regulated by the global nutrient-sensing regulators GlnR, DasR, and CRP. Mol. Microbiol. 94, 1065–1084. doi: 10.1111/mmi.12818
Lin, W., Wang, Y., Han, X. B., Zhang, Z. L., Wang, C. Y., Wang, J., et al. (2014). Atypical OmpR/PhoB subfamily response regulator GlnR of Actinomycetes functions as a homodimer, stabilized by the Unphosphorylated conserved asp-focused charge interactions. J. Biol. Chem. 289, 15413–15425. doi: 10.1074/jbc.M113.543504
Liu, X. Q., Liu, Y. Y., Lei, C., Zhao, G. P., and Wang, J. (2020). GlnR dominates Rifamycin biosynthesis by activating the rif cluster genes transcription both directly and indirectly in Amycolatopsis mediterranei. Front. Microbiol. 11:11. doi: 10.3389/fmicb.2020.00319
Liu, W. B., Liu, X. X., Shen, M. J., She, G. L., and Ye, B. C. (2019). The nitrogen regulator GlnR directly controls transcription of the prpDBC operon involved in Methylcitrate cycle in Mycobacterium smegmatis. J. Bacteriol. 201:e00099-19. doi: 10.1128/JB.00099-19
Ma, H., Liu, W. B., Zhang, X. P., Hu, H. Q., Gu, S. D., Yuan, H., et al. (2022). GlnR-mediated regulation of KstR controls cholesterol catabolism in Mycobacterium smegmatis. Biotechnol. Appl. Biochem. 69, 1209–1216. doi: 10.1002/bab.2197
Magasanik, B. (1982). Genetic control of nitrogen assimilation in bacteria. Annu. Rev. Genet. 16, 135–168. doi: 10.1146/annurev.ge.16.120182.001031
Malm, S., Tiffert, Y., Micklinghoff, J., Schultze, S., Joost, I., Weber, I., et al. (2009). The roles of the nitrate reductase NarGHJI, the nitrite reductase NirBD and the response regulator GlnR in nitrate assimilation of Mycobacterium tuberculosis. Microbiology 155, 1332–1339. doi: 10.1099/mic.0.023275-0
Martin, J. F., Sola-Landa, A., Santos-Beneit, F., Fernandez-Martinez, L. T., Prieto, C., and Rodriguez-Garcia, A. (2011). Cross-talk of global nutritional regulators in the control of primary and secondary metabolism in Streptomyces. Microb. Biotechnol. 4, 165–174. doi: 10.1111/j.1751-7915.2010.00235.x
MartinezHackert, E., and Stock, A. M. (1997). The DNA-binding domain of OmpR: crystal structure of a winged helix transcription factor. Structure 5, 109–124. doi: 10.1016/S0969-2126(97)00170-6
Mast, Y., Stegmann, E., and Lu, Y. H. (2020). Editorial: regulation of antibiotic production in Actinomycetes. Front. Microbiol. 11:11. doi: 10.3389/fmicb.2020.01566
Meehan, C. J., Goig, G. A., Kohl, T. A., Verboven, L., Dippenaar, A., Ezewudo, M., et al. (2019). Whole genome sequencing of Mycobacterium tuberculosis: current standards and open issues. Nat. Rev. Microbiol. 17, 533–545. doi: 10.1038/s41579-019-0214-5
Meng, S. T., Wu, H., Wang, L., Zhang, B. C., and Bai, L. Q. (2017). Enhancement of antibiotic productions by engineered nitrate utilization in actinomycetes. Appl. Microbiol. Biotechnol. 101, 5341–5352. doi: 10.1007/s00253-017-8292-7
Merrick, M. J., and Edwards, R. A. (1995). Nitrogen control in bacteria. Microbiol. Rev. 59, 604–622. doi: 10.1128/mr.59.4.604-622.1995
Molina-Henares, A. J., Krell, T., Eugenia Guazzaroni, M., Segura, A., and Ramos, J. L. (2006). Members of the IclR family of bacterial transcriptional regulators function as activators and/or repressors. FEMS Microbiol. Rev. 30, 157–186. doi: 10.1111/j.1574-6976.2005.00008.x
Murray, D. S., Chinnam, N., Tonthat, N. K., Whitfill, T., Wray, L. V., Fisher, S. H., et al. (2013). Structures of the Bacillus subtilis glutamine Synthetase Dodecamer reveal large Intersubunit catalytic conformational changes linked to a unique feedback inhibition mechanism. J. Biol. Chem. 288, 35801–35811. doi: 10.1074/jbc.M113.519496
Ni, L. Y., Liu, C. J., and Jin, Z. K. (1984). A positive correlation between rifamycin SV biosynthesis and the activity of glutamine synthetase. Acta Microbiol. Sin. 24, 217–223.
