- 1School of Laboratory Medicine and Medical Sciences, College of Health Sciences, University of KwaZulu-Natal, Durban, South Africa
- 2Centre for the AIDS Programme of Research in South Africa (CAPRISA), University of KwaZulu-Natal, Durban, South Africa
- 3KwaZulu-Natal Research Innovation and Sequencing Platform (KRISP), School of Laboratory Medicine & Medical Sciences, University of KwaZulu-Natal, Durban, South Africa
With approximately 38 million people living with HIV/AIDS globally, and a further 1.5 million new global infections per year, it is imperative that we advance our understanding of all factors contributing to HIV infection. While most studies have focused on the influence of host genetic factors on HIV pathogenesis, epigenetic factors are gaining attention. Epigenetics involves alterations in gene expression without altering the DNA sequence. DNA methylation is a critical epigenetic mechanism that influences both viral and host factors. This review has five focal points, which examines (i) fluctuations in the expression of methylation modifying factors upon HIV infection (ii) the effect of DNA methylation on HIV viral genes and (iii) host genome (iv) inferences from other infectious and non-communicable diseases, we provide a list of HIV-associated host genes that are regulated by methylation in other disease models (v) the potential of DNA methylation as an epi-therapeutic strategy and biomarker. DNA methylation has also been shown to serve as a robust therapeutic strategy and precision medicine biomarker against diseases such as cancer and autoimmune conditions. Despite new drugs being discovered for HIV, drug resistance is a problem in high disease burden settings such as Sub-Saharan Africa. Furthermore, genetic therapies that are under investigation are irreversible and may have off target effects. Alternative therapies that are nongenetic are essential. In this review, we discuss the potential role of DNA methylation as a novel therapeutic intervention against HIV.
Introduction
In the nuclei of eukaryotes, the chromatin is subject to intense epigenetic events resulting in either condensed repressive heterochromatin or transcriptionally permissive euchromatin (1). These epigenetic events include posttranslational modifications to histones and methylation of DNA (1). DNA methylation involves the covalent addition of methyl groups to the fifth carbon in the nitrogenous base of cytosine (5mC) bases that are usually followed by guanine bases (CpG site) in DNA (2–5). Methylation of CpG sites found in the cis-regulatory regions of genes is generally associated with silencing genes (5–7). Methylation can also occur in intergenic regions, where it prevents the expression of potentially harmful genetic elements (4) as well as within the gene body, where a positive correlation with gene expression occurs (8–10).
DNA methylation is strongly involved in the physiological control of gene expression (4). It plays a key role in normal development (11), compaction of chromatin (12), genomic imprinting (13), X chromosome inactivation (14) and the bulk silencing of viral and transposable elements (15). However, aberrant methylation patterns are associated with a multitude of diseases [reviewed in (16–19)]. Several studies have shown that viral infections can induce aberrant methylation patterns within the host genome (20–22). On the other hand, the integrated proviral genome is also influenced by the epigenetic environment of the host (20, 23, 24). Thus virus-host interaction induces an altered epigenetic environment that affects both the virus and the infected host cell.
The human immune deficiency virus (HIV) is no exception to this phenomenon. The effect of HIV infection on DNA methylation has been characterised in HIV positive individuals (25). These effects have been associated with accelerated aging and abnormalities in gene expression, especially in immune regulating genes (25–30). Furthermore, methylation of HIV provirus by the host’s methylation machinery can control HIV-1 transcription, replication, and persistence (31–35).
We review the current literature on viral and human genes affected by methylation as well as address gaps in knowledge that are yet to be explored with regards to DNA methylation and HIV. This review will focus on five aspects: (i) the fluctuations of host DNA methylation modifying factors post HIV infection, (ii) the contribution of methylation on viral genes, (iii) the contribution of human genomic methylation on HIV disease, (iv) the influence of methylation on host genes observed in other diseases and models, and (v) the potential of DNA methylation as an epi-therapeutic strategy and precision medicine biomarker.
DNA Methylation Modifying Factors Post HIV Infection
DNA methylation is not a random event. Several proteins are involved in establishing, removing, and recognising methylation marks at specific CpG sites within the eukaryotic genome (4). DNA methylation is established by a family of DNA methyltransferases (DNMTs – DNMT1, DNMT3a and DNMT3b). DNMT1 is responsible for maintaining methylation patterns following DNA replication (36), while DNMT3a and DNMT3b regulate de novo methylation (37). Therefore, alternations in DNMT expression usually leads to changes in DNA methylation levels within cells. Previous studies have highlighted the increase in expression of DNMTs in HIV infected CD4+ T cells (38–41). HIV-1 was shown to induce the expression of DNMT1 in a non-specific tissue manner, and that overexpression of the viral genes: nef, tat and rev, induced DNMT1 promoter activity (40, 42, 43). In regulatory T cells, the effect of X4-tropic HIV infection demonstrated no significant change in the expression of DNMT1 and DNMT3a, while there was a substantial increase in expression of DNMT3b (41); however, increased expression of DNMT1, DNMT3a and DNMT3b was observed in CEM*174 T cells with significantly higher expression of DNMT3b (44). Similarly, HIV-1 replication enhanced DNMT3b levels in patients receiving antiretroviral therapy (ART) (45). The expression of DNMT3b was directly correlated to patient HIV viral load, while an inverse relation was observed for DNMT1 (45). Furthermore, proteomic analysis of primary oral epithelial cells revealed significantly lower DNMT1 and DNMT3a levels in HIV patients on ART. Additionally, DNMT activity and global DNA methylation illustrated a direct correlation (46).
The effect of HIV on DNMTs has incited interest in its effect on DNA demethylase enzymes. Conversion of the methyl group from 5-methyl-cytosine are mediated by a group of ten-eleven translocation methylcytosine dioxygenase (TET) enzymes to generate 5-hydroxymethyl-cytosine. 5-hydroxymethyl-cytosine can undergo further modifications such as deamination by apolipoprotein B mRNA Editing Catalytic Polypeptide-like (APOBEC) proteins. The expression of DNMT1 and TET1 was found to be increased in HIV-1 infected individuals without ART (47). Recently, the HIV-1 Vpr, which increases HIV-1 replication in macrophages, was shown to target TET2 for degradation, exacerbating HIV-1 infection (48, 49). The status of other TET enzymes (such as TET2 and TET3) has not been explored in an HIV setting.
Interestingly, recent studies have highlighted the importance of TET2 and TET3 for regulatory T cell stability and immune homeostasis (50). The loss of TET3 gene expression may be a pivotal contributor to locus hypermethylation (51). The effect of the TET family in an HIV setting is vastly unexplored; thus, the future investigation may unearth potential mechanisms of action, as seen in non-communicable diseases (52–54). However, much interest has been given to the cytidine deamination functioning of APOBEC (especially APOBEC3G and APOBEC3F). They have been shown to extensively deaminate viral cytosine to uracil resulting in the potent inhibition of HIV-1 infections (55, 56).
Another key multifunctional epigenetic regulator associated with HIV is methyl CpG-binding protein-2 (MeCP2), which recognizes methylated CpG sites and modulates transcription and chromatin structure (57, 58). The HIV gene tat is known to induce miR-132 expression, which subsequently down-regulates the expression of MeCP2 (59). However, Periyasamy et al. (60) discovered that the HIV-1 tat protein downregulated miR-124, which increased MeCP2 and its phosphorylated (Ser80) analogue in microglial cells. Interestingly, phosphorylated MeCP2 (Ser80) blocks miRNA biogenesis machinery, subsequently down regulating miR-124. These contradictory observations suggest that the effect of HIV-1 on host genes desires more attention (60).
DNA methylation is also known to be recognized by methyl-CpG binding domains (MBDs) and Ubiquitin Like with PHD and Ring Finger Domains 1 (UHRF1), which recruits DNA methylation modifying enzymes to chromatin (61, 62). Evidence from Kauder et al. (31) showed that HIV latency is regulated epigenetically via methylation of proviral DNA by DNMTs and its recognition by MBD2 (31). UHRF1 was also shown to facilitate latency as it was recruited to the HIV-1 5’LTR in a methylation/integration dependent fashion, where UHRF1 mediates the repression of HIV-1 gene expression (63).
Contribution of Methylation on Viral Genes
Methylation of both the HIV-1 proviral genome and host genome facilitates the integration, replication, and latency of HIV-1. The integration of proviral DNA into the host chromosome is not random as it is preferentially inserted into the euchromatin or actively transcribing regions of the host (64–66). Once integrated, it becomes indistinguishable from the host genome and exploits host cellular machinery for the transcription of its genes (67). However, this also puts proviral DNA at risk for epigenetic silencing events such as DNA methylation. In most cases, presence of methylation within the viral DNA, which has been integrated into the host genome, results in the reduction of new viral particles. In contrast, when integrated viral DNA is not methylated, viral transcription and viral production proceeds as usual (Figure 1).
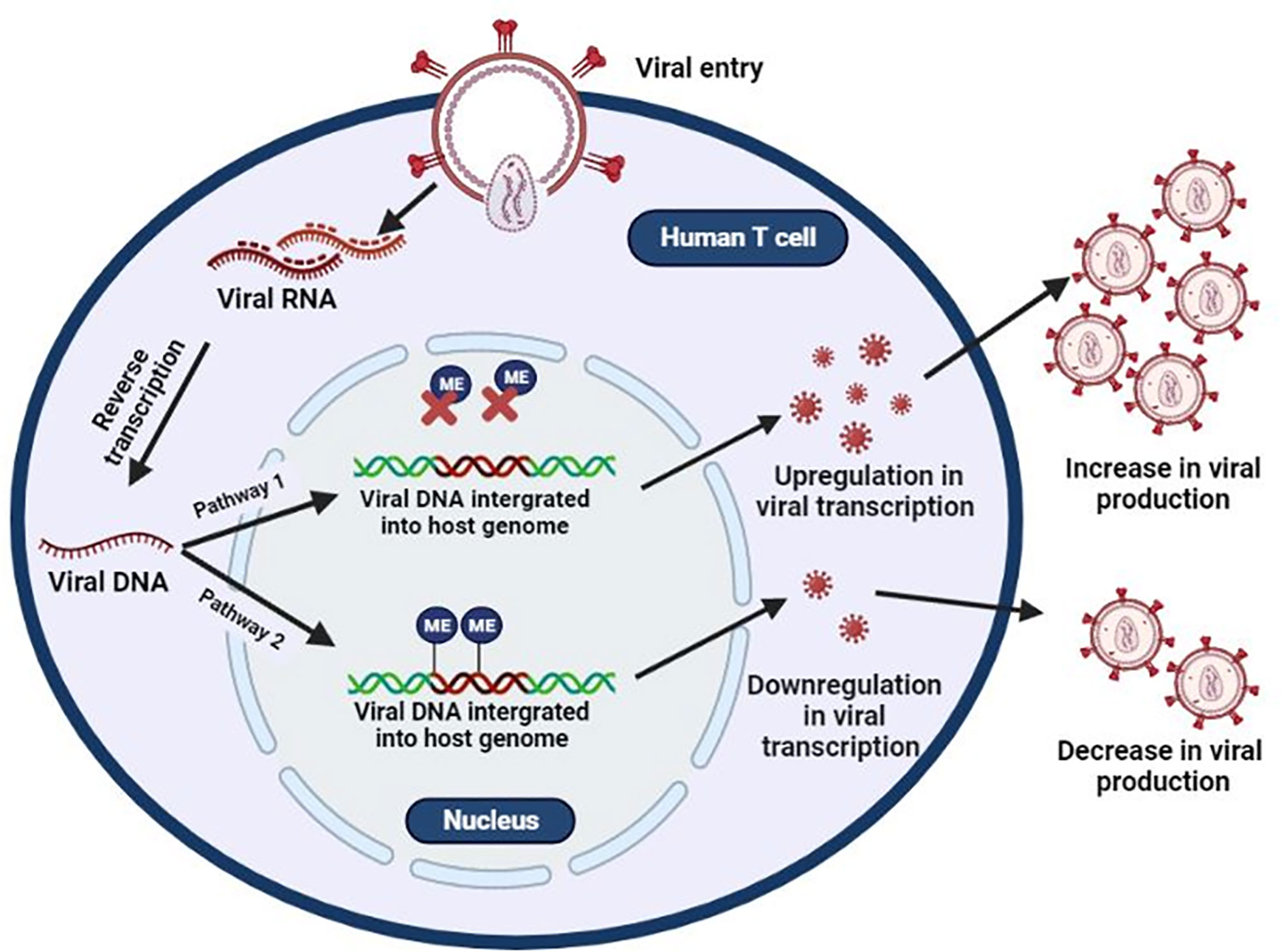
Figure 1 Epigenetic silencing of HIV transcription via methylation of integrated provirus. HIV binds to host receptors, entering the cell. HIV viral RNA is converted to single-stranded viral DNA and integrates into the host genome. HIV uses the host’s machinery to create new HIV copies (Pathway 1). However, methylation of integrated provirus results in the downregulation of HIV viral transcription, resulting in latency (Pathway 2).
