- 1State Key Laboratory of Genetic Engineering, School of Life Sciences and Huashan Hospital, MOE Engineering Research Center of Gene Technology, Shanghai Engineering Research Center of Industrial Microorganisms, Fudan University, Shanghai, China
- 2Shanghai Zelixir Biotech Company Ltd., Shanghai, China
- 3School of Basic Medical Sciences, Fudan University, Shanghai, China
- 4Key Laboratory of Synthetic Biology, CAS Center for Excellence in Molecular Plant Sciences, Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai, China
- 5Shanghai Key Laboratory of Infectious Diseases and Biosafety Emergency Response, National Medical Center for Infectious Diseases, Huashan Hospital, Fudan University, Shanghai, China
The housecleaning enzyme of Mycobacterium tuberculosis (Mtb), MazG, is a nucleoside triphosphate pyrophosphohydrolase (NTP-PPase) and can hydrolyze all canonical or non-canonical NTPs into NMPs and pyrophosphate. The Mycobacterium tuberculosis MazG (Mtb-MazG) contributes to antibiotic resistance in response to oxidative or nitrosative stress under dormancy, making it a promising target for treating TB in latent infection patients. However, the structural basis of Mtb-MazG is not clear. Here we describe the crystal structure of Mtb-MazG (1–185) at 2.7 Å resolution, composed of two similar folded spherical domains in tandem. Unlike other all-α NTP pyrophosphatases, Mtb-MazG has an N-terminal extra region composed of three α-helices and five β-strands. The second domain is global, with five α-helices located in the N-terminal domain. Gel-filtration assay and SAXS analysis show that Mtb-MazG forms an enzyme-active dimer in solution. In addition, the metal ion Mg2+ is bound with four negative-charged residues Glu119, Glu122, Glu138, and Asp141. Different truncations and site-directed mutagenesis revealed that the full-length dimeric form and the metal ion Mg2+ are indispensable for the catalytic activity of Mtb-MazG. Thus, our work provides new insights into understanding the molecular basis of Mtb-MazG.
Introduction
Tuberculosis (TB), caused by Mycobacterium tuberculosis (Mtb), is one of the infectious killers globally that accounts for about 1.4 million deaths worldwide each year (Dartois and Rubin, 2022; Yang et al., 2022). Introducing the Mycobacterium bovis bacille Calmette-Guérin (BCG) vaccine in newborn babies has dramatically decreased the threat of Mtb (Lange et al., 2022). However, BCG is less effective for preventing pulmonary tuberculosis in adults and may have side effects in the immunocompromised hosts (Furin et al., 2019). In addition, Mtb can evade the host immune system in a dormant way in latently infected patients, making the antibacterial drug development challenging (de Wet et al., 2019). Therefore, it is particularly important to understand the molecular events of growth control and metabolic adaptation of non-growing Mtb for developing new therapeutic strategies.
Cellular metabolism is precisely regulated by various housecleaning enzymes, especially the NTP pyrophosphatases, which hydrolyze wasted compounds into cellular metabolites, therefore preventing the non-canonical NTPs-triggered mutagenesis and DNA damage (Gad et al., 2014; Fan et al., 2018). Housecleaning NTP pyrophosphatases include four structural superfamilies: trimeric dUTPase, ITPase (Maf/HAM1), Nudix-box containing hydrolases, and all-α NTP pyrophosphatases (Galperin et al., 2006). In addition, structure-based analysis reveals that the all-α NTP pyrophosphatases include the dimeric dUTPase, the phosphoribosyl-ATP pyrophosphatase HisE, and the NTP pyrophosphatase MazG (Moroz et al., 2005). All the enzymes specifically target non-canonical NTPs, including 5-OH-dCTP, dUTP, dITP, 2-oxo-dATP, and 8-oxo-dGTP, with high affinities (Galperin et al., 2006; Lu et al., 2010; Lyu et al., 2013).
MazG, identified initially as a downstream gene of the toxin-antitoxin complex MazEF in E. coli, exists in different bacteria and many phages (Zhang and Inouye, 2002; Huang et al., 2021). Previous studies showed that Mtb-MazG is able to hydrolyze all canonical (d)NTPs and 8-oxo-dGTP (Lu et al., 2010). Furthermore, MazG eliminates 5-OH-dCTP and regulates pyrimidine metabolism, safeguarding the genetic stability of Mtb during oxidative stress conditions (Lyu et al., 2013). On the other way, MazG is required for the persistence of Mtb during chronic infection of mice and contributes to antibiotic tolerance of stationary-phase culture and intracellular Mtb (Shi et al., 2019). Currently, the structures of MazG from Bacillus anthracis, E. coli, and Deinococcus radiodurans reveal a quite similar dimeric or tetrameric all-α-helical architecture (Lee et al., 2008; Goncalves et al., 2011; Kim and Hong, 2016). However, Mtb-MazG shows not only a certain extent of sequence similarity with bacterial homologs, but also has one extra region at the N-terminal domain, which may contribute to the antibacterial resistance in the dormant stage.
Here, we report the crystal structure of MazG (1–185) from M. tuberculosis at 2.7 Å resolution, forming a dimer through the interaction of two repeated MazG-like domains. Structural alignment and mutation studies revealed that the magnesium-ion-binding sites and full-length dimeric protein are necessary for the catalytic activity of MazG. The structural basis of MazG might provide insights into understanding the diverse functions of MazG in dormant Mtb.
Results
The purification and crystallization of Mtb-MazG
To reveal the structure of Mtb-MazG, we expressed the 6xHis-MazG fusion protein in Escherichia coli BL21(DE3) cells and further purified with different chromatographies as previously described (Zhan et al., 2022). The purified Mtb-MazG came out at the peak of ~71 mL on a Superdex200 16/600 column, corresponding with a molecular weight of ~75 kD. As the theoretical molecular weight of Mtb-MazG is 35 kD, it showed that MazG was a dimer in solution (Figures 1A,B). Next, we screened more than 1,000 crystallizing conditions for the high purity (>95%) full-length Mtb-MazG protein. However, no crystals were grown, which probably resulted from the intrinsic disorder property (Chen et al., 2022). Thus, to identify the suitable regions for crystallization, we performed limited protease digestion for Mtb-MazG. The results showed that Mtb-MazG was cleaved into two stable fragments by endoproteinase Glu-C (Supplementary Figure S1A). Further mass spectrometry (MS) experiment showed that the upper band and lower band in SDS-PAGE might be a partial fragment of the N-terminal part (1–185) and the C-terminal part (186–325) of Mtb-MazG, respectively (Supplementary Figures S1A,B). To check whether the two parts interacted with each other, we performed the gel-filtration analysis with the endoproteinase-digested Mtb-MazG protein in vitro. However, endoproteinase digestion resulted in more than two fragments less than 15 kDa, and this could have hindered the formation of a stable complex of the two parts of MazG (Supplementary Figures S1C,D). In addition, we constructed a series of truncated versions, including MazG (1–185), MazG (1–231), MazG (1–281), MazG (1–305), MazG (186–305), MazG (186–325), and MazG (85–325) for crystallization. Specifically, MazG (1–185) came out at the peak of ~78 mL on the Superdex200 16/600 column, corresponding with a molecular weight of ~40 kD. Therefore, MazG (1–185) was a dimer in solution, as the theoretical molecular weight of MazG (1–185) is 20 kD (Figures 1C,D). Also, the dynamic light scattering (DLS) experiment showed that the radius and estimated molecular weight of full-length Mtb-MazG and Mtb-MazG (1–185) were 3.6 Å and 3.0 Å, 69.2 kD and 41.2 kD, respectively, indicating both of they possessed good homogeneities in solution (Figures 1E,F).