Nieselt, K., Battke, F., Herbig, A., Bruheim, P., Wentzel, A., Jakobsen, O. M., et al. (2010). The dynamic architecture of the metabolic switch in Streptomyces coelicolor. BMC Genomics 11:10. doi: 10.1186/1471-2164-11-10
Oelze, J., and Klein, G. (1996). Control of nitrogen fixation by oxygen in purple nonsulfur bacteria. Arch. Microbiol. 165, 219–225. doi: 10.1007/s002030050319
Ossai, J., Khatabi, B., Nybo, S. E., and Kharel, M. K. (2022). Renewed interests in the discovery of bioactive actinomycete metabolites driven by emerging technologies. J. Appl. Microbiol. 132, 59–77. doi: 10.1111/jam.15225
Pei, J. F., Li, Y. X., Tang, H., Wei, W., and Ye, B. C. (2022). PhoP- and GlnR-mediated regulation of metK transcription and its impact upon S-adenosyl-methionine biosynthesis in Saccharopolyspora erythraea. Microb. Cell Factories 21, 1–11. doi: 10.1186/s12934-022-01846-w
PerezMartin, J., and deLorenzo, V. (1997). Clues and consequences of DNA bending in transcription. Annu. Rev. Microbiol. 51, 593–628. doi: 10.1146/annurev.micro.51.1.593
Petridis, M., Vickers, C., Robson, J., McKenzie, J. L., Bereza, M., Sharrock, A., et al. (2016). Structure and function of AmtR in Mycobacterium smegmatis: implications for post-transcriptional regulation of urea metabolism through a small antisense RNA. J. Mol. Biol. 428, 4315–4329. doi: 10.1016/j.jmb.2016.09.009
Pullan, S. T., Chandra, G., Bibb, M. J., and Merrick, M. (2011). Genome-wide analysis of the role of GlnR in Streptomyces venezuelae provides new insights into global nitrogen regulation in actinomycetes. BMC Genomics 12:12. doi: 10.1186/1471-2164-12-175
Qi, N., She, G. L., Du, W., and Ye, B. C. (2021). Mycobacterium smegmatis GlnR regulates the Glyoxylate cycle and the Methylcitrate cycle on fatty acid metabolism by repressing icl transcription. Front. Microbiol. 12:603835. doi: 10.3389/fmicb.2021.603835
Qiu, H. L., Chang, X. Y., Luo, Y., Shen, F. F., Yin, A. G., Miao, T. T., et al. (2022). Regulation of Nir gene in Lactobacillus plantarum WU14 mediated by GlnR. Front. Microbiol. 13:13. doi: 10.3389/fmicb.2022.983485
Qu, S., Kang, Q. J., Wu, H., Wang, L., and Bai, L. Q. (2015). Positive and negative regulation of GlnR in validamycin a biosynthesis by binding to different loci in promoter region. Appl. Microbiol. Biotechnol. 99, 4771–4783. doi: 10.1007/s00253-015-6437-0
Rakovitsky, N., Bar Oz, M., Goldberg, K., Gibbons, S., Zimhony, O., and Barkan, D. (2018). The unexpected essentiality of glnA2 in Mycobacterium smegmatis is salvaged by overexpression of the global nitrogen regulator glnR, but not by L-, D- or Iso-glutamine. Front. Microbiol. 9:9. doi: 10.3389/fmicb.2018.02143
Reitzer, L., and Schneider, B. L. (2001). Metabolic context and possible physiological themes of sigma(54)-dependent genes in Escherichia coli. Microbiol. Mol. Biol. Rev. 65, 422–444. doi: 10.1128/MMBR.65.3.422-444.2001
Reuther, J., and Wohlleben, W. (2007). Nitrogen metabolism in Streptomyces coelicolor: transcriptional and post-translational regulation. J. Mol. Microb. Biotechnol. 12, 139–146. doi: 10.1159/000096469