The association of proviral methylation and the transcriptional inactivation of HIV-1 was introduced as early as 1990 (68, 69). Since then, several in vitro studies have reported that methylation of CpG sites found within the proximal proviral promoter, located in 5’ long terminal repeat (5’LTR), silences transcription of HIV-1 genes resulting in latency. This allows HIV to evade host immune responses and ART (31–35). However, in vivo analysis of methylation patterns in the 5’LTR with regards to latency is conflicting. High methylation patterns were found in the 5’ LTR of memory CD4+ T cells isolated from aviraemic HIV positive individuals on long term ART therapy (32). However, in a subsequent study, CpG sites were poorly methylated in resting CD4+ cells from HIV infected individuals (70). Trejbalova et al. (33) also observed low methylation levels in the 5’LTR of resting CD4+ cells isolated from individuals on effective ART; however, methylation levels appeared to increase with prolonged ART use (33). A comparison of methylation levels in the 5’LTR of long term non-progressors/elite controllers and virally suppressed individuals on ART found that methylation was virtually absent in individuals in the latter groups compared to the non-progressor/elite controller groups (71). These observations suggest that latency and ART-induced suppression might have different methylation patterns. The apparent difference in methylation patterns between in vivo and in vitro studies can be attributed to the pressure of “natural” selection in HIV-1-infected individuals. In contrast, under in vitro cell culture conditions, HIV-1 proviral genomes are not subject to the selective pressure exerted by host immune defence (72).
5’LTR methylation levels were also shown to associate with the expression of HIV-1 genes. Decreasing levels of methylation in the 5’LTR corresponded with increasing expression of HIV-1 gag in HIV-1 infected spermatozoa. Furthermore, gag protein was expressed in 2 cell embryos transfected with infected spermatozoa suggesting that 5’LTR methylation regulates the expression of HIV-1 gag in the vertical transmission from sperm to embryo (73).
Regarding methylation patterns outside the LTR, Weber et al. (72) found that CpG sites remain in a predominantly unmethylated state in the 5’LTR, 3’LTR and portions of HIV-1 gag, env, nef and tat genes. They also observed slight variations in the methylation state of the HIV-1 genome in one long term non-progressor over 11 years, although viral load and CD4+ levels remained stable (72). A recent study examined methylation of intragenic regions of the proviral genome across four groups of HIV infected individuals [i.e. long term non-progressors, early combination ART (cART) treated, late cART treated and cART naïve, acutely infected] (74). As a whole, methylation of promoter regions was reduced in all four groups, while high levels of methylation were observed in the intragenic env region. In the ART naïve acutely infected group, a distinct increase in 5’LTR and a decrease in intragenic env methylation was observed (74). Taken together, these observations suggest that intragenic methylation could be a late event during infection as well as intragenic methylation was positively associated with CD4+ counts and viral loads (74).
It is important to take into consideration the high mutation rates of HIV (75). Based on levels of mononucleotide C and Gs, the frequency of CpG sites within the HIV-1 genome is much lower than one would expect. The methylation of viral CpG sites may result in the spontaneous deamination of cytosine to thymine which increases the mutation rate of HIV (76). Moreover, coding regions such as the env region are highly variable (75). It would be interesting if future studies would evaluate whether CpG sites within these regions are lost, retained, or gained over a period of time and whether these mutations are beneficial or harmful to the virus.
Thus far, all studies on HIV DNA methylation have focused only on CpG methylation; however, non-CpG methylation was reported in other retroviral infections (77, 78). The lack of appropriate techniques that include non-CpG methylation may be why it has not been evaluated, as most of the studies discussed used nested PCR-based methods that exclude most non-CpG methylation (79).
The variability in existing data may be due to several factors. For instance, the integrated HIV provirus is subject to its immediate chromatin environment; thus, different integration sites may influence methylation status accordingly (79). Several pitfalls arise from the amplification of HIV from bisulphite-converted DNA: (i) the high mutation rates of actively replicating HIV hinders designing PCR primers that can amplify all HIV targets, (ii) longer primers are needed for bisulphite converted DNA which can worsen the biased amplification of variable sequences, (iii) multiple rounds of amplification of multiple variants can introduce stochastic bias and variable results are obtained from different methods even when the same conditions are applied (79). There is a significant need for an approach in which HIV amplification of the provirus is reproducible across different primer sets and experiments. Furthermore, attempts to establish and measure latency are unconvincing. It has previously been shown that cell lines harbouring viruses are not genuinely latent but are instead in an incapacitated state (80). Thus, in vitro studies are not an accurate measurement of methylation or latency. The development of appropriate methods for specific assessment of the replication-competent HIV reservoir in clinical samples and techniques of studying DNA methylation in the context of HIV may be helpful. Furthermore, the examination of non-CpG methylation of the provirus should be undertaken (79).
The Contribution Human Genomic Methylation on HIV Pathogenesis
While DNA methylation may influence the replication of HIV and transcription of crucial HIV genes, the integration of HIV-1 DNA into the host genome is also associated with aberrant methylation of host genes. Altered DNA methylation across the host genome has been shown to contribute to HIV disease. Previous studies have identified this via two different mechanisms. The first mechanism is a non-hypothesis driven approach which characterizes epigenome-wide methylation patterns. The second method is a hypothesis driven approach which measures methylation of specific/candidate genes. We will discuss each approach more thoroughly in the two sections which follows.
Assessing the Epigenome-Wide Methylation Patterns of the Host
Given that methylation of specific CpG sites found in either the promoter or gene body may impact gene expression, the use of epigenome-wide characterisation of DNA methylation provides a powerful approach in identifying epigenetic variations associated with disease acquisition, severity, and predictive outcomes (81, 82). Several high-throughput methods have been established for the genome-wide profiling of methylation at single-nucleotide resolution. These methods usually require the treatment of genomic DNA with sodium bisulphite, which deaminates unmethylated cysteine residues to uracil, leaving methylated cysteine residues unaffected (83). The most used techniques include whole genome-wide bisulphite sequencing and microarrays. Whole genome-wide bisulphite sequencing involves PCR amplification of bisulphite converted DNA coupled with next-generation sequencing, which allows for the methylation profiling of every cytosine in the genome (84). Methylation arrays such as Illumina’s Infinium arrays involves amplifying bisulphite converted DNA followed by its hybridisation to arrays containing probes that distinguish methylated and unmethylated cytosine and covers CpG islands, shores, and shelves (85). Other methods include methylated DNA immunoprecipitation, comprehensive high-throughput arrays for relative methylation and reduced-representation bisulphite sequencing. Most of the studies pertaining to genome-wide methylation profiling in HIV infected hosts use either methylation arrays or whole genome-wide bisulphite sequencing.
The first large scale study to characterise altered DNA methylation patterns of the host genome associated with HIV infection was conducted on DNA extracted from whole blood collected from 261 HIV infected and 117 uninfected individuals (30). The epigenome-wide association study (EWAS) identified 20 CpG sites to be significantly associated with HIV infection. Among them, 14 CpG sites were found to be hypomethylated, and six were found hypermethylated in HIV-infected individuals. These 20 CpG sites that were significantly associated with HIV infection were found within genes involved in immune activation (30). The most significant was 2 CpG sites located in the promoter region of NOD-like receptor family CARD domain containing 5 (NLRC5), an important transcriptional regulator of the Human Leucocyte Antigen (HLA) class-I genes and genes related to HLA class I antigen presentation and processing, such as TAP1 β2M, and LMP2 (86). Hypomethylation of the 2 CpG sites (cg16411857 and cg07839457) within the promoter region of the NLRC5 inversely correlated with viral load implying that DNA methylation of NLRC5 is associated with HIV disease outcome (30). In a recent study, similar results were observed in HIV infected and uninfected individuals who are injectable drug users during 6-month abstinence from drug injections. HIV infection was associated with 49 differentially methylated (DM) CpG sites. The top CpG sites identified were associated with immune and viral response pathways that are associated with HIV pathogenesis, with NRLC5 being the top-ranked gene associated with HIV status (87). Strong evidence of differential methylation within the MHC region (HLA-F, PSORS1C2, PSORS1C3 and Notch4) and NLRC5 region was also observed in children with perinatally acquired HIV. HIV was also shown to stunt B cell development and maturation via hypermethylation of EBF4, FOXP1 and DLL1 in perinatally infected children (29).
While studies on adult populations found that most DM CpG sites were hypomethylated in HIV infected individuals (30, 87), 97% of DM CpG sites tend to be hypermethylated in perinatally infected children. These differences suggest childhood acquisition of HIV alters the epigenome differently than acquisition as an adult (29). Differential methylation also occurs between perinatally infected and uninfected children (44, 88). Seeing as genetic and environmental factors influence the methylome, studies comparing the epigenetic profile of the general population is less than ideal. The use of discordant monozygotic twins with perfectly matched genetic profiles and similar lifestyles eliminates potential genetic confounders when unrelated individuals are used. Thus, variations in the methylome could be accurately attributed to exogenous factors such as viral infection (89). In a study conducted on a pair of 15-year-old monozygotic twins with discordant HIV statuses, significantly higher levels of methylated differentially methylated regions (DMRs) were observed in the infected twin compared to the uninfected sibling, further suggesting that HIV infection would cause the increase of global methylation level in perinatally infected children (44, 88). DMRs were located in chromosomes 17, 19 and 22, which are known HIV integration sites as they contain actively transcribing genes (44, 90, 91). It is possible that hypermethylation of regions in these chromosomes may be a mechanism employed by the host to suppress viral propagation. Twenty-five hyper-methylated genes in the HIV infected twin were validated at the transcriptional level. The expression of 72% of genes were downregulated by more than 50% in the HIV infected twin with IGFBP6 and SATB2 being the most significantly reduced genes. However, information on the role of IGFBP6 and SATB2 in HIV pathogenesis is limited (44). The use of HIV discordant monozygotic twins by Zhang et al. (44, 88) was an admirable attempt to account for the influence of genetic factors; however, it failed to account for environmental effects (44, 88). Further, only a single pair of twins were used in the study and the twins were recruited seven years after the acquisition of HIV infection. Thus, methylation changes cannot be used to distinguish between cause and consequence (44, 88).
While most studies have focused on variations in global DNA methylation among uninfected and infected individuals, the disparity has also been established in individuals with variable levels of HIV-1 viral load. Oriol-Tordera et al. (92) evaluated host genome methylation patterns of chronically HIV-1 infected individuals with high (>50,000 HIV-1-RNA copies/ml) and low (<10,000 HIV-1-RNA copies/ml) viral loads. Fifty-five DMRs were found to differentiate individuals with high viral load from those with low viral loads (92). Functional analysis showed genes involved in anti-viral activity and type I interferon γ (IFNγ) signalling to be hypermethylated in HIV infected individuals with low viral loads. Of particular interest, DMRs associated with IFNγ signalling included: PARP9/DTX3L, MX1, USP18, IFI44L and PLSCR1. In contrast, genes involved in general immune activation, such as T cell activation and differentiation, were found to be hypomethylated compared to individuals with a high HIV viral load (92). Thus, the epigenetic repression of IFNγ stimulating genes may assist in achieving control of HIV.
The studies described thus far provide valuable information on the association of aberrant methylation patterns and HIV infection at an epigenome-wide level; however, the use of whole blood, which consists of various cell types, has been used in these studies tend to be problematic. DNA methylation profiles differ strongly by cell type; therefore, variations in cell-type composition and proportions between samples can confound analysis (93). Furthermore, HIV mainly affects CD4 T cells which represents a small proportion of the tissue sampled; thus, the variation may not be detected. HIV further destroys CD4+ T cells levels; hence, measured epigenetic differences between cases and controls may only reflect differences in cell type composition and not true epigenetic differences (94). The use of homogeneous cell populations may provide a more accurate estimation of epigenome-wide methylation patterns and associated differential gene expression profiles between HIV infected and uninfected cells. CD4+ T lymphocytes are significant targets of HIV, with their progressive death culminating in acquired immune deficiency syndrome (AIDS). The use of the DNMT inhibitor, 5-azacytidine (5-azaC), can reverse T cell depletion, suggesting that DNA methylation may impact T cell apoptosis during HIV infection (95). Zeng et al. (96) transfected two T-cell lines (MT-2 and Jurkat cells lines) with the T-cell-tropic HIV strain, HIV-1 pNL4-3. Whole-genome methylation analysis found 1,428 hypermethylated and 1,227 hypomethylated DMRs in HIV infected MT-2 cell line compared with the uninfected controls as well as 1,231 hypermethylated and 1,833 hypomethylated DMRs in HIV infected Jurkat cells compared to uninfected control cells (96). Hypermethylated DMRs were significantly enriched in promoter and enhancer regions, suggesting that methylation changes are prone to occur in coding and transcriptional regulatory regions during HIV-1 infection (96). Hypomethylation of DMRs in 147 transcription factor binding motifs occurred in HIV infected Jurkat cells, 94 of which overlapped with the hypomethylated DMRs in the MT-2 cell line (96). HIV infected MT-2 cell lines, and Jurkat cell lines contained 83 and 53 transcription factor binding motifs found in hypermethylated DMRs. In the MT-2 cell line, five hypermethylated transcription factor binding motifs (WT1, HIF1A, EGR1, IRF1, and MEF2C) were associated with transcription factors that have been previously associated in HIV-1 induced apoptosis (96). These results suggest that the depletion of T cells during HIV infection results from aberrant DNA methylation at the binding sites of apoptosis-related transcription factors (96). Differences in epigenome-wide methylation were observed in CD4+ T cells isolated from individuals with varying degrees of control, suggesting that methylation status differs according to the progression of diseases state and control of infection. Furthermore, hypermethylation of TNF was characteristic in viremic individuals while TRIM69 and ITTGB2 were found to be hypomethylated in elite controllers (97). While the use of a homogenous in vitro models may provide more accurate methylation patterns, in vitro studies are not accurate representation of cells systems and are unable to account for ethnic differences.
Epigenome-wide characterisation reveals that global hypomethylation is prominent in HIV infected adults (30, 87), whereas global hypermethylation is prominent in HIV infected children compared to uninfected children (29, 88). Top hits include genes associated with anti-viral responses, immune defence, immune cell development and apoptosis (29, 30, 87, 96). However, the use of PBMCs and the comparison between unrelated, unmatched infected and uninfected individuals confounds results and thus, it is imperative to account for these factors. More studies should evaluate epigenetic events in monozygotic twins with discordant statuses, or a more desirable approach would be the longitudinal analysis of individuals pre- and post-HIV infection.