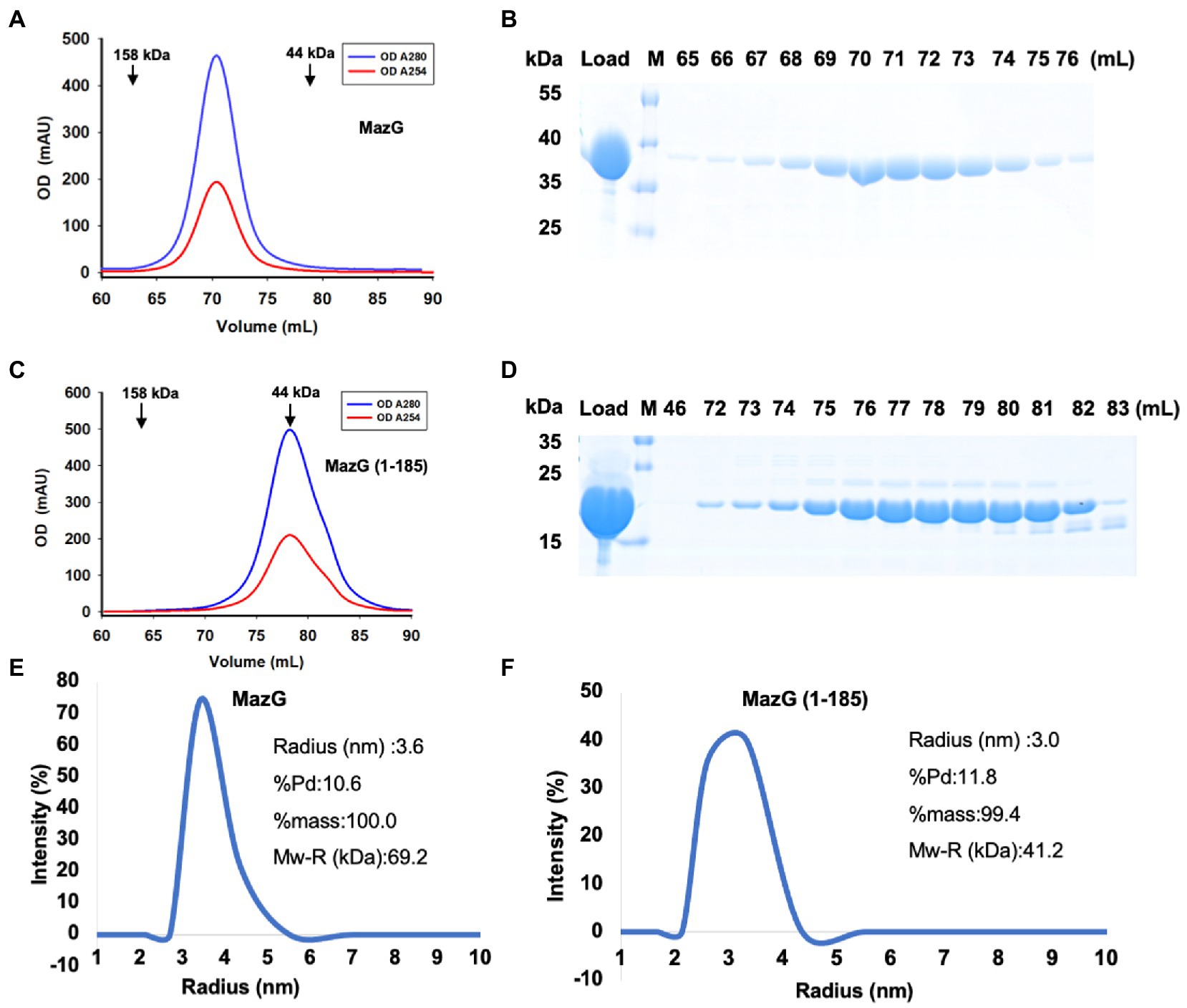
Figure 1. Characterization of Mtb-MazG and mutant (1–185) by gel-filtration and DLS methods. (A,C) Gel-filtration profiles of MazG and MazG (1–185) on a Superdex 200 16/600 column. (B,D) SDS-PAGE of the corresponding elution fractions in A,C. M: protein markers. (E,F) The DLS distributions of MazG and MazG (1–185).
The overall structure of MazG (1–185)
Mtb-MazG mainly contains three domains, an extra N-terminal region (NE, residues 1–85) that is absent in other bacterial homologs, the N-terminal domain (NTD, residues 86–253), and the C-terminal domain (CTD, residues 254–325; Figure 2A; Mota et al., 2016). Despite failing in the crystallization of full-length MazG, we succeeded in obtaining the MazG (1–185) protein crystals and solved the 2.7 Å resolution structure using molecular replacement of the AlphaFold2-predicted structure (AF-P96379-F1). There are six MazG molecules in an asymmetrical unit with a cell dimension of 109 × 109 × 242 Å, which includes two monomers and two dimers consisting of two identical chains. The Mtb-MazG (1–185) monomer contained eight α-helices and five β-stands, comprising two globular domains, including a primary three-layered α/β/α sandwich domain (named domain 1) and an entirely α-helices-composed domain (named domain 2; Figure 2B). Domain 1 was composed of three α-helices (α1, α2, and α3) and five β-sheet strands (β1, β2, β3, β4, and β5), which was absent in other NTP pyrophosphatases, including B. anthracis, E. coli, and D. radiodurans MazGs (Lee et al., 2008; Goncalves et al., 2011; Kim and Hong, 2016). Domain 2 included five α-helices (α4, α5, α6, α7, and α8), which showed a higher B-factor than that in domain 1, indicating this region exhibited more flexibility (Figure 2C). The two domains were connected with a short linker (Ala78-Gly84). Helices α3, α4, α5, α6, and α7 flanked one side of the β-sheet, while α1, α2, and α8 were located on the opposite side (Figure 2B). Also, Mtb-MazG (1–185) forms a dimer in crystal packing, with a stable conformation in the interaction region (Figure 2C). The dimeric Mtb-MazG (1–185) is with an approximate dimension of 45 × 47 × 60 Å (Figure 2D). The interface-involved residues of the swapped-dimeric MazG were located in α1 of domain 1, and α4, α6, and α8 of domain 2, of which fifteen residues formed multiple hydrogen bonds and salt bridges in relative distances within 3.5 Å (Figure 2D). These residues included D7, R10, T12, V14, V16, I19, R23, G84, E85, R86, T120, Y121, R136, E154, D164, D168, and T169 (Figure 2D).
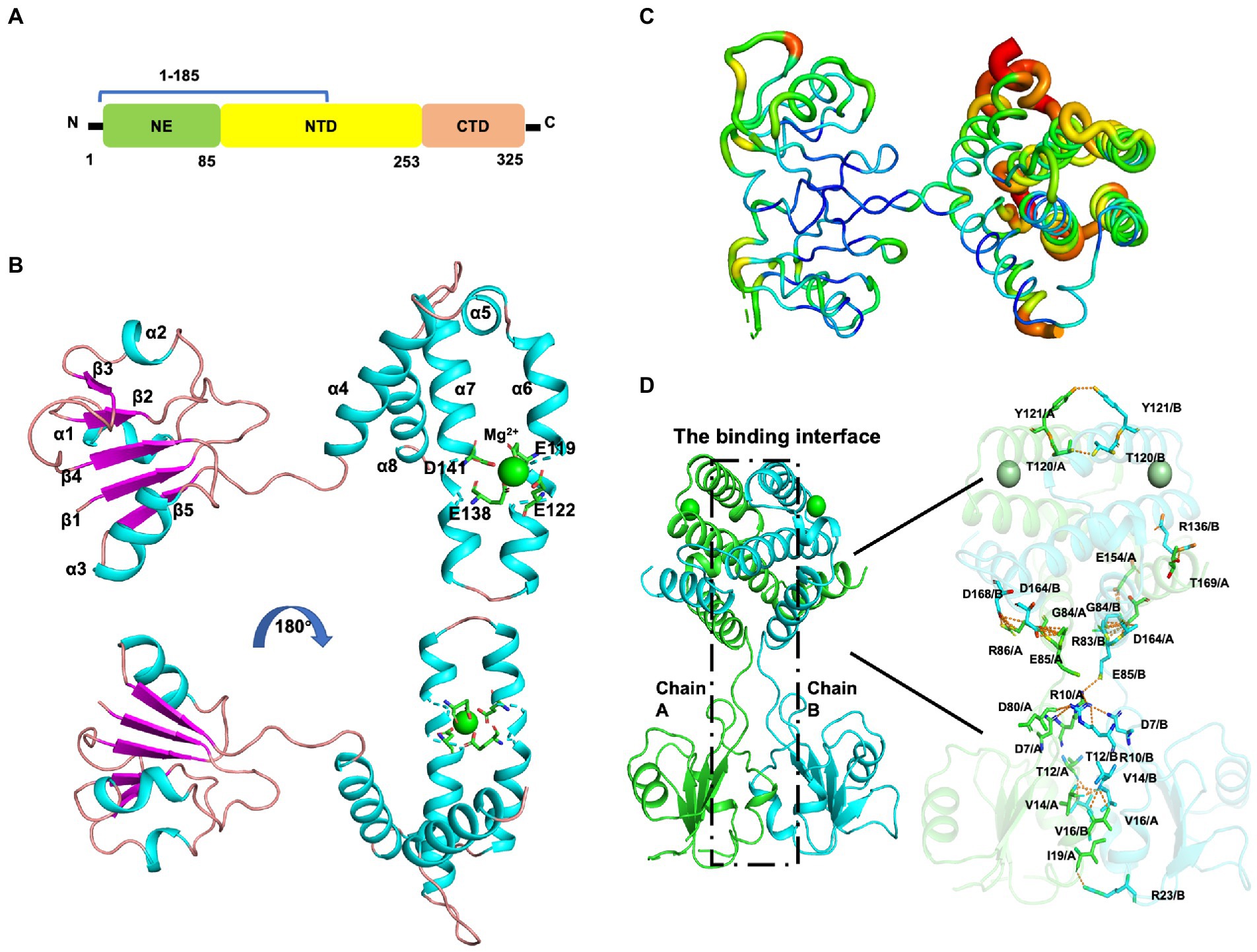
Figure 2. The overall structure of MazG (1–185) from Mycobacterium tuberculosis. (A) Schematic of the Mtb-MazG protein. Mtb-MazG is composed of the N-terminal domain (NTD), the C-terminal domain (CTD), and the N-terminal extra region (NE). (B) The crystal structure of Mtb-MazG (1–185) is represented by a cartoon model, which is composed of eight α-helices and five β-strand. The magnesium is bound with four residues that are located on α6 and α7 of MazG (1–185). Cyan: α-helix; magenta: β-strand; green: magnesium ion. (C) The B-factor distribution of Mtb-MazG protein. The wider and redder tubing indicated a higher B-factor. (D) The binding interface between the swapped-dimeric Mtb-MazG (1–185). The dashed black box (left) denotes the interface between the swapped dimeric structure. The key residues (right) involved in the interface are shown in stick models. Magnesium ions are shown in green spheres.