Rexer, H. U., Schaberle, T., Wohlleben, W., and Engels, A. (2006). Investigation of the functional properties and regulation of three glutamine synthetase-like genes in Streptomyces coelicolor A3(2). Arch. Microbiol. 186, 447–458. doi: 10.1007/s00203-006-0159-8
Rochefort, D. A., and Benson, D. R. (1990). Molecular cloning, sequencing, and expression of the glutamine synthetase II (glnII) gene from the actinomycete root nodule symbiont Frankia sp. strain CpI1. J. Bacteriol. 172, 5335–5342. doi: 10.1128/jb.172.9.5335-5342.1990
Rodriguez-Garcia, A., Barreiro, C., Santos-Beneit, F., Sola-Landa, A., and Martin, J. F. (2007). Genome-wide transcriptomic and proteomic analysis of the primary response to phosphate limitation in Streptomyces coelicolor M145 and in a Delta phoP mutant. Proteomics 7, 2410–2429. doi: 10.1002/pmic.200600883
Rodriguez-Garcia, A., Sola-Landa, A., Apel, K., Santos-Beneit, F., and Martin, J. F. (2009). Phosphate control over nitrogen metabolism in Streptomyces coelicolor: direct and indirect negative control of glnR, glnA, glnII and amtB expression by the response regulator PhoP. Nucleic Acids Res. 37, 3230–3242. doi: 10.1093/nar/gkp162
Santamarta, I., Lopez-Garcia, M. T., Perez-Redondo, R., Koekman, B., Martin, J. F., and Liras, P. (2007). Connecting primary and secondary metabolism: AreB, an IclR-like protein, binds the ARE(ccaR) sequence of S-clavuligerus and modulates leucine biosynthesis and cephamycin C and clavulanic acid production. Mol. Microbiol. 66, 511–524. doi: 10.1111/j.1365-2958.2007.05937.x
Santos-Beneit, F., Rodriguez-Garcia, A., and Martin, J. F. (2011). Complex transcriptional control of the antibiotic regulator afsS in Streptomyces: PhoP and AfsR are overlapping, competitive activators. J. Bacteriol. 193, 2242–2251. doi: 10.1128/JB.01462-10
Santos-Beneit, F., Rodriguez-Garcia, A., and Martin, J. F. (2012). Overlapping binding of PhoP and AfsR to the promoter region of glnR in Streptomyces coelicolor. Microbiol. Res. 167, 532–535. doi: 10.1016/j.micres.2012.02.010
Santos-Beneit, F., Rodriguez-Garcia, A., Sola-Landa, A., and Martin, J. F. (2009). Cross-talk between two global regulators in Streptomyces: PhoP and AfsR interact in the control of afsS, pstS and phoRP transcription. Mol. Microbiol. 72, 53–68. doi: 10.1111/j.1365-2958.2009.06624.x
Schreier, H. J., Brown, S. W., Hirschi, K. D., Nomellini, J. F., and Sonenshein, A. L. (1989). Regulation of Bacillus subtilis glutamine synthetase gene expression by the product of the glnR gene. J. Mol. Biol. 210, 51–63. doi: 10.1016/0022-2836(89)90290-8
Schuenemann, V. J., Singh, P., Mendum, T. A., Krause-Kyora, B., Jager, G., Bos, K. I., et al. (2013). Genome-wide comparison of medieval and modern Mycobacterium leprae. Science 341, 179–183. doi: 10.1126/science.1238286
Shao, Z. H., Deng, W. X., Li, S. Y., He, J. M., Ren, S. X., Huang, W. R., et al. (2015). GlnR-mediated regulation of ectABCD transcription expands the role of the GlnR regulon to osmotic stress management. J. Bacteriol. 197, 3041–3047. doi: 10.1128/JB.00185-15
Shao, Z. H., Gao, J., Ding, X. M., Wang, J., Chiao, J. S., and Zhao, G. P. (2011). Identification and functional analysis of a nitrate assimilation operon nasACKBDEF from Amycolatopsis mediterranei U32. Arch. Microbiol. 193, 463–477. doi: 10.1007/s00203-011-0690-0