Candidate Host Gene Methylation
While EWAS characterisation provides a holistic view of methylation patterns during HIV infection, it is not feasible. Thus, many researchers opt for a targeted approach by analysing the epigenetic regulation of specific genes. The four most common techniques used to determine the methylation status of specific CpG sites includes: (i) methylation-specific restriction endonucleases (MSRE) followed by qPCR using primers surrounding the sequence of interest, (ii) pyrosequencing, (iii) methylation-specific high-resolution DNA melting analysis and (iv) quantitative methylation-specific polymerase chain reaction (98). Several studies have investigated the effect of HIV infection on specific HIV associated genes.
The surface expression of C-C chemokine receptor type 5 (CCR5) influences HIV-1 acquisition and disease progression by facilitating HIV-1 viral entry into T cells (99, 100). A common determinant of CCR5 expression is specific polymorphisms in open reading frames and cis-regulatory regions of CCR5 (101). One such polymorphism is a 32 base pair deletion in the open reading frame of CCR5 (CCR5-Δ32). Individuals homozygous for the CCR5-Δ32 mutation cannot produce complete CCR5 proteins; thus, their T cells surface is devoid of the receptor, providing them with protection against HIV (102, 103). However, polymorphisms do not account for the variation in CCR5 expression between subsets of T cells and altered expression upon T cell activation (104–106). In vivo and ex vivo analysis by Gornalusse et al. (107) showed that methylation levels within the CCR5 gene might account for these variations (107). Sorted T cells with higher methylation content within the cis-region of CCR-5 correlated with low CCR5 surface levels. CpG sites in the regulatory region of CCR5 were mostly methylated in naïve T cells, whereas hypomethylation was prevalent in memory T cells (107). In vitro activation of naïve T cells was associated with demethylation of CCR5 and concomitant increase in CCR5 expression. These results were confirmed in a cohort of individuals with primary HIV infection and two cohorts of individuals with untreated chronic infection. However, viral load suppression during ART was associated with increased methylation in CCR5-cis regions and low CCR5 levels during primary infection (107). Furthermore, the authors demonstrated that specific CCR5 haplotypes contain polymorphism, which may remove CpG sites, resulting in cis-regions resistant to undergoing activation-induced demethylation and are thus constitutively expressed. Therefore, CCR5 surface levels and HIV susceptibility depend on both genetic and epigenetic mechanisms (107).
Genetic variations in the HLA region are known to influence host control of HIV infection (108, 109). HLA molecules present intracellularly derived peptides to immune cells, which elicits immune response upon recognising pathogenic peptides (110). Several previously discussed EWAS have identified differential methylation within the HLA loci in HIV positive individuals (29, 86). The elevated levels of the class I HLA-A molecules are associated with higher HIV viral load and poor HIV control. In contrast, low expression of HLA-A is associated with improved control of viremia and slower progression to AIDS (111). Methylation of the HLA-A promoter results in the reduced expression of HLA-A (112). Moreover, allelic lineage-specific methylation patterns within the HLA-A promoter region are inversely related to HLA expression. Increased DNA methylation levels correlated significantly with reduced HLA-A expression levels (112). Gross et al. (26) found that an entire HLA locus had notably reduced methylation levels in HIV infected individuals compared to uninfected individuals (26). Furthermore, several differentially methylated markers were found surrounding a single nucleotide polymorphism (SNP), rs2395029, within the HLA region (26). This variant is predictive for the presence of HLA-B*5701 and is common in HIV positive non-progressors. Further examination of this locus in neutrophils and CD4+ T cells found that the gene body of HLA Complex P5 (HCP5) was differentially methylated in neutrophils, and the methylation level of HCP5 correlated with CD4+:CD8+ T cell ratio (26). Thus, methylation dynamics plays a critical role in HIV control through its regulation of the HLA system (26, 111, 112).
A specialised subset of CD4 T lymphocytes known as regulatory T cells or Tregs plays an essential role in suppressing hyperactive immune responses that may occur during the course of HIV infection (113). However, Tregs are also susceptible to HIV infection as they contain receptors that participate in viral entry (114, 115). The maintenance of Treg functioning is heavily dependent on the surface expression of Forkhead Box Protein 3 (FOXP3) (116). In vitro transfection of Tregs with HIV-1 was shown to impair Treg functioning through the methylation of CpG sites found in FOX3P regulatory regions (41). However, in vivo analysis of FOXP3 promoters from Tregs isolated from PBMCs and colon mucosa of chronic HIV infected patients was demethylated, resulting in the increased expression of FOX3P (117). In both studies, FOX3P promoter methylation was associated with altered levels of DNA methylation regulating enzymes (41). High levels of DNMT3B were associated with the elevated methylation in the in vitro study while a significant reduction in DNMT1, DMAP1, METTL7B, and METTL1 was responsible for the reduced methylation in the in vivo study (117).
DNMTs were also shown to influence interferon-gamma (IFNγ) levels (38). INFγ, a cytokine produced by type 1 T helper cells, CD8+ cytotoxic T cells and natural killer cells, facilitates inflammation and regulates antigen presentation and macrophage differentiation upon viral infections (80). High levels of DNMTs in HIV infected T helper cells were shown to induce methylation at the SnaBI site in INFγ promoters resulting in low levels on IFNγ (38) The aberrant expression is due to methylation silencing and may play a role in the gradual loss of type 1 helper cell response seen in AIDS patients.
HIV positive women have an increased risk of developing cervical cancer and precursor lesions [cervical intraepithelial neoplasia (CIN)] (118–120). Hypermethylation and subsequent silencing of tumour suppressor genes result in gene silencing and represents an essential step for cervical cancer development (121, 122). Methylation levels of the tumour suppressor EPB41L3 were significantly higher in HIV seropositive women with moderate grade neoplasia compared to HIV seronegative women (123). Methylation levels of microRNA-124–2 (miR-124–2), was significantly associated with HIV positive women with low, moderate and severe grade neoplasia compared to HIV negative women (124); however, no association was found between the methylation content of the tumour suppressor genes CADM1, MAL RARB, DAPK1 and PAX1 in HIV (124, 125). The methylation of ASCL1, LHX8 and ST6GALNAC5 was significantly higher in HIV seropositive women with low to moderate grade neoplasia than HIV seronegative women. However, methylation levels were comparable between HIV seropositive and HIV seronegative women with high-grade neoplasia (126).
Most recently, Singh et al. (127) found that methylation levels within the gene promoter of the host anti-viral restriction factor, bone marrow stromal cell antigen 2 (BST2 or tetherin) was associated with BST2 expression and HIV disease state. Methylation levels were significantly elevated in all nine CpG sites within HIV infected individuals compared to the uninfected group. Within the HIV positive group, CpG promoter methylation of BST2 was further evaluated across four different time points (pre-infection, 3-months. 12-months and 36-months post-infection). An inverse correlation between BST2 methylation and expression was observed at all time points. Furthermore, in an in vitro HIV replication assay, treatment with the DNA hypomethylation drug, 5’-Aza-CdR corresponded with an increased expression of BST2 and lower viral load, suggesting that controlling regulation may be an important strategy in controlling HIV infection (127).
While DNA methylation is an epigenetic modification, candidate gene methylation may be influenced by variations in the DNA sequence. Several studies have mapped the interactions between genetic differences and variations in DNA methylation across numerous tissue and cell types (128–131). The methylation quantitative trait loci showed that up to 48% of inter-individual variation in DNA methylation was related to CpG sites that were associated with nearby single nucleotide polymorphisms (SNPs) found in cis regulatory regions (132, 133). SNPs located near or in CpG sites found in the promoter region of genes can either produce or remove CpG site methylation, leading to an alteration in the expression of the genes (Figure 2A). DNA methylation can also differ among alleles of a given gene. This is referred to allele-specific methylation (Figure 2B). For example, the promoter region of HLA-A*24 (highest HLA-A expressing lineage) and -A*03 (lowest HLA-A expressing linage) contain a similar number of CpG sites; however, only one CpG site was found methylated in the promoter of the HLA-A*24 lineage, while most CpG sites were found to be methylated in the HLA-A*03 linage (112). The influence of genetic variation on promoter methylation of specific host genes in relation to HIV pathogenesis has yet to be investigated. As discussed in this section, we, however, do know that increased promoter methylation generally lowers mRNA expression of specific genes affecting HIV disease progression (Figure 2C).
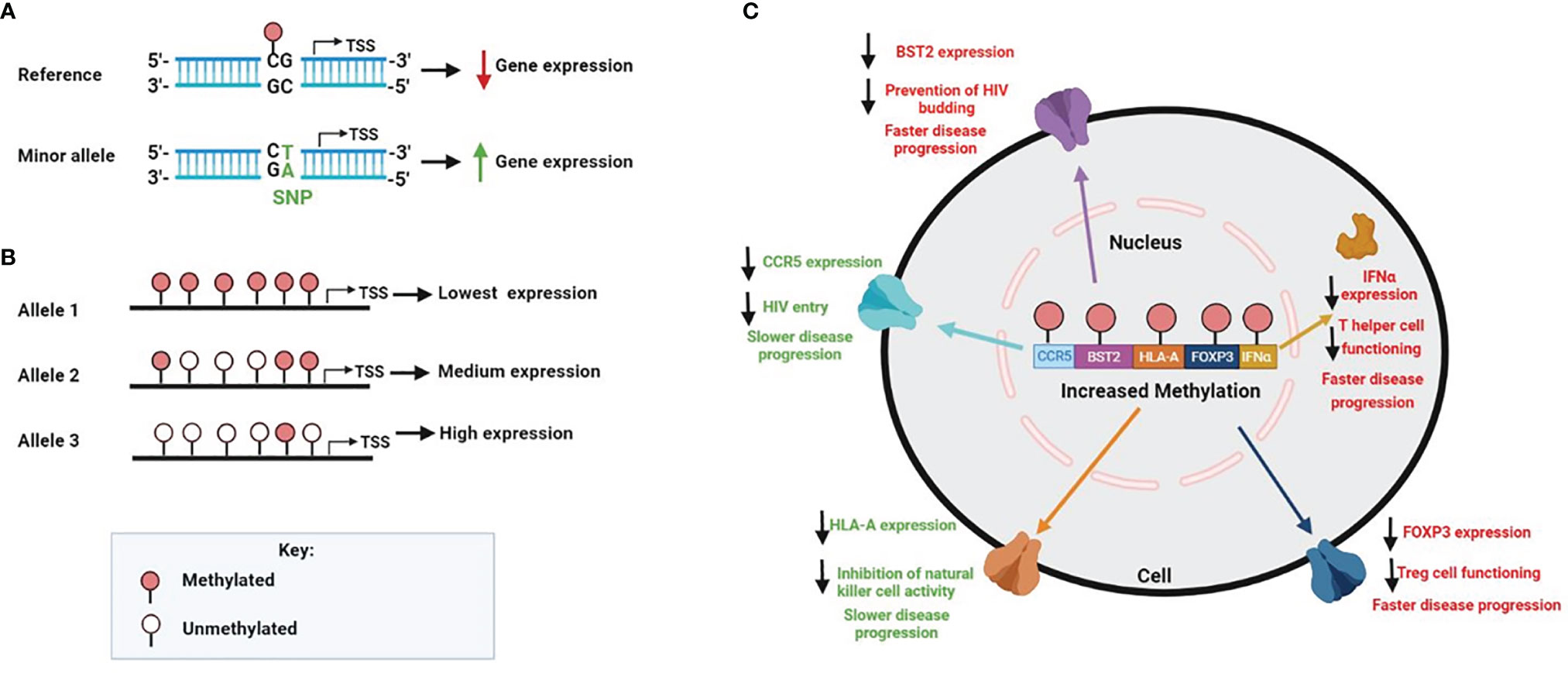
Figure 2 Factors that contribute to human genomic methylation on HIV disease. (A) SNPs found in regulatory gene regions can create or abolish CPG sites, which in turn may affect methylation and gene expression. In the genomic sequence, if C is followed by a G, the C can be methylated; however, when the SNP is mutated from a G to a T, it removes the CpG site and methylation cannot occur (B) DNA methylation can also differ among alleles of a given gene. The number of methylated CpG sites on each allele can affect expression accordingly (C) Increased promoter methylation generally results in decreased mRNA gene expression, which is specifically observed within the HIV setting for the following genes CCR5, BST2, HLA-A, FOX3P and IFNα. Since expression variability of these genes are directly linked to HIV pathogenesis, a change in methylation levels have shown to alter HIV disease progression. Higher levels of methylation for CCR5 and HLA-A results in slower HIV disease progression, however, higher levels of methylation for BST2, FOX3P and IFNα results in faster disease progression.
Methylation Controlled Host Genes Observed in Other Diseases and Models
With only a few studies evaluating the influence of HIV on the methylation of specific host genes, further examination is essential (26, 38, 41, 107, 117, 123, 125, 126). Henceforth, we discuss potential host genes whose methylation status should be investigated with regard to HIV. These genes have been previously shown to associate with HIV disease and were shown to be controlled by DNA methylation in conditions other than HIV. Based on the principle that these genes have been regulated by DNA methylation for a particular disease association, we assume that they may also be regulated similarly in an HIV setting.
For instance, the co-receptor C-X-C chemokine receptor type 4 (CXCR4), like CCR5, mediates the entry of HIV into host cells. Low surface expression of CXCR4 confers with reduced viral entry, while increased expression is associated with the elevated viral entry. Therefore alternations of CXCR4 expression has a significant influence on HIV progression (134, 135). DNA methylation has been shown to regulate CXCR4 expression in pancreatic cancer (136), sporadic breast cancer (137), and primary myelofibrosis (138).