Next, we investigated the solution status of MazG (1–185) and full-length MazG by the small-angle X-ray scattering (SAXS) method (Figure 3). MazG (1–185) and full-length MazG behaved well in solution, evidenced by the Guinier plots and intensity profiles (Figures 3A,B,F,G). The maximum dimensions (Dmax) from the distance distribution function p(r) of MazG (1–185) and full-length MazG were around 68 Å and 120 Å, respectively (Figures 3C,H). Moreover, when superimposed the crystal structure of MazG (1–185) with the ab initio envelope obtained from the de-novo DAMMIN model of SAXS, MazG (1–185) showed high similarities, confirming that the purified MazG (1–185) in vitro was indeed the active dimeric form (Figures 3D,E). For full-length MazG, we constructed the dimeric structure from the AlphaFold2 model (AF-P96379-F1; Figure 3I). Also, the dimeric structure overlapped well with the ab initio envelope obtained from the de-novo DAMMIN model of SAXS (Figure 3J).
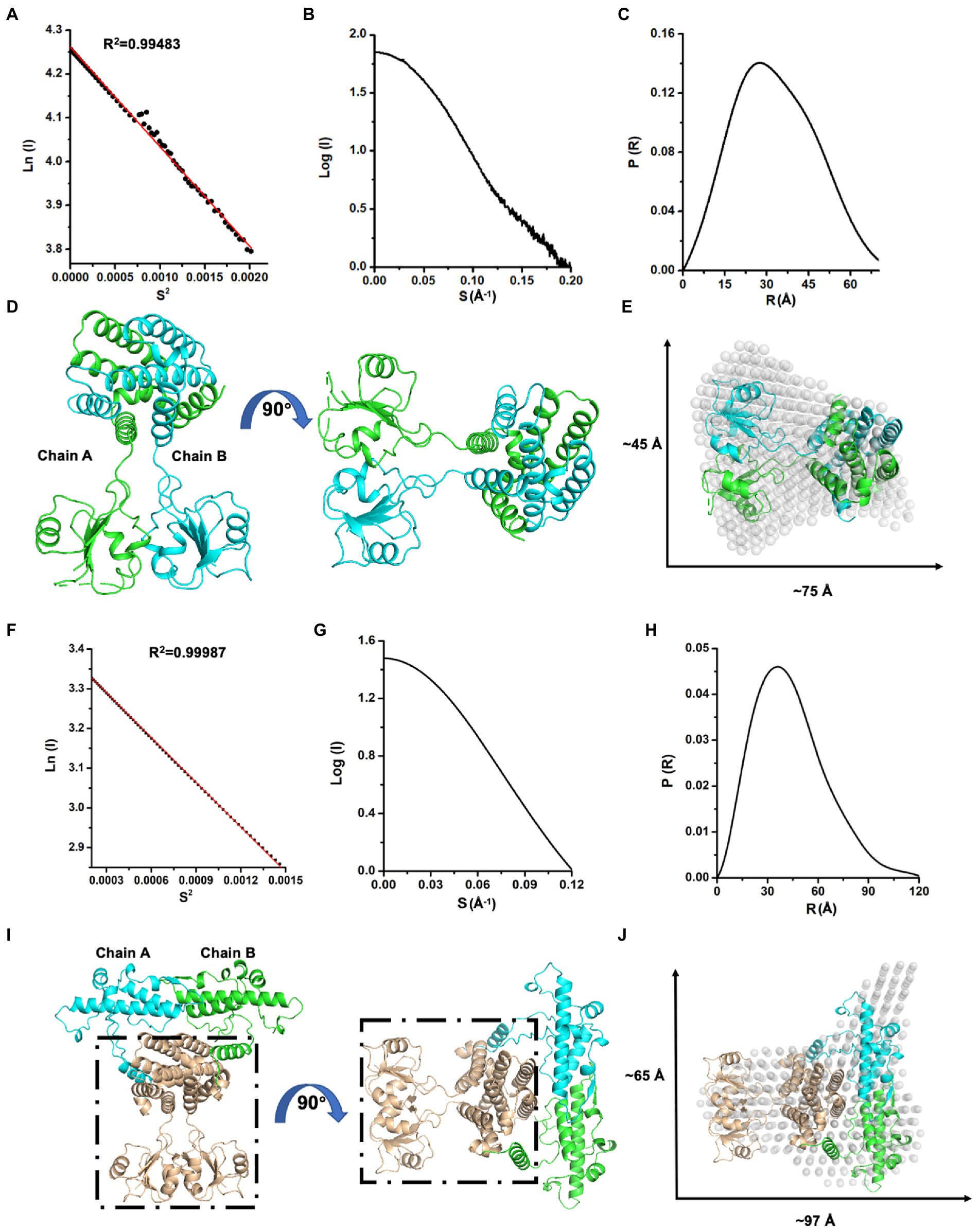
Figure 3. SAXS analysis of Mtb-MazG and its truncation MazG (1–185). (A–C) Guinier plot, intensity profile, and the P(R) curve of MazG (1–185). (D) The dimeric structure of MazG (1–185). The two subunits are colored cyan and green, respectively. (E) The de-novo DAMMIN model was overlapped with the crystal structure of MazG (1–185; PDB code: 7YH5, grey). (F–H) Guinier plot, intensity profile, and the P(R) curve of full-length Mtb-MazG. (I) The AlphaFold2 predicted-dimeric structure of full-length MazG. Chains A and B are shown in cyan and green, respectively. The region (1–185) is shown in wheat. (J) The de-novo DAMMIN model was overlapped with the AlphaFold2-predicted structure of full-length MazG (AF-P96379-F1).