Shi, J., Feng, Z., Xu, J., Li, F., Zhang, Y., Wen, A., et al. (2023). Structural insights into the transcription activation mechanism of the global regulator GlnR from actinobacteria. Proc. Natl. Acad. Sci. 120:e2300282120. doi: 10.1073/pnas.2300282120
Sola-Landa, A., Rodriguez-Garcia, A., Amin, R., Wohlleben, W., and Martin, J. F. (2013). Competition between the GlnR and PhoP regulators for the glnA and amtB promoters in Streptomyces coelicolor. Nucleic Acids Res. 41, 1767–1782. doi: 10.1093/nar/gks1203
Sprusansky, O., Zhou, L. Q., Jordan, S., White, J., and Westpheling, J. (2003). Identification of three new genes involved in morphogenesis and antibiotic production in. J. Bacteriol. 185, 6147–6157. doi: 10.1128/JB.185.20.6147-6157.2003
Stock, J. B., Ninfa, A. J., and Stock, A. M. (1989). Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol. Rev. 53, 450–490. doi: 10.1128/mr.53.4.450-490.1989
Storz, G., and Toledano, M. B. (1994). Regulation of bacterial gene-expression in response to oxidative stress. Methods Enzymol. 236, 196–207. doi: 10.1016/0076-6879(94)36017-0
Sun, Y. H., De Vos, P., and Willems, A. (2017). Nitrogen assimilation in denitrifier Bacillus azotoformans LMG 9581(T). Anton Leeuw. Int. J. G. 110, 1613–1626. doi: 10.1007/s10482-017-0911-x
Tan, W., Liao, T. H., Wang, J., Ye, Y., Wei, Y. C., Zhou, H. K., et al. (2020). A recently evolved diflavin-containing monomeric nitrate reductase is responsible for highly efficient bacterial nitrate assimilation. J. Biol. Chem. 295, 5051–5066. doi: 10.1074/jbc.RA120.012859
Tiffert, Y., Franz-Wachtel, M., Fladerer, C., Nordheim, A., Reuther, J., Wohlleben, W., et al. (2011). Proteomic analysis of the GlnR-mediated response to nitrogen limitation in Streptomyces coelicolor M145. Appl. Microbiol. Biotechnol. 89, 1149–1159. doi: 10.1007/s00253-011-3086-9
Tiffert, Y., Supra, P., Wurm, R., Wohlleben, W., Wagner, R., and Reuther, J. (2008). The Streptomyces coelicolor GlnR regulon: identification of new GlnR targets and evidence for a central role of GlnR in nitrogen metabolism in actinomycetes. Mol. Microbiol. 67, 861–880. doi: 10.1111/j.1365-2958.2007.06092.x
Travis, B. A., Peck, J. V., Salinas, R., Dopkins, B., Lent, N., Nguyen, V. D., et al. (2022). Molecular dissection of the glutamine synthetase-GlnR nitrogen regulatory circuitry in gram-positive bacteria. Nat. Commun. 13:3793. doi: 10.1038/s41467-022-31573-0
Tullius, M. V., Harth, G., and Horwitz, M. A. (2003). Glutamine synthetase GlnA1 is essential for growth of Mycobacterium tuberculosis in human THP-1 macrophages and guinea pigs. Infect. Immun. 71, 3927–3936. doi: 10.1128/IAI.71.7.3927-3936.2003
Wang, R., Cao, Y., Kong, F., Hou, B., Zhao, J., Kang, Y., et al. (2022). Developmental regulator RamRsl controls both morphological development and lincomycin biosynthesis in Streptomyces lincolnensis. J. Appl. Microbiol. 133, 400–409. doi: 10.1111/jam.15568
Wang, Y., Cen, X. F., Zhao, G. P., and Wang, J. (2012). Characterization of a new GlnR binding box in the promoter of amtB in Streptomyces coelicolor inferred a PhoP/GlnR competitive binding mechanism for transcriptional regulation of amtB. J. Bacteriol. 194, 5237–5244. doi: 10.1128/JB.00989-12