Another example is the host restriction factor, sterile alpha motif and histidine/aspartic acid domain-containing protein 1 (SAMHD1) which limits HIV reverse transcription by depleting the intracellular pool of deoxynucleotide triphosphates (139, 140). De Silva et al. (141) used CD4+ T cell lines as a model to identify mechanisms that regulate SAMHD1 gene expression. The results indicated that the SAMHD1 promoter contains a CpG island proximal to the initiation codon of the SAMHD1 gene, which, upon DNA methylation, leads to transcriptional repression in certain CD4+ T cell lines (142). Regarding disease association, reduced levels of SAMHD1 expression corresponded with SAMHD1 promoter methylation in lung cancer (143) and patients with Sezary syndrome (141).
The tumour suppressor, p53 and its downstream gene, p21, were shown to hinder early-stage replication of HIV-1 (144). p21, a cyclin dependant kinase, promotes cell cycle arrest by downregulating G1/S transition (144, 145). p21 is also shown to regulate SAMHD1 in HIV-1 infection (145). Epigenetic alterations, including promoter DNA methylation and histone deacetylation, have long been established as crucial mechanisms of carcinogenesis (146–148). p53 promoter methylation leads to downregulation of p53 in several cancers (149–151). Loss of p21 has been shown to occur in colorectal cancer (152). Additionally, the p21 gene is frequently methylated and is an essential factor in predicting the clinical outcome of acute lymphoblastic leukaemia patients (153). The loss of p21 expression was commonly observed in lung cancer and malignant pleural mesothelioma, and aberrant methylation was one of the mechanisms of suppression of p21 (154).
Methylation of several other host factors such as CCR2, CCL2, CXCR6, CCL5, TSG101, PD-1, PD-L1, TIM3, LAG-3, CTLA-4, TRIM22, DC-SIGN (CD209), IL-10, IL-32, IRF1, Perforin, ICAM-1, and PCSK9 could potentially play a role in HIV disease. Table 1 provides a list of host factors associated with HIV pathogenesis which should be examined in future methylation studies. Although these disease-methylation associations have been shown in other diseases, it is yet to be proven in HIV disease. Based on the principle that these genes have been regulated by DNA methylation for a particular disease association, we assume that they may also be regulated similarly in an HIV setting. These listed genes may be potential host gene targets that may provide an alternative approach towards precision medicine or personalised therapeutic interventions against HIV and other diseases.

Table 1 HIV-associated host genes that are regulated by methylation in other diseases or in vitro models.
DNA Methylation: A Valuable Tool for Epi-Therapeutics and Precision Medicine
‘The Berlin patient’ and ‘the London patient’ were the first two individuals reportedly “cured” of HIV. They both received a stem cell transplant containing the CCR5 Δ-32 mutation to treat their leukaemia which consequentially eliminated the virus from their bodies (221, 222). Such cases provided proof that HIV-1 can be eradicated in those already living with the virus. Given that this approach is not feasible for most people living with HIV, other therapeutic strategies are essential. Furthermore, recent studies have shown that early treatment with ART, is ineffective against returning the altered DNA methylation profile of HIV positive individuals during acute infection (223). Therefore, there is a need for epigenetic strategies for the treatment of HIV.
Recently, Shrivastava et al. (224) developed a zinc finger protein (ZFP-362) that specifically targeted the HIV-1 promoter region. The ZFP-362 fuses to active domains of DNMT3A and induces a long-term stable epigenetic repression of HIV-1. This suppression was found to be driven by DNA methylation (224). Like ART, this intervention may repress viral transcription and control viral replication in HIV positive individuals; however, it is ineffective against latent HIV reservoirs. Thus, efforts have mainly been focused on targeting the latent HIV-1 reservoir responsible for viral persistence and strengthening immunological defences against HIV. Many researchers are adopting the “shock and kill” approach to targeting HIV. This strategy involves the forced reversal of HIV latency (shock) followed by the robust elimination of infected cells by viral or host immune-mediated cytolysis (kill). Therefore novel approaches for the development of latency-reversing agents (LRA) are needed (225). Much interest has been given to the development of epi-LRA – agents that disrupt latency by interfering with the epigenetic silencing mechanism of the 5’LTR (226). In the instance of methylation of 5’LTR, the use of DNMT inhibitors have been considered (31).
Bouchat et al. (227) found that the DNMT inhibitor, 5‐AzaC, combined with histone deacetylase inhibitors panobinostat or romidepsin, was potent in reducing HIV-1 latent reservoirs in ART-treated patients (227). The 5-AzaC analogue, 5-aza-2′ deoxycytidine (5‐AzadC), alone and in combination with TNFα and prostratin, significantly increased HIV gene expression through altered methylation levels (31, 227). Both 5-AzaC and 5-AzadC, commercially known as Vidaza® and Dacogen®, respectively, have been approved by the FDA to treat myelodysplastic syndrome and in phase II clinical trials for chronic myelomonocytic leukaemia (227–229). Treatment with either 5-AzaC or 5-AzadC was shown to increase the overall survival of patients with higher-risk myelodysplastic syndromes and prolong time to leukaemia transformation and death compared to conventional care regimens (230–232). According to clinicaltrials.gov, 389 clinical trials are actively investigating 5-AzaC and 5-AzadC as interventions for various cancers and conditions. These include: ependymoma, breast cancers, lymphomas, osteosarcoma, and pancreatic cancer, as well as other conditions such as immune thrombocytopenia, sickle cell disease, myelofibrosis, and COVID-19. Therefore, the inclusion of DNMT inhibitors with ART could represent a significant step towards the elimination of the latent HIV-1 reservoir and clearance of virus from infected patients.
Other novel technologies, such as Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR), have great potential in eradicating viral genomes from infected individuals by editing genes as well as the methylation levels associated with HIV. Ebina et al. (233) successfully excised the latently integrated provirus from the host genome and restricted transcriptionally active provirus using the CRISPR/Cas9 approach (233). CRISPR-Cas9 editing of the host genome has also been investigated as an intervention against HIV. Silencing of CCR5 and CXCR4 genes by CRISPR have already been shown as effective towards a functional cure for HIV-1 infection (234–236). While the conventional CRISPR approach may have revolutionised genetic therapies, it permanently switches off host genes and may have unwanted consequences such as off-target gene mutations (237, 238). Therefore, approaches that edit the epigenome rather than the genome may be a more suitable and safer strategy. CRISPR-based epigenome technologies involve the fusion of inactivated Cas9 (dCas9) with DNA methyltransferase or demethylase enzymes, allowing for manipulating methylation levels at specific CpG sites. Because this approach targets the epigenome and uses inactivated Cas9, it will enable reversible editing and prevents the formation of double-strand breaks (239–241). Therefore, this approach may be ideal in prospective studies that evaluate host gene regulation as a treatment strategy against HIV (240).
As the medical field rapidly moves towards precision medicine and theragnostic approaches, DNA methylation profiling can play a tremendous role in these strategies. DNA methylations can serve as biomarkers for diagnosis, prognosis, monitoring and predicting treatment response and disease outcome (242). Due to its dynamic and stable nature, it is more reliable and suitable than genetic and protein-based biomarkers. Methylation levels can be easily measured in circulating cell-free DNA, which is the preferable method in clinical settings as it is minimally invasive (243). Several DNA methylation-based in vitro diagnostic tests have been developed and commercialised for profiling DNA methylation (241). Tests may be specific for a disease such as Epi proColon® 2.0 CE, which detects methylated Septin9 to diagnose colon cancer and Bladder EpiCheck®, which measures changes in methylation of 15 genes associated with bladder cancer (244, 245). The utilisation of the EpiSign assay has been well established in clinical diagnostic laboratories and uses genome-wide methylation patterns to diagnose up to 42 rare neurodevelopmental Mendelian syndromes (246, 247). Many of the commercialised clinical DNA methylation assays implement practical and cost-effective assays such as qPCR and microarrays. The use of DNA methylation-based biomarkers for precision medicine has been extensively studied with regards to cancer; however, its application has great potential in other diseases, including HIV. For instance, DNA methylation has been shown to be a potentially effective prognostic biomarker for predicting risk and type of HIV-associated lymphomas and HIV associated cognitive impairment; however, these results are yet to be translated to a clinical setting (94, 248). There is still a lot to be investigated regarding the epigenetic signature of HIV for precision medicine. Future studies should focus on using well-characterised clinical cohorts to evaluate methylation profiling as a biomarker for predicting HIV disease course, development of HIV associated comorbidities, monitoring patient response to ARVs and personalised therapy.
The Epi-therapeutic interventions, either through LRA or CRISPR technologies and DNA methylation in precision medicine and theragnostics, provides a novel and powerful approach against HIV. However, there is much-needed research to be done to translate these approaches into a clinical setting.
Conclusion and Future Perspectives
Since the beginning of the HIV epidemic, the impact of host genetic variations on HIV susceptibility and disease outcomes has attracted a vast amount of attention, while epigenetic changes have long been neglected. This review provided a comprehensive overview of the intricate interplay between DNA methylation and viral and host genome. Once integrated, the HIV viral genome is subject to the intense epigenetic environment of the host genome. This includes silencing of HIV transcription via DNA methylation. Integration of the proviral genome also induces aberrant methylation of the host genome, influencing HIV disease progression. Several host genes involved in viral entry, anti-viral responses and immune defences are altered by DNA methylation in HIV infected individuals.
However, many of the studies discussed are limited by the study designs used. Many of the studies discussed failed to account for the influence of genetic and/or environmental factors on promoter methylation. Another drawback of most studies reviewed is the type of sample that was used. The type of sample selected for a study involving DNA methylation is crucial as methylation patterns differ substantially according to cell type (93). Studies using mixed cell samples such as whole blood or PBMCs need to account for cell type composition and variation in the methylation patterns of different cells. Some studies have tried to account for account for cell type heterogeneity by transfecting homogenous T cell lines (95–97). However in vitro studies are not accurate representation of cells systems and are unable to account for ethnic differences. Increased susceptibility to HIV and varying responses to ARVs have been noted amongst different ethnic groups [extensively reviewed in (249)]. Disparities regarding DNA methylation have also been observed between diverse ethnic populations, including Caucasians, Hispanics, Middle Eastern, and African populations and may serve as a biomarker for underlying ethnic health disparities between human populations (250). Thus far, very little is known about the contribution of DNA methylation on ethnic differences to HIV acquisition, disease and treatment outcomes. Seeing that aberrant methylation patterns have been associated with HIV and that the rate of incidence differs amongst different ethnic groups, it is vital ethnic differences are taken into consideration when conducting studies and clinical trials therefore researchers should also take ethnicity into consideration (249, 250). The results of trials on one ethnic group may not necessarily be applicable to another ethnic, therefore researchers should also take ethnicity into consideration. We believe that the ideal model for epigenetic studies related to HIV disease are sorted PBMCs or CD4+ T cells that are isolated form a prospectively obtained longitudinal cohort consisting of different ethnic groups. Admittedly, it will be challenging to recruit and maintain such a cohort, nonetheless, more accurate and useful information can be gained from such a study design.
There is still a lot of gaps in knowledge regarding the relationship between methylation and HIV. But once we have a complete picture, the knowledge gained will contribute substantially to understanding HIV disease. Moreover, the use of epigenetic interventions such as DNMTs inhibitors as LRA, CRISPR editing, and methylation biomarkers may revolutionise our fight against HIV and the AIDS pandemic.
Author Contributions
Conceptualization and conceiving of idea, VR. Additional input with regards to conceptualization, ThiA. Writing, ThiA, TheA, and UR. Research, ThiA, TheA, UR, and RC. Figure design, ThiA. Editing of manuscript, VR. All authors contributed to the article and approved the submitted version.