The magnesium-binding sites and enzymatic activity
The two-metal-ion mechanism is conserved in the MazG family across bacteria to phages (Mota et al., 2016; Huang et al., 2021; Wood et al., 2021). In the MazG (1–185) structure, one magnesium ion was present per subunit. The Mg2+ was coordinated by three glutamate residues (Glu119, Glu122, and Glu138) and one aspartate (Asp141; Figure 2B). Surface charge and electron density map analysis showed that Mg2+ was surrounded by negative charges, which are conserved in the NTP pyrophosphatase superfamily (Figures 4A,B). The four residues were mutated into alanines to unveil further the critical roles for enzymatic activity (Figure 4C). Compared with the wild-type (WT) MazG protein, the mutants came out at the same peak on gel-filtration profiles, indicating they were also dimer in solution (Figure 4D). Also, given that mutants were essential for catalysis, to exclude the possible reason that mutations affect the enzyme activity by changing the original structural conformation of the protein, we performed circular dichroism spectra (CD) analysis with the WT MazG and mutant proteins. The CD results showed that the four mutants did not change the structural conformation of the MazG (Figure 4E).
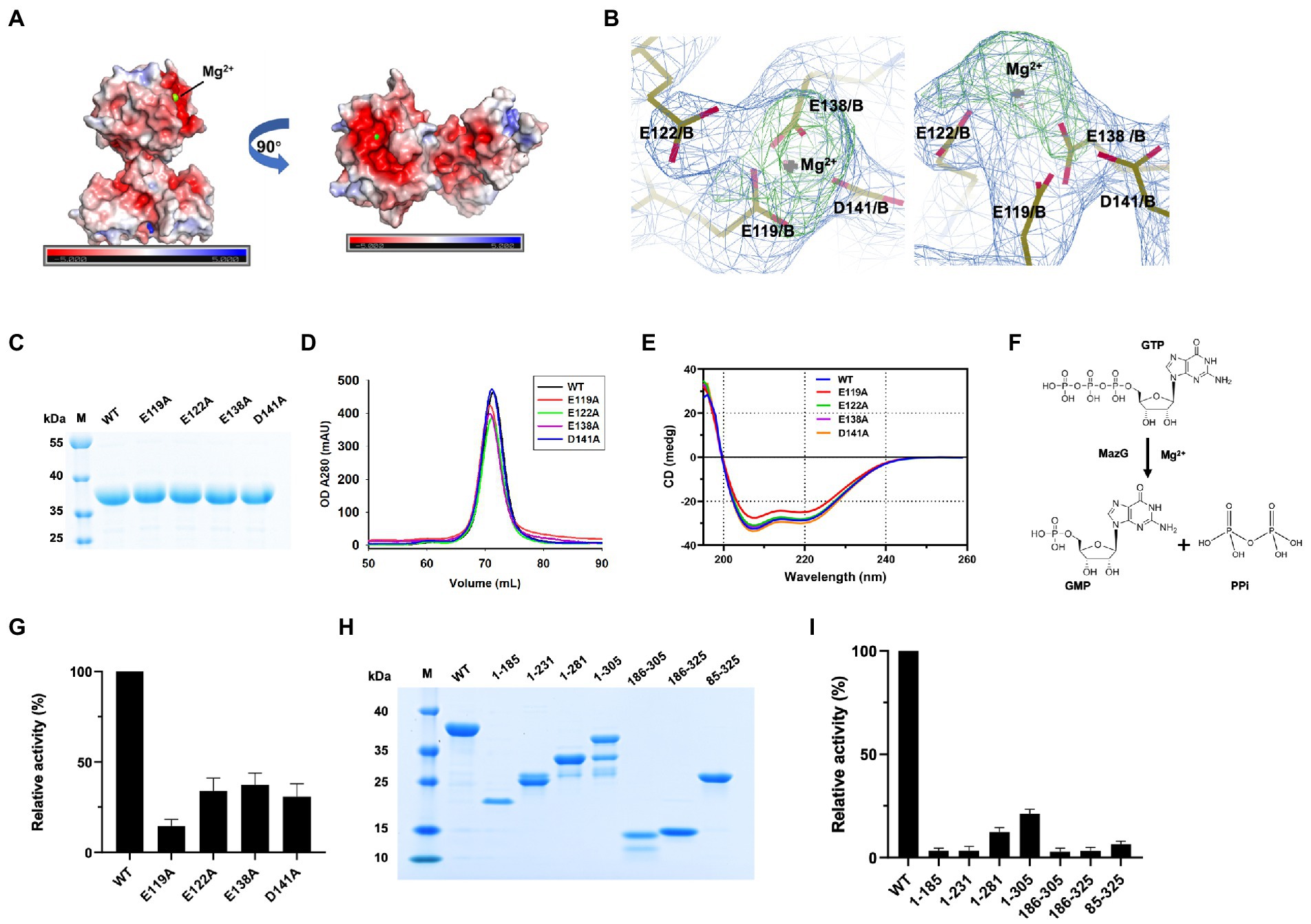
Figure 4. The magnesium binding sites and enzymatic characterizations of Mtb-MazG. (A) The surface potential charge of MazG (1–185) structure. Red: negative charge; blue: positive charge. (B) The electron density map (2Fo-Fc, level = 1.0) of magnesium-bound residues in MazG. (C) SDS-PAGE of MazG and different mutants. (D) The gel filtration profiles of MazG and different mutants. (E) CD spectra of MazG and its mutants measured at 18°C. Each spectrum reported here is an average of three scans. (F) Schematic diagram of the enzymatic reaction. MazG hydrolyzes GTP into GMP and pyrophosphoric acid. (G) The relative activities of WT and different mutants of Mtb-MazG. The activity of WT was taken as 100%. (H) SDS-PAGE of WT MazG and its various truncations. Notably, MazG (1–281), MazG (1–305), and MazG (186–305) showed degradation during the purification process. (I) The relative activities of WT MazG and different truncations. The activity of WT was taken as 100%.
The hydrolase activity of Mtb-MazG was assayed with a spectrophotometric method using GTP as the substrate, as previously described (Lyu et al., 2013). The wild type Mtb-MazG possessed a kcat value of (0.83 ± 0.06) s−1 and a Km value of (0.4 ± 0.1) mM, respectively. The catalytic efficiency (kcat/Km) of the MazG protein toward GTP was 2.1 mM−1 s−1. Also, the NTP-PPase activity was significantly lowered for the mutants of E119A, E122A, E138A, and D141A (Figures 4F,G). Among the four mutants, E119A has the lowest enzymatic activity, whereas E122A, E138A, and D141A possess 20 ~ 40% relative activities with that in WT MazG. Moreover, the enzyme activities toward different truncations (1–185, 1–231, 1–281, 1–305, 186–305, 186–325, and 85–325) were measured and found to be significantly lower in all cases compared to WT MazG (Figures 4H,I), which showed similar enzyme activity to our previous reported (Lyu et al., 2013). Taken together, the metal ion and full-length protein are indispensable for the activity of MazG in Mtb.
Structural comparison of Mtb-MazG with its homologs
To further identify the critical configuration elements of MazG in cell metabolism, we performed a structural-based alignment for Mtb-MazG with its different bacteria orthologs with Clustal X software (Figure 5). The results showed that the NTD and CTD of MazG are conserved in different bacteria, including M. tuberculosis, M. marinum, M. smegmatis, M. avium, and E. coli. However, the mycobaterial MazGs possess one specific N-terminal extra region, while E. coli MazG does not (Figures 2A,B, 5). When superimposing the crystal structure of Mtb-MazG with the AlphaFold2-predicted model, the RMSD value was only 1.207 Å, indicating the two structures were very similar (Figures 6A,B). Also, Mtb-MazG showed different overall folds with homologs from different species. Although the amino acid sequence showed high similarity (~39%) between E. coli MazG (EcMazG) and Mtb-MazG, the overall structure was dramatically different, evidenced by that the RMSD value was 32.275 Å when superimposing the Mtb-MazG structure with the EcMazG (PDB ID: 3CRC; Figure 6C; Lee et al., 2008). Interestingly, MazG proteins from different mycobacterial species might have conserved structures, as superimposing the Alphafold-predicative MazG models of the M. marinum, the M. avium, and the M. smegmatis with Mtb-MazG showed that the RMSD values were 1.723 Å, 0.670 Å, and 0.500 Å, respectively. Moreover, superimposing the C. jejun dUTPase structure (PDB ID: 1W2Y; Moroz et al., 2004), the S. solfataricus MazG (PDB ID: 1VMG), and the D. radiodurans MazG (PDB ID: 2YFD; Goncalves et al., 2011) with Mtb-MazG showed that the RMSD values were 2.165 Å, 5.332 Å, and 13.52 Å, respectively (Figures 6D,F).