Wang, Y., Li, C., Duan, N., Li, B., Ding, X. M., Yao, Y. F., et al. (2014). GlnR negatively regulates the transcription of the alanine dehydrogenase encoding gene ald in Amycolatopsis mediterranei U32 under nitrogen limited conditions via specific binding to its major transcription initiation site. PLoS One 9:e104811. doi: 10.1371/journal.pone.0104811
Wang, R., Mast, Y., Wang, J., Zhang, W. W., Zhao, G. P., Wohlleben, W., et al. (2013). Identification of two-component system AfsQ1/Q2 regulon and its cross-regulation with GlnR in Streptomyces coelicolor. Mol. Microbiol. 87, 30–48. doi: 10.1111/mmi.12080
Wang, Y., Wang, J. Z., Shao, Z. H., Yuan, H., Lu, Y. H., Jiang, W. H., et al. (2013). Three of four GlnR binding sites are essential for GlnR-mediated activation of transcription of the Amycolatopsis mediterranei nas operon. J. Bacteriol. 195, 2595–2602. doi: 10.1128/JB.00182-13
Wang, J., Wang, Y., and Zhao, G. P. (2015). Precise characterization of GlnR box in actinomycetes. Biochem. Biophys. Res. Commun. 458, 605–607. doi: 10.1016/j.bbrc.2015.02.010
Wang, J., and Zhao, G. P. (2009). GlnR positively regulates nasA transcription in Streptomyces coelicolor. Biochem. Biophys. Res. Commun. 386, 77–81. doi: 10.1016/j.bbrc.2009.05.147
Wang, T. S., Zhao, X. Y., Wu, X. Y., and Chen, S. F. (2023). Genome-wide mapping of GlnR-binding sites reveals the global regulatory role of GlnR in controlling the metabolism of nitrogen and carbon in Paenibacillus polymyxa WLY78. Bmc. Genomics 24:85. doi: 10.1186/s12864-023-09147-1
Wang, R. D., Zhou, T. Y., Kong, F. J., Hou, B. B., Ye, J., Wu, H. Z., et al. (2023). AflQ1-Q2 represses lincomycin biosynthesis via multiple cascades in Streptomyces lincolnensis. Appl. Microbiol. Biotechnol. 107, 2933–2945. doi: 10.1007/s00253-023-12429-z
Weiss, V., Kramer, G., Dunnebier, T., and Flotho, A. (2002). Mechanism of regulation of the bifunctional histidine kinase NtrB in Escherichia coli. J. Mol. Microb. Biotechnol. 4, 229–233.
Weissschuh, N., Fink, D., Vierling, S., Bibb, M. J., Wohlleben, W., and Engels, A. (2000). Transcriptional analysis of the gene for glutamine synthetase II and two upstream genes in Streptomyces coelicolor A3(2). Mol. Gen. Genet. 264, 461–469. doi: 10.1007/s004380000315
Welch, M., Chinardet, N., Mourey, L., Birck, C., and Samama, J. P. (1998). Structure of the CheY-binding domain of histidine kinase CheA in complex with CheY. Nat. Struct. Biol. 5, 25–29. doi: 10.1038/nsb0198-25
West, A. H., and Stock, A. M. (2001). Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 26, 369–376. doi: 10.1016/S0968-0004(01)01852-7
Williams, K. J., Jenkins, V. A., Barton, G. R., Bryant, W. A., Krishnan, N., and Robertson, B. D. (2015). Deciphering the metabolic response of Mycobacterium tuberculosis to nitrogen stress. Mol. Microbiol. 97, 1142–1157. doi: 10.1111/mmi.13091
Wray, L. V., Atkinson, M. R., and Fisher, S. H. (1991). Identification and cloning of the glnR locus, which is required for transcription of the glnA gene in Streptomyces coelicolor A3(2). J. Bacteriol. 173, 7351–7360. doi: 10.1128/jb.173.22.7351-7360.1991