Funding
This publication was supported by the South African Medical Research Council with funds received from the South African Department of Science and Technology. VR was funded as a FLAIR Research Fellow [the Future Leader in African Independent Research (FLAIR) Fellowship Programme was a partnership between the African Academy of Sciences (AAS) and the Royal Society that was funded by the UK Government as part of the Global Challenge Research Fund (GCRF) Grant # FLAIR-FLR\R1\190204]; supported by the South African Medical Research Council (SAMRC) with funds from the Department of Science and Technology (DST); and VR was also supported in part through the Sub-Saharan African Network for TB/HIV Research Excellence (SANTHE), a DELTAS Africa Initiative (Grant # DEL-15-006) by the AAS.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Grewal SIS, Moazed D. Heterochromatin and Epigenetic Control of Gene Expression. Science (2003) 301(5634):798–802. doi: 10.1126/science.1086887
2. Singal R, Ginder GD. DNA Methylation. Blood (1999) 93(12):4059–70. doi: 10.1182/blood.V93.12.4059
3. Bird A. The Essentials of DNA Methylation. Cell (1992) 70(1):5–8. doi: 10.1016/0092-8674(92)90526-I
4. Moore LD, Le T, Fan G. DNA Methylation and Its Basic Function. Neuropsychopharmacology (2013) 38(1):23–38. doi: 10.1038/npp.2012.112
5. Bird AP. CpG-Rich Islands and the Function of DNA Methylation. Nature (1986) 321(6067):209–13. doi: 10.1038/321209a0
6. Antequera F. Structure, Function and Evolution of CpG Island Promoters. Cell Mol Life Sci (2003) 60(8):1647–58. doi: 10.1007/s00018-003-3088-6
7. Saxonov S, Berg P, Brutlag DL. A Genome-Wide Analysis of CpG Dinucleotides in the Human Genome Distinguishes Two Distinct Classes of Promoters. Proc Natl Acad Sci USA (2006) 103(5):1412–7. doi: 10.1073/pnas.0510310103
8. Hellman A, Chess A. Gene Body-Specific Methylation on the Active X Chromosome. Science (2007) 315(5815):1141–3. doi: 10.1126/science.1136352
9. Aran D, Toperoff G, Rosenberg M, Hellman A. Replication Timing-Related and Gene Body-Specific Methylation of Active Human Genes. Hum Mol Genet (2011) 20(4):670–80. doi: 10.1093/hmg/ddq513
10. Jjingo D, Conley AB, Yi SV, Lunyak VV, Jordan IK. On the Presence and Role of Human Gene-Body DNA Methylation. Oncotarget (2012) 3(4):462–74. doi: 10.18632/oncotarget.497
11. Li E, Bestor TH, Jaenisch R. Targeted Mutation of the DNA Methyltransferase Gene Results in Embryonic Lethality. Cell (1992) 69(6):915–26. doi: 10.1016/0092-8674(92)90611-F
12. Geiman TM, Sankpal UT, Robertson AK, Chen Y, Mazumdar M, Heale JT, et al. Isolation and Characterization of a Novel DNA Methyltransferase Complex Linking DNMT3B With Components of the Mitotic Chromosome Condensation Machinery. Nucleic Acids Res (2004) 32(9):2716–29. doi: 10.1093/nar/gkh589
13. Li E, Beard C, Jaenisch R. Role for DNA Methylation in Genomic Imprinting. Nature (1993) 366(6453):362–5. doi: 10.1038/366362a0
14. Csankovszki G, Nagy A, Jaenisch R. Synergism of Xist RNA, DNA Methylation, and Histone Hypoacetylation in Maintaining X Chromosome Inactivation. J Cell Biol (2001) 153(4):773–84. doi: 10.1083/jcb.153.4.773
15. Schulz WA, Steinhoff C, Florl AR. Methylation of Endogenous Human Retroelements in Health and Disease. Curr Top Microbiol Immunol (2006) 310:211–50. doi: 10.1007/3-540-31181-5_11
16. Robertson KD. DNA Methylation and Human Disease. Nat Rev Genet (2005) 6(8):597–610. doi: 10.1038/nrg1655
17. Conerly M, Grady WM. Insights Into the Role of DNA Methylation in Disease Through the Use of Mouse Models. Dis Models Mech (2010) 3(5-6):290–7. doi: 10.1242/dmm.004812
18. Richardson B. DNA Methylation and Autoimmune Disease. Clin Immunol (2003) 109(1):72–9. doi: 10.1016/S1521-6616(03)00206-7
19. Vukic M, Daxinger L. DNA Methylation in Disease: Immunodeficiency, Centromeric Instability, Facial Anomalies Syndrome. Essays Biochem (2019) 63(6):773–83. doi: 10.1042/EBC20190035
20. Vivekanandan P, Daniel HD-J, Kannangai R, Martinez-Murillo F, Torbenson M. Hepatitis B Virus Replication Induces Methylation of Both Host and Viral DNA. J Virol (2010) 84(9):4321–9. doi: 10.1128/JVI.02280-09
21. Cicchini L, Blumhagen RZ, Westrich JA, Myers ME, Warren CJ, Siska C, et al. High-Risk Human Papillomavirus E7 Alters Host DNA Methylome and Represses HLA-E Expression in Human Keratinocytes. Sci Rep (2017) 7(1):1–13. doi: 10.1038/s41598-017-03295-7
22. Matsusaka K, Funata S, Fukayama M, Kaneda A. DNA Methylation in Gastric Cancer, Related to Helicobacter Pylori and Epstein-Barr Virus. World J Gastroenterol (2014) 20(14):3916–26. doi: 10.3748/wjg.v20.i14.3916
23. Shalginskikh N, Poleshko A, Skalka A, Katz RA. Retroviral DNA Methylation and Epigenetic Repression Are Mediated by the Antiviral Host Protein Daxx. J Virol (2013) 87(4):2137–50. doi: 10.1128/JVI.02026-12
24. Gillet NA, Malani N, Melamed A, Gormley N, Carter R, Bentley D, et al. The Host Genomic Environment of the Provirus Determines the Abundance of HTLV-1–Infected T-Cell Clones. Blood (2011) 117(11):3113–22. doi: 10.1182/blood-2010-10-312926
25. Rickabaugh TM, Baxter RM, Sehl M, Sinsheimer JS, Hultin PM, Hultin LE, et al. Acceleration of Age-Associated Methylation Patterns in HIV-1-Infected Adults. PloS One (2015) 10(3):e0119201. doi: 10.1371/journal.pone.0119201
26. Gross AM, Jaeger PA, Kreisberg JF, Licon K, Jepsen KL, Khosroheidari M, et al. Methylome-Wide Analysis of Chronic HIV Infection Reveals Five-Year Increase in Biological Age and Epigenetic Targeting of HLA. Mol Cell (2016) 62(2):157–68. doi: 10.1016/j.molcel.2016.03.019
27. Shiau S, Brummel SS, Kennedy EM, Hermetz K, Spector SA, Williams PL, et al. Longitudinal Changes in Epigenetic Age in Youth With Perinatally Acquired HIV and Youth Who Are Perinatally HIV-Exposed Uninfected. AIDS (2021) 35(5):811–9. doi: 10.1097/QAD.0000000000002805
28. Horvath S, Stein DJ, Phillips N, Heany SJ, Kobor MS, Lin DTS. Perinatally Acquired HIV Infection Accelerates Epigenetic Aging in South African Adolescents. AIDS (London England) (2018) 32(11):1465. doi: 10.1097/QAD.0000000000001854
29. Shiau S, Strehlau R, Wang S, Violari A, Do C, Patel F, et al. Distinct Epigenetic Profiles in Children With Perinatally-Acquired HIV on Antiretroviral Therapy. Sci Rep (2019) 9(1):1–15. doi: 10.1038/s41598-019-46930-1
30. Zhang X, Justice AC, Hu Y, Wang Z, Zhao H, Wang G, et al. Epigenome-Wide Differential DNA Methylation Between HIV-Infected and Uninfected Individuals. Epigenetics (2016) 11(10):750–60. doi: 10.1080/15592294.2016.1221569
31. Kauder SE, Bosque A, Lindqvist A, Planelles V, Verdin E. Epigenetic Regulation of HIV-1 Latency by Cytosine Methylation. PloS Pathog (2009) 5(6):e1000495. doi: 10.1371/journal.ppat.1000495
32. Blazkova J, Trejbalova K, Gondois-Rey F, Halfon P, Philibert P, Guiguen A, et al. CpG Methylation Controls Reactivation of HIV From Latency. PloS Pathog (2009) 5(8):e1000554. doi: 10.1371/journal.ppat.1000554
33. Trejbalová K, Kovářová D, Blažková J, Machala L, Jilich D, Weber J, et al. Development of 5 ‘LTR DNA Methylation of Latent HIV-1 Provirus in Cell Line Models and in Long-Term-Infected Individuals. Clin Epigenet (2016) 8(1):1–20. doi: 10.1186/s13148-016-0185-6
34. Chávez L, Kauder S, Verdin E. In Vivo, In Vitro, and in Silico Analysis of Methylation of the HIV-1 Provirus. Methods (2011) 53(1):47–53. doi: 10.1016/j.ymeth.2010.05.009
35. Ishida T, Hamano A, Koiwa T, Watanabe T. 5' Long Terminal Repeat (LTR)-Selective Methylation of Latently Infected HIV-1 Provirus That Is Demethylated by Reactivation Signals. Retrovirology (2006) 3(1):69. doi: 10.1186/1742-4690-3-69
36. Schermelleh L, Haemmer A, Spada F, Rösing N, Meilinger D, Rothbauer U, et al. Dynamics of Dnmt1 Interaction With the Replication Machinery and Its Role in Postreplicative Maintenance of DNA Methylation. Nucleic Acids Res (2007) 35(13):4301–12. doi: 10.1093/nar/gkm432
37. Okano M, Bell DW, Haber DA, Li E. DNA Methyltransferases Dnmt3a and Dnmt3b Are Essential for De Novo Methylation and Mammalian Development. Cell (1999) 99(3):247–57. doi: 10.1016/S0092-8674(00)81656-6
38. Mikovits JA, Young HA, Vertino P, Issa J-PJ, Pitha PM, Turcoski-Corrales S, et al. Infection With Human Immunodeficiency Virus Type 1 Upregulates DNA Methyltransferase, Resulting in De Novo Methylation of the Gamma Interferon (IFN-γ) Promoter and Subsequent Downregulation of IFN-γ Production. Mol Cell Biol (1998) 18(9):5166–77. doi: 10.1128/MCB.18.9.5166
39. Fang J-Y, Mikovits JA, Bagni R, Petrow-Sadowski CL, Ruscetti FW. Infection of Lymphoid Cells by Integration-Defective Human Immunodeficiency Virus Type 1 Increases De Novo Methylation. J Virol (2001) 75(20):9753–61. doi: 10.1128/JVI.75.20.9753-9761.2001
40. Youngblood B, Reich NO. The Early Expressed HIV-1 Genes Regulate DNMT1 Expression. Epigenetics (2008) 3(3):149–56. doi: 10.4161/epi.3.3.6372
41. Pion M, Jaramillo-Ruiz D, Martínez A, Muñoz-Fernández MA, Correa-Rocha R. HIV Infection of Human Regulatory T Cells Downregulates Foxp3 Expression by Increasing DNMT3b Levels and DNA Methylation in the FOXP3 Gene. Aids (2013) 27(13):2019–29. doi: 10.1097/QAD.0b013e32836253fd
42. Luzzi A, Morettini F, Gazaneo S, Mundo L, Onnis A, Mannucci S, et al. HIV-1 Tat Induces DNMT Over-Expression Through microRNA Dysregulation in HIV-Related non Hodgkin Lymphomas. Infect Agents Cancer (2014) 9(1):1–18. doi: 10.1186/1750-9378-9-41
43. Doke M, Jeganathan V, McLaughlin JP, Samikkannu T. HIV-1 Tat and Cocaine Impact Mitochondrial Epigenetics: Effects on DNA Methylation. Epigenetics (2020) p:1–20. doi: 10.1080/15592294.2020.1834919
44. Zhang Y, Li S-K, Yang KY, Liu M, Lee N, Tang X, et al. Whole Genome Methylation Array Reveals the Down-Regulation of IGFBP6 and SATB2 by HIV-1. Sci Rep (2015) 5(1):1–14. doi: 10.1038/srep10806
45. Rosca A, Anton G, Ene L, Iancu I, Temereanca A, Achim CL, et al. Immunoassay and Molecular Methods to Investigate DNA Methylation Changes in Peripheral Blood Mononuclear Cells in HIV Infected Patients on cART. J Immunoassay Immunochem (2017) 38(3):299–307. doi: 10.1080/15321819.2016.1260587
46. Ghosh SK, McCormick TS, Eapen BL, Yohannes E, Chance MR, Weinberg A, et al. Comparison of Epigenetic Profiles of Human Oral Epithelial Cells From HIV-Positive (on HAART) and HIV-Negative Subjects. Epigenetics (2013) 8(7):703–9. doi: 10.4161/epi.25028
47. Marimani M, Ahmad A, Stacey S, Duse A. Examining the Levels of Acetylation, DNA Methylation and Phosphorylation in HIV-1 Positive and Multidrug-Resistant TB-HIV Patients. J Global Antimicrob Resist (2020) 23:232–42. doi: 10.1016/j.jgar.2020.09.023
48. Wang Q, Su L. Vpr Enhances HIV-1 Env Processing and Virion Infectivity in Macrophages by Modulating TET2-Dependent IFITM3 Expression. MBio (2019) 10(4):e01344–19. doi: 10.1128/mBio.01344-19
49. Lv L, Wang Q, Xu Y, Tsao L-C, Nakagawa T, Guo H, et al. Vpr Targets TET2 for Degradation by CRL4VprBP E3 Ligase to Sustain IL-6 Expression and Enhance HIV-1 Replication. Mol Cell (2018) 70(5):961–970. e5. doi: 10.1016/j.molcel.2018.05.007
50. Yue X, Lio C-WJ, Samaniego-Castruita D, Li X, Rao A. Loss of TET2 and TET3 in Regulatory T Cells Unleashes Effector Function. Nat Commun (2019) 10(1):1–14. doi: 10.1038/s41467-019-09541-y
51. Mudd JC, Lai S, Shah S, Rahmberg A, Flynn JK, Starke CE, et al. Epigenetic Silencing of CD4 Expression in Nonpathogenic SIV Infection in African Green Monkeys. JCI Insight (2020) 5(18):e139043. doi: 10.1172/jci.insight.139043
52. van Wijnen AJ, Westendorf JJ. Epigenetics as a New Frontier in Orthopedic Regenerative Medicine and Oncology. J Orthop Res (2019) 37(7):1465–74. doi: 10.1002/jor.24305
53. Ko M, An J, Pastor WA, Koralov SB, Rajewsky K, Rao A, et al. TET Proteins and 5-Methylcytosine Oxidation in Hematological Cancers. Immunol Rev (2015) 263(1):6–21. doi: 10.1111/imr.12239
54. Yang L, Yu S-J, Hong Q, Yang Y, Shao Z-M. Reduced Expression of TET1, TET2, TET3 and TDG mRNAs Are Associated With Poor Prognosis of Patients With Early Breast Cancer. PloS One (2015) 10(7):e0133896. doi: 10.1371/journal.pone.0133896
55. Harris RS. Enhancing Immunity to HIV Through APOBEC. Nat Biotechnol (2008) 26(10):1089–90. doi: 10.1038/nbt1008-1089
56. Goila-Gaur R, Strebel K. HIV-1 Vif, APOBEC, and Intrinsic Immunity. Retrovirology (2008) 5(1):51. doi: 10.1186/1742-4690-5-51
57. Stuss DP, Cheema M, Ng MK, Martinez de Paz A, Williamson B, Missiaen K, et al. Impaired In Vivo Binding of MeCP2 to Chromatin in the Absence of Its DNA Methyl-Binding Domain. Nucleic Acids Res (2013) 41(9):4888–900. doi: 10.1093/nar/gkt213
58. Gos M. Epigenetic Mechanisms of Gene Expression Regulation in Neurological Diseases. Acta Neurobiol Exp (2013) 73:19–37.