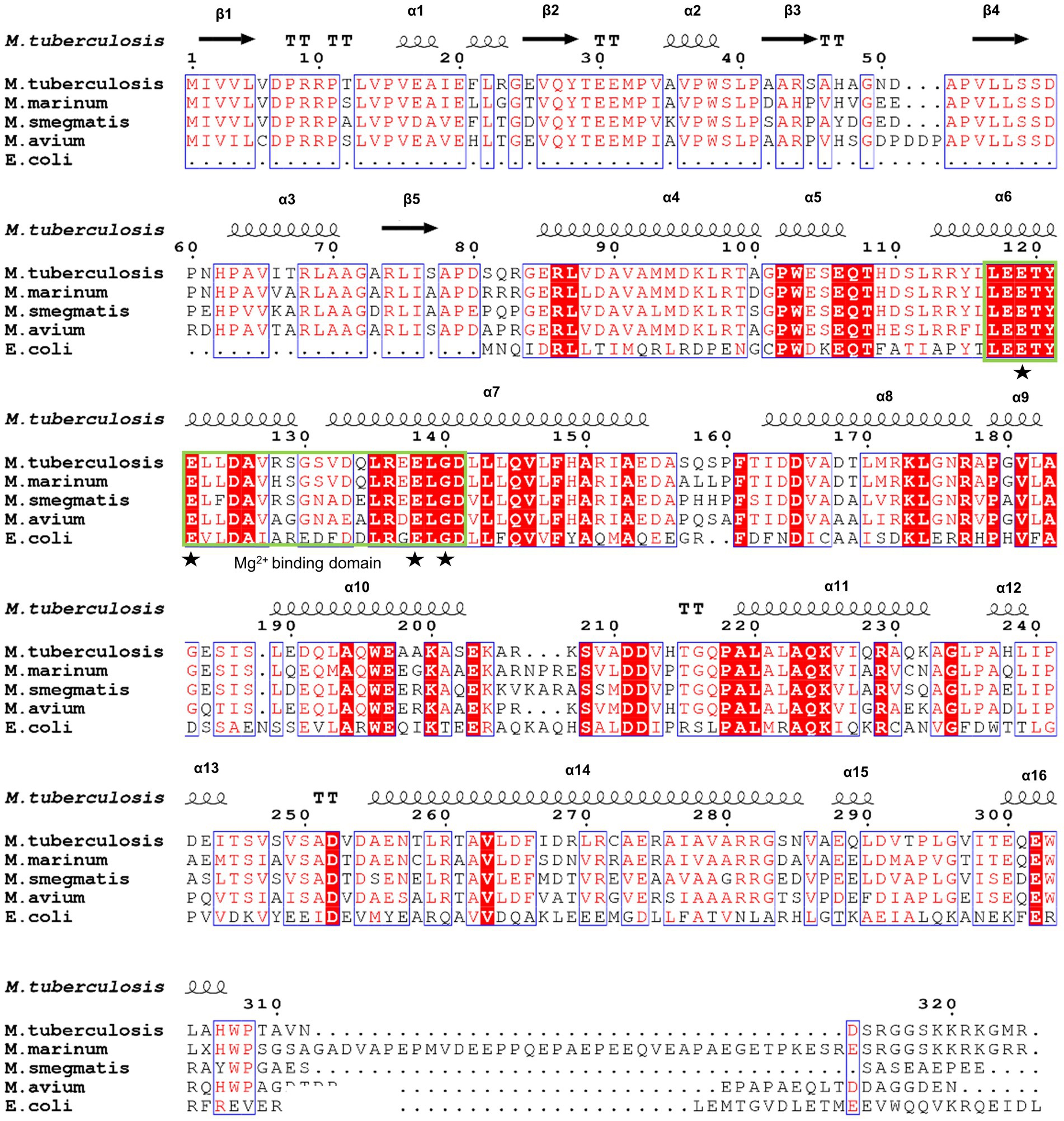
Figure 5. Sequence alignment of Mtb-MazG with its orthologs. Sequence alignment was performed using the ClustalX and ESpript v.3.0 programs. Identical and similar residues are shown in white text on a red background and in red text on a white background, respectively. The magnesium binding domain was highlighted with a green box. The star denotes residues that are bound with magnesium.
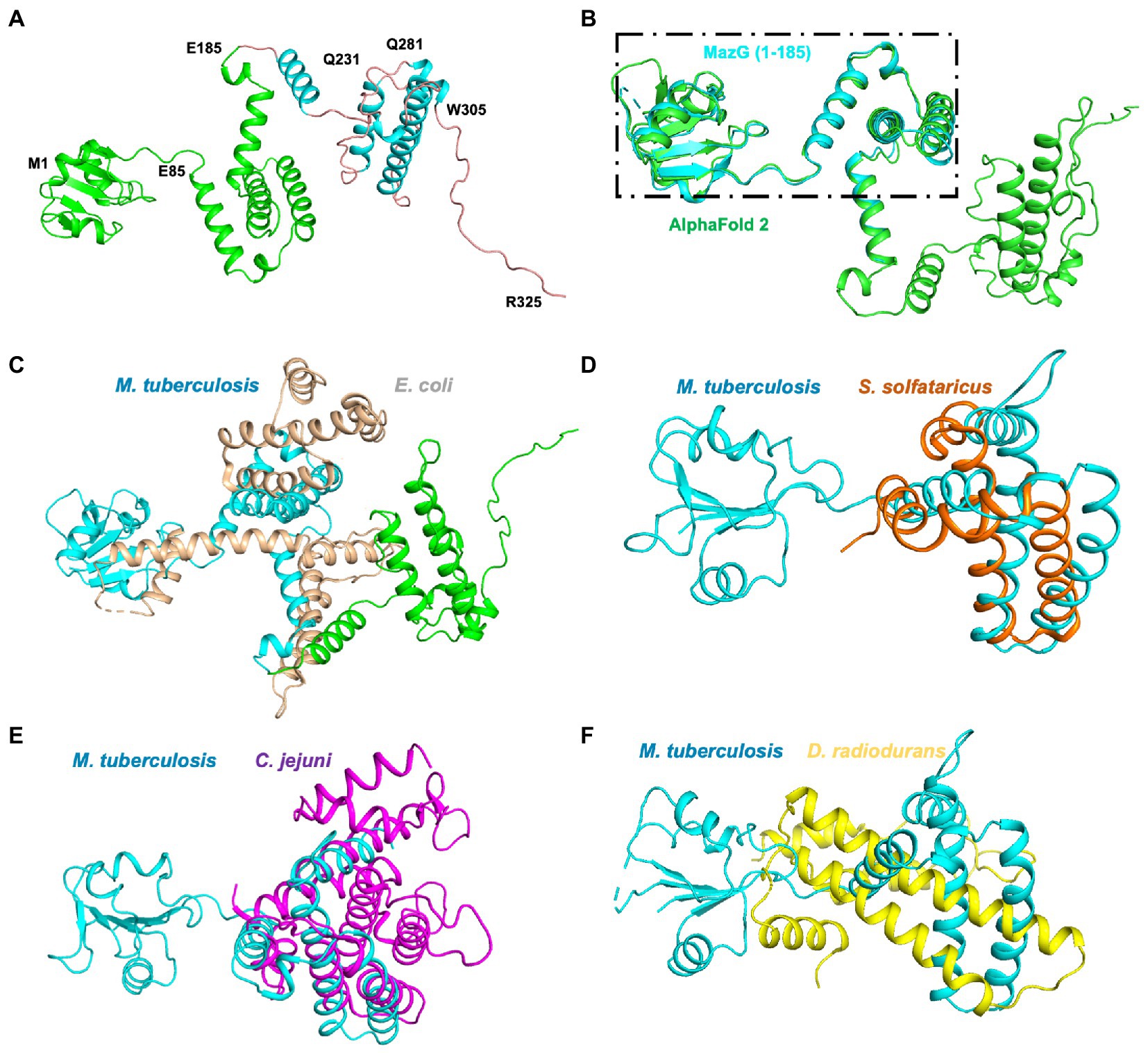
Figure 6. Structure superimposition of Mtb-MazG and its homologous proteins. (A) The AlphaFold2-predicted structure of MazG is shown in the cartoon model with different colors. The crystal structure of MazG (1–185) is shown in green color. Different numbers denote the truncated fragments. (B) The structural superposition of Mtb MazG (1–185; cyan) and AlphaFold2-predicted full-length MazG (green). (C) The AlphaFold2-predicted full-length MazG (cyan and green) was superimposed with E. coli MazG (PDB code 3CRC; wheat). (D–F) Structural superimposition of Mtb MazG (1–185; cyan) with S. solfataricus MazG (PDB code 1VMG; orange), C. jejuni MazG (PDB code 1W2Y; purple), and D. radiodurans MazG (PDB code 2YFD; yellow).