Wray, L. V., Ferson, A. E., Rohrer, K., and Fisher, S. H. (1996). TnrA, a transcription factor required for global nitrogen regulation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 93, 8841–8845. doi: 10.1073/pnas.93.17.8841
Wray, L. V., and Fisher, S. H. (1993). The Streptomyces coelicolor glnR gene encodes a protein similar to other bacterial response regulators. Gene 130, 145–150. doi: 10.1016/0378-1119(93)90359-B
Wray, L. V. Jr., and Fisher, S. H. (1988). Cloning and nucleotide sequence of the Streptomyces coelicolor gene encoding glutamine synthetase. Gene 71, 247–256. doi: 10.1016/0378-1119(88)90041-8
Xu, Y., Liao, C. H., Yao, L. L., Ye, X., and Ye, B. C. (2016). GlnR and PhoP directly regulate the transcription of genes encoding starch-degrading, Amylolytic enzymes in Saccharopolyspora erythraea. Appl. Environ. Microbiol. 82, 6819–6830. doi: 10.1128/AEM.02117-16
Xu, Y., and Ye, B. C. (2018). GlnR and PhoP regulate beta-glucosidases involved in cellulose digestion in response to nitrogen and phosphate availability. Microbiology 164, 779–789. doi: 10.1099/mic.0.000654
Xu, Y., You, D., Yao, L. L., Chu, X. H., and Ye, B. C. (2019). Phosphate regulator PhoP directly and indirectly controls transcription of the erythromycin biosynthesis genes in Saccharopolyspora erythraea. Microb. Cell Factories 18:206. doi: 10.1186/s12934-019-1258-y
Yang, Y. H., Song, E., Kim, E. J., Lee, K., Kim, W. S., Park, S. S., et al. (2009). NdgR, an IclR-like regulator involved in amino-acid-dependent growth, quorum sensing, and antibiotic production in Streptomyces coelicolor. Appl. Microbiol. Biotechnol. 82, 501–511. doi: 10.1007/s00253-008-1802-x
Yao, L. L., Liao, C. H., Huang, G., Zhou, Y., Rigali, S., Zhang, B. C., et al. (2014). GlnR-mediated regulation of nitrogen metabolism in the actinomycete Saccharopolyspora erythraea. Appl. Microbiol. Biotechnol. 98, 7935–7948. doi: 10.1007/s00253-014-5878-1
Yao, L. L., and Ye, B. C. (2016). Reciprocal regulation of GlnR and PhoP in response to nitrogen and phosphate limitations in Saccharopolyspora erythraea. Appl. Environ. Microbiol. 82, 409–420. doi: 10.1128/AEM.02960-15
Yoshida, K., Yamaguchi, H., Kinehara, M., Ohki, Y., Nakaura, Y., and Fujita, Y. (2003). Identification of additional TnrA-regulated genes of Bacillus subtilis associated with a TnrA box. Mol. Microbiol. 49, 157–165. doi: 10.1046/j.1365-2958.2003.03567.x
You, D., Wang, M. M., and Ye, B. C. (2017). Acetyl-CoA synthetases of Saccharopolyspora erythraea are regulated by the nitrogen response regulator GlnR at both transcriptional and post-translational levels. Mol. Microbiol. 103, 845–859. doi: 10.1111/mmi.13595
You, D., Xu, Y., Yin, B. C., and Ye, B. C. (2019). Nitrogen regulator GlnR controls redox sensing and lipids anabolism by directly activating the whiB3 in Mycobacterium smegmatis. Front. Microbiol. 10:74. doi: 10.3389/fmicb.2019.00074
You, D., Yin, B. C., Li, Z. H., Zhou, Y., Yu, W. B., Zuo, P., et al. (2016). Sirtuin-dependent reversible lysine acetylation of glutamine synthetases reveals an autofeedback loop in nitrogen metabolism. Proc. Natl. Acad. Sci. USA 113, 6653–6658. doi: 10.1073/pnas.1525654113