59. Rahimian P, He JJ. HIV-1 Tat-Shortened Neurite Outgrowth Through Regulation of microRNA-132 and Its Target Gene Expression. J Neuroinflammation (2016) 13(1):1–17. doi: 10.1186/s12974-016-0716-2
60. Periyasamy P, Thangaraj A, Guo M-L, Hu G, Callen S, Buch S. Epigenetic Promoter DNA Methylation of miR-124 Promotes HIV-1 Tat-Mediated Microglial Activation via MECP2-STAT3 Axis. J Neurosci (2018) 38(23):5367–83. doi: 10.1523/JNEUROSCI.3474-17.2018
61. Du Q, Luu P-C, Stirzaker C, Clark SJ. Methyl-CpG-Binding Domain Proteins: Readers of the Epigenome. Epigenomics (2015) 7(6):1051–73. doi: 10.2217/epi.15.39
62. Bronner C, Alhosin M, Hamiche A, Mousli M. Coordinated Dialogue Between UHRF1 and DNMT1 to Ensure Faithful Inheritance of Methylated DNA Patterns. Genes (Basel) (2019) 10(1):65. doi: 10.3390/genes10010065
63. Bouchat S, Verdikt R, Delacourt N, Vanhulle C, Vandriessche B, Darcis G, et al. OA2-1 Identification of a New Factor Involved in DNA Methylation-Mediated Repression of Latent HIV-1. J Virus Erad (2017) 3:2. doi: 10.1016/S2055-6640(20)30835-9
64. Schröder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F, et al. HIV-1 Integration in the Human Genome Favors Active Genes and Local Hotspots. Cell (2002) 110(4):521–9. doi: 10.1016/S0092-8674(02)00864-4
65. Rezaei SD, Cameron PU. Human Immunodeficiency Virus (HIV)-1 Integration Sites in Viral Latency. Curr HIV/AIDS Rep (2015) 12(1):88–96. doi: 10.1007/s11904-014-0241-9
66. Han Y, Lassen K, Monie D, Sedaghat AR, Shimoji S, Liu X, et al. Resting CD4+ T Cells From Human Immunodeficiency Virus Type 1 (HIV-1)-Infected Individuals Carry Integrated HIV-1 Genomes Within Actively Transcribed Host Genes. J Virol (2004) 78(12):6122–33. doi: 10.1128/JVI.78.12.6122-6133.2004
67. Wu Y. HIV-1 Gene Expression: Lessons From Provirus and Non-Integrated DNA. Retrovirology (2004) 1(1):13. doi: 10.1186/1742-4690-1-13
68. Bednarik DP, Cook JA, Pitha PM. Inactivation of the HIV LTR by DNA CpG Methylation: Evidence for a Role in Latency. EMBO J (1990) 9(4):1157–64. doi: 10.1002/j.1460-2075.1990.tb08222.x
69. Singh MK, Pauza CD. Extrachromosomal Human Immunodeficiency Virus Type 1 Sequences Are Methylated in Latently Infected U937 Cells. Virology (1992) 188(2):451–8. doi: 10.1016/0042-6822(92)90498-E
70. Blazkova J, Murray D, Justement J, Funk EK, Nelson AK, Moir S, et al. Paucity of HIV DNA Methylation in Latently Infected, Resting CD4+ T Cells From Infected Individuals Receiving Antiretroviral Therapy. J Virol (2012) 86(9):5390–2. doi: 10.1128/JVI.00040-12
71. Palacios JA, Pérez-Piñar T, Toro C, Sanz-Minguela B, Moreno V, Valencia E, et al. Long-Term Nonprogressor and Elite Controller Patients Who Control Viremia Have a Higher Percentage of Methylation in Their HIV-1 Proviral Promoters Than Aviremic Patients Receiving Highly Active Antiretroviral Therapy. J Virol (2012) 86(23):13081–4. doi: 10.1128/JVI.01741-12
72. Weber S, Weiser B, Kemal KS, Burger H, Ramirez CM, Korn K, et al. Epigenetic Analysis of HIV-1 Proviral Genomes From Infected Individuals: Predominance of Unmethylated CpG's. Virology (2014) 449:181–9. doi: 10.1016/j.virol.2013.11.013
73. Li F, Li L, Zhong Y, Xie Q, Huang J, Kang X, et al. Relationship Between LTR Methylation and Gag Expression of HIV-1 in Human Spermatozoa and Sperm-Derived Embryos. PloS One (2013) 8(1):e54801. doi: 10.1371/journal.pone.0054801
74. Kint S, Trypsteen W, Spiegelaere WD, Malatinkova E, Kinloch-de Loes S, Meyer T, et al. Underestimated Effect of Intragenic HIV-1 DNA Methylation on Viral Transcription in Infected Individuals. Clin Epigenet (2020) 12(1):1–11. doi: 10.1186/s13148-020-00829-1
75. Cuevas JM, Geller R, Garijo R, López-Aldeguer J, Sanjuán R. Extremely High Mutation Rate of HIV-1 In Vivo. PloS Biol (2015) 13(9):e1002251. doi: 10.1371/journal.pbio.1002251
76. Alinejad-Rokny H, Anwar F, Waters SA, Davenport MP, Ebrahimi D. Source of CpG Depletion in the HIV-1 Genome. Mol Biol Evol (2016) 33(12):3205–12. doi: 10.1093/molbev/msw205
77. Lorincz MC, Schübeler D, Goeke SC, Walters M, Groudine M, Martin DI, et al. Dynamic Analysis of Proviral Induction and De Novo Methylation: Implications for a Histone Deacetylase-Independent, Methylation Density-Dependent Mechanism of Transcriptional Repression. Mol Cell Biol (2000) 20(3):842–50. doi: 10.1128/MCB.20.3.842-850.2000
78. Dodge JE, Ramsahoye BH, Wo ZG, Okano M, Li E. De Novo Methylation of MMLV Provirus in Embryonic Stem Cells: CpG Versus Non-CpG Methylation. Gene (2002) 289(1-2):41–8. doi: 10.1016/S0378-1119(02)00469-9
79. LaMere SA, Chaillon A, Huynh C, Smith DM, Gianella S. Challenges in Quantifying Cytosine Methylation in the HIV Provirus. mBio (2019) 10(1):e02268–18. doi: 10.1128/mBio.02268-18
80. Whitney JB, Brad Jones R. In Vitro and In Vivo Models of HIV Latency. In: Zhang L, Lewin SR, editors. HIV Vaccines and Cure : The Path Towards Finding an Effective Cure and Vaccine. Singapore: Springer Singapore (2018). p. 241–63.
81. Flanagan JM. Epigenome-Wide Association Studies (EWAS): Past, Present, and Future. Methods Mol Biol (2015) 1238:51–63. doi: 10.1007/978-1-4939-1804-1_3
82. Fazzari MJ, Greally JM. Introduction to Epigenomics and Epigenome-Wide Analysis. Methods Mol Biol (2010) 620:243–65. doi: 10.1007/978-1-60761-580-4_7
83. Hayatsu H. Discovery of Bisulfite-Mediated Cytosine Conversion to Uracil, the Key Reaction for DNA Methylation Analysis—a Personal Account. Proc Jpn Acad Ser B (2008) 84(8):321–30. doi: 10.2183/pjab.84.321
84. Xi Y, Li W. BSMAP: Whole Genome Bisulfite Sequence MAPping Program. BMC Bioinformatics (2009) 10(1):232. doi: 10.1186/1471-2105-10-232
85. Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, et al. High Density DNA Methylation Array With Single CpG Site Resolution. Genomics (2011) 98(4):288–95. doi: 10.1016/j.ygeno.2011.07.007
86. Meissner TB, Li A, Biswas A, Lee K-H, Liu Y-J, Bayir E, et al. NLR Family Member NLRC5 Is a Transcriptional Regulator of MHC Class I Genes. Proc Natl Acad Sci USA (2010) 107(31):13794–9. doi: 10.1073/pnas.1008684107
87. Shu C, Jaffe AE, Sabunciyan S, Ji H, Astemborski J, Sun J, et al. Epigenome-Wide Association Scan Identifies Methylation Sites Associated With HIV Infection. Epigenomics (2020) 12(21):1917–27. doi: 10.2217/epi-2020-0123
88. Zhang Y, Li S-K, Tsui SK-W. Genome-Wide Analysis of DNA Methylation Associated With HIV Infection Based on a Pair of Monozygotic Twins. Genomics Data (2015) 6:12–5. doi: 10.1016/j.gdata.2015.07.024
89. Castillo-Fernandez JE, Spector TD, Bell JT. Epigenetics of Discordant Monozygotic Twins: Implications for Disease. Genome Med (2014) 6(7):60. doi: 10.1186/s13073-014-0060-z
90. Nowrouzi A, Dittrich M, Klanke C, Heinkelein M, Rammling M, Dandekar T, et al. Genome-Wide Mapping of Foamy Virus Vector Integrations Into a Human Cell Line. J Gen Virol (2006) 87(Pt 5):1339–47. doi: 10.1099/vir.0.81554-0
91. Wellensiek BP, Ramakrishnan R, Sundaravaradan V, Mehta R, Harris DT, Ahmad N, et al. Differential HIV-1 Integration Targets More Actively Transcribed Host Genes in Neonatal Than Adult Blood Mononuclear Cells. Virology (2009) 385(1):28–38. doi: 10.1016/j.virol.2008.10.052
92. Oriol-Tordera B, Berdasco M, Llano A, Mothe B, Gálvez C, Martinez-Picado J, et al. Methylation Regulation of Antiviral Host Factors, Interferon Stimulated Genes (ISGs) and T-Cell Responses Associated With Natural HIV Control. PloS Pathog (2020) 16(8):e1008678. doi: 10.1371/journal.ppat.1008678
93. McGregor K, Bernatsky S, Colmegna I, Hudson M, Pastinen T, Labbe A, et al. An Evaluation of Methods Correcting for Cell-Type Heterogeneity in DNA Methylation Studies. Genome Biol (2016) 17:84–4. doi: 10.1186/s13059-016-0935-y
94. Corley MJ, Dye C, D'Antoni ML, Byron MM, Leite-Ah K, Lum-Jones A, et al. Comparative DNA Methylation Profiling Reveals an Immunoepigenetic Signature of HIV-Related Cognitive Impairment. Sci Rep (2016) 6(1):33310. doi: 10.1038/srep33310
95. Chandel N, Husain M, Goel H, Salhan D, Lan X, Malhotra A, et al. VDR Hypermethylation and HIV-Induced T Cell Loss. J Leukoc Biol (2013) 93(4):623–31. doi: 10.1189/jlb.0812383
96. Zeng X, Tsui JC-C, Shi M, Peng J, Cao CY, Kan L, et al. Genome-Wide Characterization of Host Transcriptional and Epigenetic Alterations During HIV Infection of T Lymphocytes. Front Immunol (2020) 11:2131. doi: 10.3389/fimmu.2020.02131
97. Moron-Lopez S, Urrea V, Dalmau J, Lopez M, Puertas MC, Ouchi D, et al. The Genome-Wide Methylation Profile of CD4+ T Cells From Individuals With Human Immunodeficiency Virus (HIV) Identifies Distinct Patterns Associated With Disease Progression. Clin Infect Dis (2020) 72(9):e256–64. doi: 10.1093/cid/ciaa1047
98. Kurdyukov S, Bullock M. DNA Methylation Analysis: Choosing the Right Method. Biology (2016) 5(1):3. doi: 10.3390/biology5010003
99. Reynes J, Baillat V, Portales P, Clot J, Corbeau P. Low CD4+ T-Cell Surface CCR5 Density as a Cause of Resistance to In Vivo HIV-1 Infection. J Acquir Immune Defic Syndr (2003) 34(1):114–6. doi: 10.1097/00126334-200309010-00018
100. Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. Effects of CCR5 and CD4 Cell Surface Concentrations on Infections by Macrophagetropic Isolates of Human Immunodeficiency Virus Type 1. J Virol (1998) 72(4):2855–64. doi: 10.1128/JVI.72.4.2855-2864.1998
101. de Roda Husman A-M, Koot M, Cornelissen M, Keet IP, Brouwer M, Broersen SM, et al. Association Between CCR5 Genotype and the Clinical Course of HIV-1 Infection. Ann Internal Med (1997) 127(10):882–90. doi: 10.7326/0003-4819-127-10-199711150-00004
102. Mummidi S, Ahuja SS, Gonzalez E, Anderson SA, Santiago EN, Stephan KT, et al. Genealogy of the CCR5 Locus and Chemokine System Gene Variants Associated With Altered Rates of HIV-1 Disease Progression. Nat Med (1998) 4(7):786–93. doi: 10.1038/nm0798-786
103. de Silva E, Stumpf MPH. HIV and the CCR5-Δ32 Resistance Allele. FEMS Microbiol Lett (2004) 241(1):1–12. doi: 10.1016/j.femsle.2004.09.040
104. Wu L, Paxton WA, Kassam N, Ruffing N, Rottman JB, Sullivan N, et al. CCR5 Levels and Expression Pattern Correlate With Infectability by Macrophage-Tropic HIV-1, In Vitro. J Exp Med (1997) 185(9):1681–92. doi: 10.1084/jem.185.9.1681
105. Bleul CC, Wu L, Hoxie JA, Springer TA, Mackay CR. The HIV Coreceptors CXCR4 and CCR5 Are Differentially Expressed and Regulated on Human T Lymphocytes. Proc Natl Acad Sci (1997) 94(5):1925–30. doi: 10.1073/pnas.94.5.1925
106. Ostrowski MA, Justement SJ, Catanzaro A, Hallahan CA, Ehler A, Mizell SB, et al. Expression of Chemokine Receptors CXCR4 and CCR5 in HIV-1-Infected and Uninfected Individuals. J Immunol (1998) 161(6):3195–201.