Discussion
MazG belongs to the all-α NTP pyrophosphatases (Galperin et al., 2006), which exist from viruses and bacteria to humans (Song et al., 2015; Rihtman et al., 2019; Zaide et al., 2020; Huang et al., 2021; Wood et al., 2021; Han et al., 2022). As a housecleaning enzyme, MazG functions at the cell nucleotide metabolism by degrading the non-canonical NTPs, preventing mutagenesis and DNA damage. Moreover, MazG helps bacteria in response to oxidative stress in E. coli (Han and Eiteman, 2018), B. anthracis (Zaide et al., 2020), and Mycobacterium (Shi et al., 2019). In addition, many studies show that MazG is critical in regulating the DNA damage response in mycobacterium (Lu et al., 2010; Lyu et al., 2013; Fan et al., 2018; Shi et al., 2019). Deletion of MazG in mycobacteria resulted in a 20-fold increase in the frequency of genomic CG-TA mutation both under oxidative stress and the stationary phase of growth (Fan et al., 2018). This suggests that MazG plays an important function in Mycobacterium tuberculosis infection.
In the current study, we solved the crystal structure of Mtb-MazG (1–185), which shows distinct overall architecture with other all-α NTP pyrophosphatases. The specific NE domain of Mtb-MazG comprises α-helices and β-stands (Figure 2), which is indispensable for its enzymatic activity (Figure 4). We found that the 1–85 sequence deletion significantly impacts MazG enzyme activity. The formation of dimeric MazG is related to multiple amino acid sites (Figure 2D). Therefore, we speculated that the deletion of the 1–85 sequence might have a negative impact on MazG dimeric formation, thus affecting the overall enzyme activity. The NTD domain is composed of α-helices and has a typical EEXX (E/D) motif, which forms the magnesium ion binding sites with four residues Glu119, Glu122, Glu138, and Asp141 (Figure 4). The four potential active site residues, the EEXX(E/D) motif, are frequently found in different enzymes, requiring magnesium or manganese ions for their activities (Peters and Croteau, 2002; Lee et al., 2008). Also, Campylobacter jejuni dUTPase coordinates a magnesium ion with acidic Glu46, Glu49, Glu74, and Asp77, and hydrolyzes the substrate using basic residues of Lys175, Arg182, and Lys194 (Moroz et al., 2004). The active site of EcMazG contains six conserved acidic residues (Glu171, Glu172, Glu175, Glu192, Glu193, and Asp196). However, only three (Glu172, Glu193, and Asp196) are involved in the magnesium coordination for NTPase activity. The NTD and CTD of Mtb-MazG show a high similarity with other bacterial homologs in amino acid sequences. All of them are composed of α-helices that play essential roles for (d)NTPs hydrolysis. However, the overall structures are quite different (Figure 6). Thus, the specific 3D structure of Mtb-MazG may contribute to the antibacterial lethality and coordinate the metabolic adaption of dormant Mtb. Further studies targeting MazG could benefit us in identifying potential compounds in the treatment of TB.
Materials and methods
Protein expression and purification
The gene encoding Mtb-MazG (NP_215537) of M.tuberculosis H37Rv was synthesized and constructed into the pSMT3 vector to produce the N-terminal 6× His-SUMO tagged fusion protein. The expression and purification processes were similar to previous works (Chen et al., 2020; Gao et al., 2021). In brief, Mtb-MazG was expressed in Escherichia coli BL21(DE3) cells. Cell cultures in LB medium were induced with 0.5 mM isopropyl-β-D-thiogalactoside (IPTG) at 16°C for 20 h when OD600 reached 0.6. Then, cells were suspended in buffer A (50 mM Tris–HCl, pH 8.0, 300 mM NaCl, 5% glycerol, 10 mM Imidazole) and lysed by a high-pressure homogenizer. Mtb-MazG was then purified by Ni-NTA affinity chromatography, and the His-sumo tag was removed by ULP1 enzyme cleavage, followed by an additional Ni-NTA affinity chromatography. The target protein was then applied to a Superdex 200 16/600 gel filtration column pre-equilibrated in buffer B (20 mM Tris–HCl, pH 8.0, 150 mM NaCl, 2 mM DTT). Finally, 10 ~ 20 mg per liter LB medium of the target protein with purity above 95% was obtained.
Site-directed mutants were constructed according to the standard QuikChange Site-Directed Mutagenesis protocol (Stratagene, United States) using the wild-type (WT) Mtb-MazG as the template. All the constructs were confirmed by DNA sequencing. The expression and purification of truncations and mutants were the same with the WT Mtb-MazG.
Limited proteolysis
The full-length MazG was incubated with different proteases (Hampton Research, Proti-Ace • Proti-Ace 2, HR2-432) at 37°C for 2 h in the reaction buffer (20 mM Tris–HCl, pH 8.0, 150 mM NaCl, 5 mM MgCl2). The reaction was stopped by heating at 95°C for 10 min. The ratio of enzyme to protein is 1:1000 (mol: mol).
Dynamic light scattering measurement
The Dynamic light scattering (DLS) data were collected on the DYNAMICS software from DynaPro NanoStar (Wyatt Technology), operating at a light source wavelength of 658 nm and a fixed scattering angle of 90°. The fresh proteins were diluted to 1 mg/mL with a buffer containing 20 mM Tris–HCl (pH 8.0), 150 mM NaCl, 2 mM DTT at 25°C.
Crystallization and data collection
Mtb-MazG (1–185) protein (~20 mg/mL) in a buffer with 20 mM Tris pH 8.0, 150 mM NaCl, 2 mM DTT, and 5 mM MgCl2 was crystallized at 18°C using the vapor-diffusion method by mixing with equal volume reservoir solution (0.1 M HEPES pH 7.5, 10% W/V PEG8000, 8% ethylene glycol). Crystals grew out at 18°C after 3 days. Diffraction data were collected with crystals flash-frozen in the crystallization buffer supplemented with 20% (v/v) glycerol. Integration, scaling, and merging of the diffraction data were performed using the HKL2000 suite.