Yu, H., Peng, W. T., Liu, Y., Wu, T., Yao, Y. F., Cui, M. X., et al. (2006). Identification and characterization of glnA promoter and its corresponding trans-regulatory protein GlnR in the rifamycin SV producing actinomycete, Amycolatopsis mediterranei u32. Acta Biochim. Biophys. Sin. 38, 831–843. doi: 10.1111/j.1745-7270.2006.00238.x
Yu, H., Yao, Y. F., Liu, Y., Jiao, R. S., Jiang, W. H., and Zhao, G. P. (2007). A complex role of Amycolatopsis mediterranei GlnR in nitrogen metabolism and related antibiotics production. Arch. Microbiol. 188, 89–96. doi: 10.1007/s00203-007-0228-7
Zeth, K. (2013). Crystal structure of glutamine synthetase from Streptomyces coelicolor. doi: 10.2210/pdb4BAX/pdb,
Zhang, P. P., Wu, L. L., Zhu, Y. P., Liu, M., Wang, Y. M., Cao, G. X., et al. (2017). Deletion of MtrA inhibits cellular development of Streptomyces coelicolor and alters expression of developmental regulatory genes. Front. Microbiol. 8:8. doi: 10.3389/fmicb.2017.02013
Zhang, M., Zheng, P., Wang, R., Li, W., Lu, H. F., and Zhang, J. Q. (2014). Nitrate-dependent anaerobic ferrous oxidation (NAFO) by denitrifying bacteria: a perspective autotrophic nitrogen pollution control technology. Chemosphere 117, 604–609. doi: 10.1016/j.chemosphere.2014.09.029
Zhang, Y., Zhou, X. L., Wang, X. M., Wang, L., Xia, M. L., Luo, J. M., et al. (2020). Improving phytosterol biotransformation at low nitrogen levels by enhancing the methylcitrate cycle with transcriptional regulators PrpR and GlnR of Mycobacterium neoaurum. Microb. Cell Factories 19:13. doi: 10.1186/s12934-020-1285-8
Zhu, Y. P., Wang, J., Su, W. Y., Lu, T., Li, A. Y., and Pang, X. H. (2022). Effects of dual deletion of glnR and mtrA on expression of nitrogen metabolism genes in Streptomyces venezuelae. Microb. Biotechnol. 15, 1795–1810. doi: 10.1111/1751-7915.14016
Zhu, Y. P., Zhang, P. P., Lu, T., Wang, X. Y., Li, A. Y., Lu, Y. H., et al. (2021). Impact of MtrA on phosphate metabolism genes and the response to altered phosphate conditions in Streptomyces. Environ. Microbiol. 23, 6907–6923. doi: 10.1111/1462-2920.15719
Zhu, Y. P., Zhang, P. P., Zhang, J., Wang, J., Lu, Y. H., and Pang, X. H. (2020). Impact on multiple antibiotic pathways reveals MtrA as a master regulator of antibiotic production in Streptomyces spp. and potentially in other Actinobacteria. Appl. Environ. Microbiol. 86:e01201-20. doi: 10.1128/AEM.01201-20
Zhu, Y. P., Zhang, P. P., Zhang, J., Xu, W. H., Wang, X. Y., Wu, L. L., et al. (2019). The developmental regulator MtrA binds GlnR boxes and represses nitrogen metabolism genes in Streptomyces coelicolor. Mol. Microbiol. 112, 29–46. doi: 10.1111/mmi.14252
Keywords: Actinomycetes, global regulator, GlnR, metabolic regulation, cross-regulation
Citation: Gao B, Li G, Gu D and Wang J (2023) Research progress on GlnR-mediated regulation in Actinomycetes. Front. Microbiol. 14:1282523. doi: 10.3389/fmicb.2023.1282523
Edited by:
Marie-Joelle Virolle, Centre National de la Recherche Scientifique (CNRS), FranceReviewed by:
Angel Manteca, Universidad de Oviedo Mieres, SpainAlberto Sola-Landa, Fundacion Cesefor, Spain
Copyright © 2023 Gao, Li, Gu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dayong Gu, wanhood@163.com; Jin Wang, wangj01@hotmail.com
 Bo Gao
Bo Gao Guoqiang Li3
Guoqiang Li3 Jin Wang
Jin Wang