107. Gornalusse GG, Mummidi S, Gaitan AA, Jimenez F, Ramsuran V, Picton A, et al. Epigenetic Mechanisms, T-Cell Activation, and CCR5 Genetics Interact to Regulate T-Cell Expression of CCR5, the Major HIV-1 Coreceptor. Proc Natl Acad Sci (2015) 112(34):E4762–71. doi: 10.1073/pnas.1423228112
108. Study, TIHC. The Major Genetic Determinants of HIV-1 Control Affect HLA Class I Peptide Presentation. Science (2010) 330(6010):1551–7. doi: 10.1126/science.1195271
109. McLaren PJ, Coulonges C, Bartha I, Lenz TL, Deutsch AJ, Bashirova A, et al. Polymorphisms of Large Effect Explain the Majority of the Host Genetic Contribution to Variation of HIV-1 Virus Load. Proc Natl Acad Sci (2015) 112(47):14658–63. doi: 10.1073/pnas.1514867112
110. Klein J, Sato A. The HLA System. N Engl J Med (2000) 343(10):702–9. doi: 10.1056/NEJM200009073431006
111. Ramsuran V, Naranbhai V, Horowitz A, Qi Y, Martin MP, Yuki Y, et al. Elevated HLA-A Expression Impairs HIV Control Through Inhibition of NKG2A-Expressing Cells. Science (2018) 359(6371):86–90. doi: 10.1126/science.aam8825
112. Ramsuran V, Kulkarni S, O'huigin C, Yuki Y, Augusto DG, Gao X, et al. Epigenetic Regulation of Differential HLA-A Allelic Expression Levels. Hum Mol Genet (2015) 24(15):4268–75. doi: 10.1093/hmg/ddv158
113. López-Abente J, Correa-Rocha R, Pion M. Functional Mechanisms of Treg in the Context of HIV Infection and the Janus Face of Immune Suppression. Front Immunol (2016) 7:192. doi: 10.3389/fimmu.2016.00192
114. Moreno-Fernandez ME, Zapata W, Blackard JT, Franchini G, Chougnet CA. Human Regulatory T Cells Are Targets for Human Immunodeficiency Virus (HIV) Infection, and Their Susceptibility Differs Depending on the HIV Type 1 Strain. J Virol (2009) 83(24):12925–33. doi: 10.1128/JVI.01352-09
115. Oswald-Richter K, Grill SM, Shariat N, Leelawong M, Sundrud MS, Haas DW, et al. HIV Infection of Naturally Occurring and Genetically Reprogrammed Human Regulatory T-Cells. PloS Biol (2004) 2(7):e198. doi: 10.1371/journal.pbio.0020198
116. Williams LM, Rudensky AY. Maintenance of the Foxp3-Dependent Developmental Program in Mature Regulatory T Cells Requires Continued Expression of Foxp3. Nat Immunol (2007) 8(3):277–84. doi: 10.1038/ni1437
117. Abdel-Hameed EA, Ji H, Sherman KE, Shata MTM. Epigenetic Modification of FOXP3 in Patients With Chronic HIV Infection. J Acquir Immune Defic Syndr (1999) (2014) 65(1):19. doi: 10.1097/QAI.0b013e3182a1bca4
118. Sun XW, Kuhn L, Ellerbrock TV, Chiasson MA, Bush TJ, Wright TC Jr, et al. Human Papillomavirus Infection in Women Infected With the Human Immunodeficiency Virus. N Engl J Med (1997) 337(19):1343–9. doi: 10.1056/NEJM199711063371903
119. Chaturvedi AK, Madeleine M, Biggar RJ, Engels EA. Risk of Human Papillomavirus–Associated Cancers Among Persons With AIDS. J Natl Cancer Inst (2009) 101(16):1120–30. doi: 10.1093/jnci/djp205
120. Denslow SA, Rositch AF, Firnhaber C, Ting J, Smith JS. Incidence and Progression of Cervical Lesions in Women With HIV: A Systematic Global Review. Int J STD AIDS (2014) 25(3):163–77. doi: 10.1177/0956462413491735
121. Yang H-J. Aberrant DNA Methylation in Cervical Carcinogenesis. Chin J Cancer (2013) 32(1):42–8. doi: 10.5732/cjc.012.10033
122. Siegel EM, Riggs BM, Delmas AL, Koch A, Hakam A, Brown KD, et al. Quantitative DNA Methylation Analysis of Candidate Genes in Cervical Cancer. PloS One (2015) 10(3):e0122495. doi: 10.1371/journal.pone.0122495
123. Kelly HA, Chikandiwa A, Warman R, Segondy M, Sawadogo B, Vasiljevic N, et al. Associations of Human Gene EPB41L3 DNA Methylation and Cervical Intraepithelial Neoplasia in Women Living With HIV-1 in Africa. Aids (2018) 32(15):2227–36. doi: 10.1097/QAD.0000000000001932
124. De Vuyst H, Franceschi S, Plummer M, Mugo NR, Sakr SR, Meijer CJLM, et al. Methylation Levels of CADM1, MAL, and MIR124-2 in Cervical Scrapes for Triage of HIV-Infected, High-Risk HPV-Positive Women in Kenya. J Acquir Immune Defic Syndr (2015) 70(3):311–8. doi: 10.1097/QAI.0000000000000744
125. Tawe L, Grover S, Zetola N, Robertson ES, Gaseitsiwe S, Moyo S, et al. Promoter Hypermethylation Analysis of Host Genes in Cervical Cancer Patients With and Without Human Immunodeficiency Virus in Botswana. Front Oncol (2021) 11:509. doi: 10.3389/fonc.2021.560296
126. Kremer WW, Van Zummeren M, Novianti PW, Richter KL, Verlaat W, Snijders PJF, et al. Detection of Hypermethylated Genes as Markers for Cervical Screening in Women Living With HIV. J Int AIDS Soc (2018) 21(8):e25165–5. doi: 10.1002/jia2.25165
127. Singh R, Ramsuran V, Naranbhai V, Yende-Zuma N, Garrett N, Mlisana K, et al. Epigenetic Regulation of BST-2 Expression Levels and the Effect on HIV-1 Pathogenesis. Front Immunol (2021) 12:1623. doi: 10.3389/fimmu.2021.669241
128. Gibbs JR, van der Brug MP, Hernandez DG, Traynor BJ, Nalls MA, Lai S-L, et al. Abundant Quantitative Trait Loci Exist for DNA Methylation and Gene Expression in Human Brain. PloS Genet (2010) 6(5):e1000952. doi: 10.1371/journal.pgen.1000952
129. Zhang D, Cheng L, Badner JA, Chen C, Chen Q, Luo W, et al. Genetic Control of Individual Differences in Gene-Specific Methylation in Human Brain. Am J Hum Genet (2010) 86(3):411–9. doi: 10.1016/j.ajhg.2010.02.005
130. Bell JT, Pai AA, Pickrell JK, Gaffney DJ, Pique-Regi R, Degner JF, et al. DNA Methylation Patterns Associate With Genetic and Gene Expression Variation in HapMap Cell Lines. Genome Biol (2011) 12(1):1–13. doi: 10.1186/gb-2011-12-1-r10
131. Kerkel K, Spadola A, Yuan E, Kosek J, Jiang L, Hod E, et al. Genomic Surveys by Methylation-Sensitive SNP Analysis Identify Sequence-Dependent Allele-Specific DNA Methylation. Nat Genet (2008) 40(7):904–8. doi: 10.1038/ng.174
132. Bonder MJ, Luijk R, Zhernakova DV, Moed M, Deelen P, Vermaat M, et al. Disease Variants Alter Transcription Factor Levels and Methylation of Their Binding Sites. Nat Genet (2017) 49(1):131–8. doi: 10.1038/ng.3721
133. Villicaña S, Bell JT. Genetic Impacts on DNA Methylation: Research Findings and Future Perspectives. Genome Biol (2021) 22(1):127. doi: 10.1186/s13059-021-02347-6
134. Olinger GG, Saifuddin M, Hart ML, Spear GT. Cellular Factors Influence the Binding of HIV Type 1 to Cells. AIDS Res Hum Retroviruses (2002) 18(4):259–67. doi: 10.1089/088922202753472838
135. Gorry PR, Ancuta P. Coreceptors and HIV-1 Pathogenesis. Curr HIV/AIDS Rep (2011) 8(1):45–53. doi: 10.1007/s11904-010-0069-x
136. Sato N, Matsubayashi H, Fukushima N, Goggins M. The Chemokine Receptor CXCR4 Is Regulated by DNA Methylation in Pancreatic Cancer. Cancer Biol Ther (2005) 4(1):77–83. doi: 10.4161/cbt.4.1.1378
137. Ramos EAS, Grochoski M, Braun-Prado K, Seniski GG, Cavalli IJ, Ribeiro EMSF, et al. Epigenetic Changes of CXCR4 and Its Ligand CXCL12 as Prognostic Factors for Sporadic Breast Cancer. PloS One (2011) 6(12):e29461. doi: 10.1371/journal.pone.0029461
138. Bogani C, Ponziani V, Guglielmelli P, Desterke C, Rosti V, Bosi A, et al. Hypermethylation of CXCR4 Promoter in CD34+ Cells From Patients With Primary Myelofibrosis. Stem Cells (2008) 26(8):1920–30. doi: 10.1634/stemcells.2008-0377
139. Baldauf H-M, Pan X, Erikson E, Schmidt S, Daddacha W, Burggraf M, et al. SAMHD1 Restricts HIV-1 Infection in Resting CD4+ T Cells. Nat Med (2012) 18(11):1682–8. doi: 10.1038/nm.2964
140. Descours B, Cribier A, Chable-Bessia C, Ayinde D, Rice G, Crow Y, et al. SAMHD1 Restricts HIV-1 Reverse Transcription in Quiescent CD4+ T-Cells. Retrovirology (2012) 9(1):1–8. doi: 10.1186/1742-4690-9-87
141. de Silva S, Wang F, Hake TS, Porcu P, Wong HK, Wu L, et al. Downregulation of SAMHD1 Expression Correlates With Promoter DNA Methylation in Sezary Syndrome Patients. J Invest Dermatol (2014) 134(2):562. doi: 10.1038/jid.2013.311
142. de Silva S, Hoy H, Hake TS, Wong HK, Porcu P, Wu L, et al. Promoter Methylation Regulates SAMHD1 Gene Expression in Human CD4+ T Cells. J Biol Chem (2013) 288(13):9284–92. doi: 10.1074/jbc.M112.447201
143. Wang J-L, Lu F-Z, Shen X-Y, Wu Y, Zhao L-T. SAMHD1 Is Down Regulated in Lung Cancer by Methylation and Inhibits Tumor Cell Proliferation. Biochem Biophys Res Commun (2014) 455(3-4):229–33. doi: 10.1016/j.bbrc.2014.10.153
144. Shi B, Sharifi HJ, DiGrigoli S, Kinnetz M, Mellon K, Hu W, et al. Inhibition of HIV Early Replication by the P53 and Its Downstream Gene P21. Virol J (2018) 15(1):1–13. doi: 10.1186/s12985-018-0959-x
145. Pauls E, Ruiz A, Riveira-Muñoz E, Permanyer M, Badia R, Clotet B, et al. P21 Regulates the HIV-1 Restriction Factor SAMHD1. Proc Natl Acad Sci (2014) 111(14):E1322–4. doi: 10.1073/pnas.1322059111
146. Miozzo M, Vaira V, Sirchia SM. Epigenetic Alterations in Cancer and Personalized Cancer Treatment. Future Oncol (2015) 11(2):333–48. doi: 10.2217/fon.14.237
147. Laird PW, Jaenisch R. DNA Methylation and Cancer. Hum Mol Genet (1994) 3(suppl_1):1487–95. doi: 10.1093/hmg/3.suppl_1.1487
148. Baylin SB, Esteller M, Rountree MR, Bachman KE, Schuebel K, Herman JG, et al. Aberrant Patterns of DNA Methylation, Chromatin Formation and Gene Expression in Cancer. Hum Mol Genet (2001) 10(7):687–92. doi: 10.1093/hmg/10.7.687
149. Chmelarova M, Krepinska E, Spacek J, Laco J, Beranek M, Palicka V, et al. Methylation in the P53 Promoter in Epithelial Ovarian Cancer. Clin Trans Oncol (2013) 15(2):160–3. doi: 10.1007/s12094-012-0894-z
150. Kang JH, Kim SJ, Noh DY, Park IA, Choe KJ, Yoo OJ, et al. Methylation in the P53 Promoter Is a Supplementary Route to Breast Carcinogenesis: Correlation Between CpG Methylation in the P53 Promoter and the Mutation of the P53 Gene in the Progression From Ductal Carcinoma in Situ to Invasive Ductal Carcinoma. Lab Invest (2001) 81(4):573–9. doi: 10.1038/labinvest.3780266
151. Pogribny I, James SJ. Reduction of P53 Gene Expression in Human Primary Hepatocellular Carcinoma Is Associated With Promoter Region Methylation Without Coding Region Mutation. Cancer Lett (2002) 176(2):169–74. doi: 10.1016/S0304-3835(01)00748-0