Structure determination and refinement
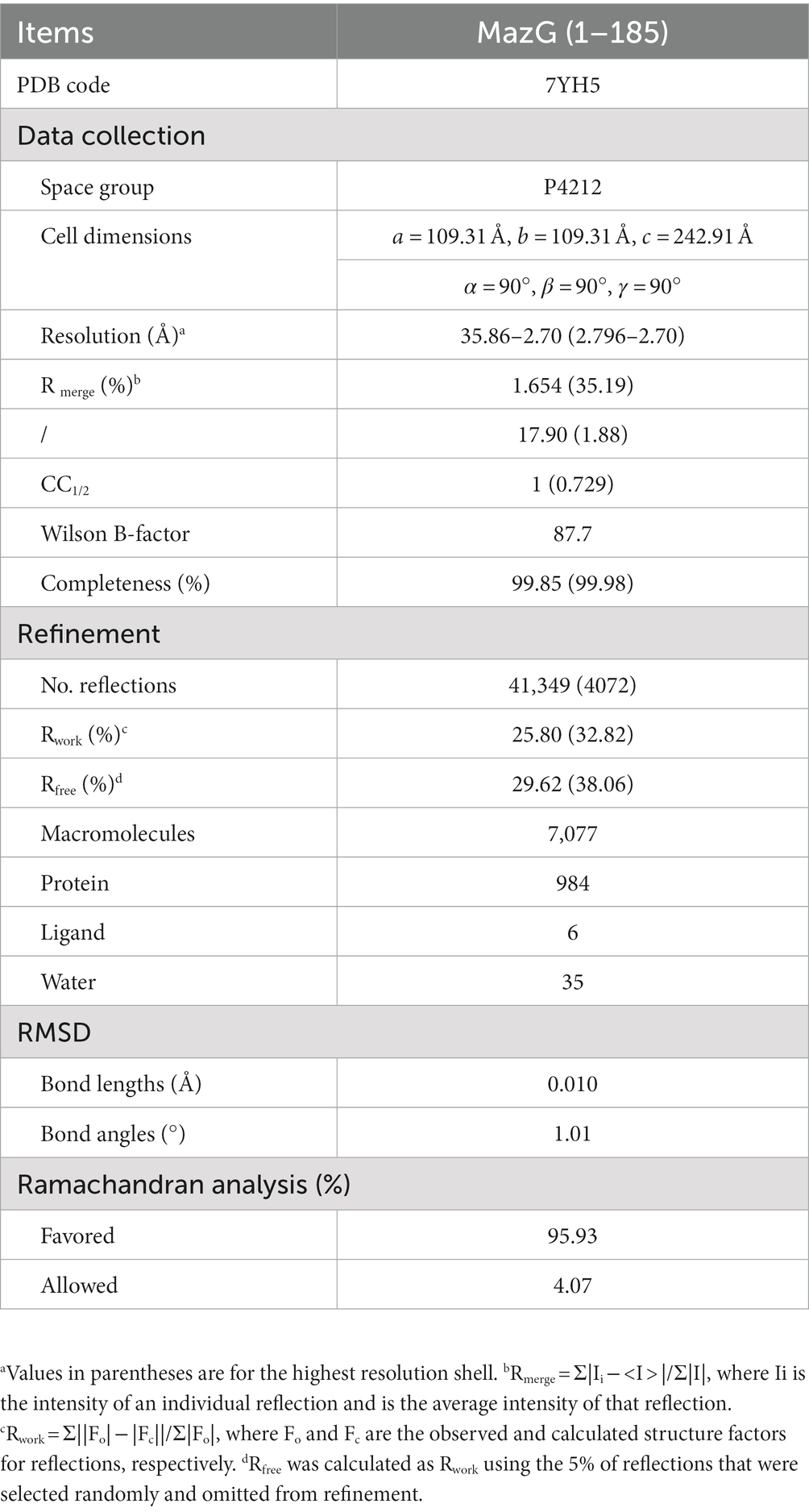
The Mtb-MazG structure was determined by the molecular replacement method using the structure (ID: AF-P96379-F1) predicted by AlphaFold2 as the search model (Tunyasuvunakool et al., 2021). Crystal structure refinements were performed with the program PHENIX (Adams et al., 2010). COOT and PyMOL software were used for model building and analysis (Emsley et al., 2010). The collected data and refinement statistics are summarized in Table 1.
Structure-based sequence alignment
Multiple alignments of amino acid sequences of different MazG proteins were performed using ClustalX v.2 program. Secondary structure alignment was generated by DSSP v.2.0 and ESpript v.3.0.1
Small-angle X-ray scattering analysis
The MazG Small-angle X-ray scattering (SAXS) data were collected at beamline BL19U2 of the Shanghai Synchrotron Radiation Facility with a radiation wavelength of 1.03 Å. The protein samples were prepared at concentrations of 1 mg/mL in 20 mM Tris–HCl (pH 8.0), 150 mM NaCl. Each blank or sample was measured in triplicate, and the sample measurements were adjusted by subtracting the scattering from the buffer alone. SAXS data analysis program ATSAS 2.84 suite was used to process and analyze the MazG scattering curve (Manalastas-Cantos et al., 2021). The radius of gyration (Rg) of globular, rod shape (Rc), and flat shape (Rf) were determined by applying the Guinier approximation equation in primusqt. GNOM software coupled with ATSAS was used for the evaluation of the maximum particle dimension (Dmax) and the distance distribution function plot (P(R)). The SAXSMow program was used to determine the molecular mass of MazG. DAMMIF program was used to construct 10 independent ab initio models. The software PyMol was used to show the model shape.
Enzymatic activity measurement
The NTP-PPase activity of Mtb-MazG was assayed by measuring the hydrolyzed product, PPi, by an enzyme-coupled colorimetric assay (Molecular Probes) with a detection limit of 0.2 μM. The standard NTP-PPase assay was carried out in a 20 μl reaction buffer (20 mM Tris–HCl, pH 8.0, 150 mM NaCl, 5 mM MgCl2) containing 1 μg of Mtb-MazG and an appropriate amount of nucleoside triphosphates at 37°C for 10 min. The reaction was stopped by heating at 70°C for 10 min, and 5 μl of the reaction product was applied for the PiPer pyrophosphate assay (Molecular Probes) according to the manufacturer’s instructions. The reaction without Mtb-MazG or substrates was carried out as a background control.
Circular dichroism spectrometry
The Circular dichroism (CD) spectra were measured on a Chirascan Plus spectropolarimeter in the far-ultraviolet region (260 nm-190 nm) in a step of 1 nm. Records on protein solutions (0.2 mg/mL in PBS) employing a cell with a path length of 1 mm at 18°C were obtained. Each spectrum reported is an average of three scans.
Statistical analysis
Each experiment was performed at least three times. All experiment data were analyzed using GraphPad Prism 7.0 (GraphPad software Inc. United States) and were presented as mean values ± SD. Statistical analysis was performed using a t-test (*: p < 0.05; **: p < 0.01; ***: p < 0.001).
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
JL and GZ conceived and supervised the study. SeW, BG, AC, ZZ, ShW, and LL performed experiments and data analysis. JL and SeW wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Key Research and Development Project of China (2021YFC2301500 and 2016YFA0500600), the National Natural Science Foundation of China (32161160323), and the Shanghai Committee of Science and Technology (20XD1400800).
Acknowledgments
We thank the staff of the Shanghai Synchrotron Radiation Facility beamlines BL17U1, BL18U, BL19U1, and BL19U2 for help with X-ray data and SAXS data collection.
Conflict of interest
ShW was employed by Shanghai Zelixir Biotech Company Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1137279/full#supplementary-material
Footnotes
References
Adams, P. D., Afonine, P. V., Bunkóczi, G., Chen, V. B., Davis, I. W., Echols, N., et al. (2010). PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221. doi: 10.1107/S0907444909052925
Chen, Z., Huang, J., and Li, J. (2022). Protein purification, crystallization, and structure determination of human DEAD-box RNA helicase DDX21 in different unwinding states. Star Protoc. 3:101642. doi: 10.1016/j.xpro.2022.101642
Chen, Z., Li, Z., Hu, X., Xie, F., Kuang, S., Zhan, B., et al. (2020). Structural basis of human helicase DDX21 in RNA binding, unwinding, and antiviral signal activation. Adv Sci (Weinh) 7:2000532. doi: 10.1002/advs.202000532
Dartois, V. A., and Rubin, E. J. (2022). Anti-tuberculosis treatment strategies and drug development: challenges and priorities. Nat. Rev. Microbiol. 20, 685–701. doi: 10.1038/s41579-022-00731-y
de Wet, T. J., Warner, D. F., and Mizrahi, V. (2019). Harnessing biological insight to accelerate tuberculosis drug discovery. Acc. Chem. Res. 52, 2340–2348. doi: 10.1021/acs.accounts.9b00275
Emsley, P., Lohkamp, B., Scott, W. G., and Cowtan, K. (2010). Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501. doi: 10.1107/S0907444910007493
Fan, X. Y., Tang, B. K., Xu, Y. Y., Han, A. X., Shi, K. X., Wu, Y. K., et al. (2018). Oxidation of dCTP contributes to antibiotic lethality in stationary-phase mycobacteria. Proc. Natl. Acad. Sci. U. S. A. 115, 2210–2215. doi: 10.1073/pnas.1719627115
Furin, J., Cox, H., and Pai, M. (2019). Tuberculosis. Lancet 393, 1642–1656. doi: 10.1016/S0140-6736(19)30308-3
Gad, H., Koolmeister, T., Jemth, A. S., Eshtad, S., Jacques, S. A., Ström, C. E., et al. (2014). MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool. Nature 508, 215–221. doi: 10.1038/nature13181
Galperin, M. Y., Moroz, O. V., Wilson, K. S., and Murzin, A. G. (2006). House cleaning, a part of good housekeeping. Mol. Microbiol. 59, 5–19. doi: 10.1111/j.1365-2958.2005.04950.x
Gao, B., Ji, R., Li, Z., Su, X., Li, H., Sun, Y., et al. (2021). Structural analysis and functional study of phosphofructokinase B (PfkB) from Mycobacterium marinum. Biochem. Biophys. Res. Commun. 579, 129–135. doi: 10.1016/j.bbrc.2021.09.051
Goncalves, A. M. D., de Sanctis, D., and McSweeney, S. M. (2011). Structural and functional insights into DR2231 protein, the MazG-like nucleoside triphosphate pyrophosphohydrolase from Deinococcus radiodurans. J. Biol. Chem. 286, 30691–30705. doi: 10.1074/jbc.M111.247999
Han, Q., and Eiteman, M. A. (2018). Enhancement of NAD(H) pool for formation of oxidized biochemicals in Escherichia coli. J. Ind. Microbiol. Biotechnol. 45, 939–950. doi: 10.1007/s10295-018-2072-y
Han, L. L., Yu, D. T., Bi, L., du, S., Silveira, C., Cobián Güemes, A. G., et al. (2022). Distribution of soil viruses across China and their potential role in phosphorous metabolism. Environ. Microbiome 17:6. doi: 10.1186/s40793-022-00401-9
Huang, X., Jiao, N., and Zhang, R. (2021). The genomic content and context of auxiliary metabolic genes in roseophages. Environ. Microbiol. 23, 3743–3757. doi: 10.1111/1462-2920.15412
Kim, M. I., and Hong, M. (2016). Crystal structure of the bacillus-conserved MazG protein, a nucleotide pyrophosphohydrolase. Biochem. Biophys. Res. Commun. 472, 237–242. doi: 10.1016/j.bbrc.2016.02.097
Lange, C., Aaby, P., Behr, M. A., Donald, P. R., Kaufmann, S. H. E., Netea, M. G., et al. (2022). 100 years of Mycobacterium bovis bacille Calmette-Guerin. Lancet Infect. Dis. 22, e2–e12. doi: 10.1016/S1473-3099(21)00403-5
Lee, S., Kim, M. H., Kang, B. S., Kim, J. S., Kim, G. H., Kim, Y. G., et al. (2008). Crystal structure of Escherichia coli MazG, the regulator of nutritional stress response. J. Biol. Chem. 283, 15232–15240. doi: 10.1074/jbc.M800479200
Lu, L. D., Sun, Q., Fan, X. Y., Zhong, Y., Yao, Y. F., and Zhao, G. P. (2010). Mycobacterial MazG is a novel NTP pyrophosphohydrolase involved in oxidative stress response. J. Biol. Chem. 285, 28076–28085. doi: 10.1074/jbc.M109.088872
Lyu, L. D., Tang, B. K., Fan, X. Y., Ma, H., and Zhao, G. P. (2013). Mycobacterial MazG safeguards genetic stability via housecleaning of 5-OH-dCTP. PLoS Pathog. 9:e1003814. doi: 10.1371/journal.ppat.1003814
Manalastas-Cantos, K., Konarev, P. V., Hajizadeh, N. R., Kikhney, A. G., Petoukhov, M. V., Molodenskiy, D. S., et al. (2021). ATSAS 3.0: expanded functionality and new tools for small-angle scattering data analysis. J. Appl. Crystallogr. 54, 343–355. doi: 10.1107/S1600576720013412
Moroz, O. V., Harkiolaki, M., Galperin, M. Y., Vagin, A. A., González-Pacanowska, D., and Wilson, K. S. (2004). The crystal structure of a complex of campylobacter jejuni dUTPase with substrate analogue sheds light on the mechanism and suggests the "basic module" for dimeric d(C/U)TPases. J. Mol. Biol. 342, 1583–1597. doi: 10.1016/j.jmb.2004.07.050
Moroz, O. V., Murzin, A. G., Makarova, K. S., Koonin, E. V., Wilson, K. S., and Galperin, M. Y. (2005). Dimeric dUTPases, HisE, and MazG belong to a new superfamily of all-alpha NTP pyrophosphohydrolases with potential "house-cleaning" functions. J. Mol. Biol. 347, 243–255. doi: 10.1016/j.jmb.2005.01.030
Mota, C. S., Goncalves, A. M., and de Sanctis, D. (2016). Deinococcus radiodurans DR2231 is a two-metal-ion mechanism hydrolase with exclusive activity on dUTP. FEBS J. 283, 4274–4290. doi: 10.1111/febs.13923
Peters, R. J., and Croteau, R. B. (2002). Abietadiene synthase catalysis: mutational analysis of a prenyl diphosphate ionization-initiated cyclization and rearrangement. Proc. Natl. Acad. Sci. U. S. A. 99, 580–584. doi: 10.1073/pnas.022627099
Rihtman, B., Bowman-Grahl, S., Millard, A., Corrigan, R. M., Clokie, M. R. J., and Scanlan, D. J. (2019). Cyanophage MazG is a pyrophosphohydrolase but unable to hydrolyse magic spot nucleotides. Environ. Microbiol. Rep. 11, 448–455. doi: 10.1111/1758-2229.12741
Shi, K. X., Wu, Y. K., Tang, B. K., Zhao, G. P., and Lyu, L. D. (2019). Housecleaning of pyrimidine nucleotide pool coordinates metabolic adaptation of nongrowing Mycobacterium tuberculosis. Emerg. Microbes Infect. 8, 40–44. doi: 10.1080/22221751.2018.1559706
Song, F. F., Xia, L. L., Ji, P., Tang, Y. B., Huang, Z. M., Zhu, L., et al. (2015). Human dCTP pyrophosphatase 1 promotes breast cancer cell growth and stemness through the modulation on 5-methyl-dCTP metabolism and global hypomethylation. Oncogenesis 4:e159. doi: 10.1038/oncsis.2015.10
Tunyasuvunakool, K., Adler, J., Wu, Z., Green, T., Zielinski, M., Žídek, A., et al. (2021). Highly accurate protein structure prediction for the human proteome. Nature 596, 590–596. doi: 10.1038/s41586-021-03828-1
Wood, W. N., Mohler, K., Rinehart, J., and Ibba, M. (2021). Deacylated tRNA accumulation is a trigger for bacterial antibiotic persistence independent of the stringent response. MBio 12:e0113221. doi: 10.1128/mBio.01132-21
Yang, L., Hu, X., Chai, X., Ye, Q., Pang, J., Li, D., et al. (2022). Opportunities for overcoming tuberculosis: emerging targets and their inhibitors. Drug Discov. Today 27, 326–336. doi: 10.1016/j.drudis.2021.09.003
Zaide, G., Elia, U., Cohen-Gihon, I., Israeli, M., Rotem, S., Israeli, O., et al. (2020). Comparative analysis of the global transcriptomic response to oxidative stress of bacillus anthracis htrA-disrupted and parental wild type strains. Microorganisms 8:896. doi: 10.3390/microorganisms8121896
Zhan, B., Gao, Y., Gao, W., Li, Y., Li, Z., Qi, Q., et al. (2022). Structural insights of the elongation factor EF-Tu complexes in protein translation of Mycobacterium tuberculosis. Commun. Biol. 5:1052. doi: 10.1038/s42003-022-04019-y
Keywords: Mycobacterium tuberculosis, MazG, NTP pyrophosphatases, crystal structure, SAXS
Citation: Wang S, Gao B, Chen A, Zhang Z, Wang S, Lv L, Zhao G and Li J (2023) Structural analysis of the housecleaning nucleoside triphosphate pyrophosphohydrolase MazG from Mycobacterium tuberculosis. Front. Microbiol. 14:1137279. doi: 10.3389/fmicb.2023.1137279
Edited by:
Alberto A. Iglesias, CONICET Coastline Agrobiotechnology Institute (IAL), ArgentinaReviewed by:
Cristiano Valim Bizarro, Pontifical Catholic University of Rio Grande do Sul, BrazilTom Desmet, Ghent University, Belgium
Copyright © 2023 Wang, Gao, Chen, Zhang, Wang, Lv, Zhao and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jixi Li, lijixi@fudan.edu.cn; Guoping Zhao, gpzhao@sibs.ac.cn
 Sen Wang
Sen Wang Baocai Gao1
Baocai Gao1 Liangdong Lv
Liangdong Lv Guoping Zhao
Guoping Zhao Jixi Li
Jixi Li