152. Ogino S, Kawasaki T, Kirkner GJ, Ogawa A, Dorfman I, Loda M, et al. Down-Regulation of P21 (CDKN1A/CIP1) Is Inversely Associated With Microsatellite Instability and CpG Island Methylator Phenotype (CIMP) in Colorectal Cancer. J Pathol (2006) 210(2):147–54. doi: 10.1002/path.2030
153. Jimenez A, Gonzalez MG, Moreno F, del Carmen Rodriguez M. 5′ CpG Island Hypermethylation Is Associated With Transcriptional Silencing of the P21cip1/WAF1/SDI1 Gene and Confers Poor Prognosis in Acute Lymphoblastic Leukemia. Blood (2002) 99(7):2291–6. doi: 10.1182/blood.V99.7.2291
154. Teramen H, Tsukuda K, Tanaka N, Ueno T, Kubo T, Ando M, et al. Aberrant Methylation of P21 Gene in Lung Cancer and Malignant Pleural Mesothelioma. Acta Med Okayama (2011) 65(3):179–84. doi: 10.18926/AMO/46629
155. Angela Covino D, Sabbatucci M, Fantuzzi L. The CCL2/CCR2 Axis in the Pathogenesis of HIV-1 Infection: A New Cellular Target for Therapy? Curr Drug Targets (2016) 17(1):76–110. doi: 10.2174/138945011701151217110917
156. Williams DW, Calderon TM, Lopez L, Carvallo-Torres L, Gaskill PJ, Eugenin EA, et al. Mechanisms of HIV Entry Into the CNS: Increased Sensitivity of HIV Infected CD14+CD16+ Monocytes to CCL2 and Key Roles of CCR2, JAM-A, and ALCAM in Diapedesis. PloS One (2013) 8(7):e69270. doi: 10.1371/journal.pone.0069270
157. Pfalzer AC, Choi S-W, Tammen SA, Park LK, Bottiglieri T, Parnell LD, et al. S-Adenosylmethionine Mediates Inhibition of Inflammatory Response and Changes in DNA Methylation in Human Macrophages. Physiol Genomics (2014) 46(17):617–23. doi: 10.1152/physiolgenomics.00056.2014
158. Campbell GR, Spector SA. CCL2 Increases X4-Tropic HIV-1 Entry Into Resting CD4+ T Cells. J Biol Chem (2008) 283(45):30745–53. doi: 10.1074/jbc.M804112200
159. Eugenin EA, Osiecki K, Lopez L, Goldstein H, Calderon TM, Berman JW, et al. CCL2/monocyte Chemoattractant Protein-1 Mediates Enhanced Transmigration of Human Immunodeficiency Virus (HIV)-Infected Leukocytes Across the Blood–Brain Barrier: A Potential Mechanism of HIV–CNS Invasion and NeuroAIDS. J Neurosci (2006) 26(4):1098–106. doi: 10.1523/JNEUROSCI.3863-05.2006
160. Li B, Chen X, Jiang Y, Yang Y, Zhong J, Zhou C, et al. CCL2 Promoter Hypomethylation Is Associated With Gout Risk in Chinese Han Male Population. Immunol Lett (2017) 190:15–9. doi: 10.1016/j.imlet.2017.06.011
161. Zheng Y, Wang Z, Wei S, Liu Z, Chen G. Epigenetic Silencing of Chemokine CCL2 Represses Macrophage Infiltration to Potentiate Tumor Development in Small Cell Lung Cancer. Cancer Lett (2021) 499:148–63. doi: 10.1016/j.canlet.2020.11.034
162. Cao Q, Wang X, Jia L, Mondal AK, Diallo A, Hawkins GA, et al. Inhibiting DNA Methylation by 5-Aza-2′-Deoxycytidine Ameliorates Atherosclerosis Through Suppressing Macrophage Inflammation. Endocrinology (2014) 155(12):4925–38. doi: 10.1210/en.2014-1595
163. Ashokkumar M, Aralaguppe SG, Tripathy SP, Hanna LE, Neogi U. Unique Phenotypic Characteristics of Recently Transmitted HIV-1 Subtype C Envelope Glycoprotein Gp120: Use of CXCR6 Coreceptor by Transmitted Founder Viruses. J Virol (2018) 92(9):e00063–18. doi: 10.1128/JVI.00063-18
164. Blaak H, Boers PHM, Gruters RA, Schuitemaker H, van der Ende ME, Osterhaus ADME, et al. CCR5, GPR15, and CXCR6 Are Major Coreceptors of Human Immunodeficiency Virus Type 2 Variants Isolated From Individuals With and Without Plasma Viremia. J Virol (2005) 79(3):1686–700. doi: 10.1128/JVI.79.3.1686-1700.2005
165. Bergmann AK, Fataccioli V, Castellano G, Martin-Garcia N, Pelletier L, Ammerpohl O, et al. DNA Methylation Profiling of Hepatosplenic T-Cell Lymphoma. Haematologica (2019) 104(3):e104–7. doi: 10.3324/haematol.2018.196196
166. Zhu H, Zhu C, Mi W, Chen T, Zhao H, Zuo X, et al. Integration of Genome-Wide DNA Methylation and Transcription Uncovered Aberrant Methylation-Regulated Genes and Pathways in the Peripheral Blood Mononuclear Cells of Systemic Sclerosis. Int J Rheumatol (2018) 2018:7342472. doi: 10.1155/2018/7342472
167. Paxton WA, Martin SR, Tse D, O'Brien TR, Skurnick J, Van Devanter NL, et al. Relative Resistance to HIV–1 Infection of CD4 Lymphocytes From Persons Who Remain Uninfected Despite Multiple High–Risk Sexual Exposures. Nat Med (1996) 2(4):412–7. doi: 10.1038/nm0496-412
168. Arenzana-Seisdedos F, Virelizier JL, Rousset D, Clark-Lewis I, Loetscher P, Moser B, et al. HIV Blocked by Chemokine Antagonist. Nature (1996) 383(6599):400–0. doi: 10.1038/383400a0
169. Tserel L, Kolde R, Limbach M, Tretyakov K, Kasela S, Kisand K, et al. Age-Related Profiling of DNA Methylation in CD8+ T Cells Reveals Changes in Immune Response and Transcriptional Regulator Genes. Sci Rep (2015) 5(1):1–11. doi: 10.1038/srep13107
170. Rastogi D, Suzuki M, Greally JM. Differential Epigenome-Wide DNA Methylation Patterns in Childhood Obesity-Associated Asthma. Sci Rep (2013) 3(1):2164. doi: 10.1038/srep02164
171. Demirov DG, Ono A, Orenstein JM, Freed EO. Overexpression of the N-Terminal Domain of TSG101 Inhibits HIV-1 Budding by Blocking Late Domain Function. Proc Natl Acad Sci (2002) 99(2):955–60. doi: 10.1073/pnas.032511899
172. Broniarczyk J, Olejnik-Schmidt AK, Luczak MW, Schmidt MT, Dabrowski M, Józefiak A, et al. Analysis of Expression and Structure of the TSG101 Gene in Cervical Cancer Cells. Int J Mol Med (2010) 25(5):777–83. doi: 10.3892/ijmm_00000404
173. Evans VA, van der Sluis RM, Solomon A, Dantanarayana A, McNeil C, Garsia R, et al. Programmed Cell Death-1 Contributes to the Establishment and Maintenance of HIV-1 Latency. AIDS (London England) (2018) 32(11):1491–7. doi: 10.1097/QAD.0000000000001849
174. Porichis F, Kaufmann DE. Role of PD-1 in HIV Pathogenesis and as Target for Therapy. Curr HIV/AIDS Rep (2012) 9(1):81–90. doi: 10.1007/s11904-011-0106-4
175. Nair VS, Toor SM, Taha RZ, Shaath H, Elkord E. DNA Methylation and Repressive Histones in the Promoters of PD-1, CTLA-4, TIM-3, LAG-3, TIGIT, PD-L1, and Galectin-9 Genes in Human Colorectal Cancer. Clin Epigenet (2018) 10(1):1–9. doi: 10.1186/s13148-018-0539-3
176. Goltz D, Gevensleben H, Dietrich J, Schroeck F, de Vos L, Droege F, et al. PDCD1 (PD-1) Promoter Methylation Predicts Outcome in Head and Neck Squamous Cell Carcinoma Patients. Oncotarget (2017) 8(25):41011. doi: 10.18632/oncotarget.17354
177. Ørskov AD, Treppendahl MB, Skovbo A, Holm MS, Friis LS, Hokland M, et al. Hypomethylation and Up-Regulation of PD-1 in T Cells by Azacytidine in MDS/AML Patients: A Rationale for Combined Targeting of PD-1 and DNA Methylation. Oncotarget (2015) 6(11):9612. doi: 10.18632/oncotarget.3324
178. Goltz D, Gevensleben H, Dietrich J, Ellinger J, Landsberg J, Kristiansen G, et al. Promoter Methylation of the Immune Checkpoint Receptor PD-1 (PDCD1) Is an Independent Prognostic Biomarker for Biochemical Recurrence-Free Survival in Prostate Cancer Patients Following Radical Prostatectomy. Oncoimmunology (2016) 5(10):e1221555. doi: 10.1080/2162402X.2016.1221555
179. Elashi AA, Nair VS, Taha RZ, Shaath H, Elkord E. DNA Methylation of Immune Checkpoints in the Peripheral Blood of Breast and Colorectal Cancer Patients. OncoImmunology (2019) 8(2):e1542918. doi: 10.1080/2162402X.2018.1542918
180. Day CL, Kaufmann DE, Kiepiela P, Brown JA, Moodley ES, Reddy S, et al. PD-1 Expression on HIV-Specific T Cells Is Associated With T-Cell Exhaustion and Disease Progression. Nature (2006) 443(7109):350–4. doi: 10.1038/nature05115
181. Zhang Y, Xiang C, Wang Y, Duan Y, Liu C, Zhang Y, et al. PD-L1 Promoter Methylation Mediates the Resistance Response to Anti-PD-1 Therapy in NSCLC Patients With EGFR-TKI Resistance. Oncotarget (2017) 8(60):101535. doi: 10.18632/oncotarget.21328
182. Goltz D, Gevensleben H, Grünen S, Dietrich J, Kristiansen G, Landsberg J, et al. PD-L1 (CD274) Promoter Methylation Predicts Survival in Patients With Acute Myeloid Leukemia. Leukemia (2017) 31(3):738–43. doi: 10.1038/leu.2016.328
183. Jones RB, Ndhlovu LC, Barbour JD, Sheth PM, Jha AR, Long BR, et al. Tim-3 Expression Defines a Novel Population of Dysfunctional T Cells With Highly Elevated Frequencies in Progressive HIV-1 Infection. J Exp Med (2008) 205(12):2763–79. doi: 10.1084/jem.20081398
184. Lu X, Yang L, Yao D, Wu X, Li J, Liu X, et al. Tumor Antigen-Specific CD8+ T Cells Are Negatively Regulated by PD-1 and Tim-3 in Human Gastric Cancer. Cell Immunol (2017) 313:43–51. doi: 10.1016/j.cellimm.2017.01.001
185. Graydon CG, Balasko AL, Fowke KR. Roles, Function and Relevance of LAG3 in HIV Infection. PloS Pathog (2019) 15(1):e1007429. doi: 10.1371/journal.ppat.1007429
186. Tian X, Zhang A, Qiu C, Wang W, Yang Y, Qiu C, et al. The Upregulation of LAG-3 on T Cells Defines a Subpopulation With Functional Exhaustion and Correlates With Disease Progression in HIV-Infected Subjects. J Immunol (2015) 194(8):3873–82. doi: 10.4049/jimmunol.1402176
187. Klümper N, Ralser DJ, Bawden G, Landsberg J, Zarbl R, Kristiansen G, et al. LAG3 (LAG-3, CD223) DNA Methylation Correlates With LAG3 Expression by Tumor and Immune Cells, Immune Cell Infiltration, and Overall Survival in Clear Cell Renal Cell Carcinoma. J immunother Cancer (2020) 8(1):e000552. doi: 10.1136/jitc-2020-000552
188. Fröhlich A, Sirokay J, Fietz S, Vogt TJ, Dietrich J, Zarbl R, et al. Molecular, Clinicopathological, and Immune Correlates of LAG3 Promoter DNA Methylation in Melanoma. EBioMedicine (2020) 59:102962. doi: 10.1016/j.ebiom.2020.102962
189. Kaufmann DE, Kavanagh DG, Pereyra F, Zaunders JJ, Mackey EW, Miura T, et al. Upregulation of CTLA-4 by HIV-Specific CD4+ T Cells Correlates With Disease Progression and Defines a Reversible Immune Dysfunction. Nat Immunol (2007) 8(11):1246–54. doi: 10.1038/ni1515
190. Cribbs AP, Kennedy A, Penn H, Read JE, Amjadi P, Green P, et al. Treg Cell Function in Rheumatoid Arthritis Is Compromised by CTLA-4 Promoter Methylation Resulting in a Failure to Activate the Indoleamine 2, 3-Dioxygenase Pathway. Arthritis Rheumatol (2014) 66(9):2344–54. doi: 10.1002/art.38715
191. Xu W, Ren M, Ghosh S, Qian K, Luo Z, Zhang A, et al. Defects of CTLA-4 Are Associated With Regulatory T Cells in Myasthenia Gravis Implicated by Intravenous Immunoglobulin Therapy. Mediators Inflamm (2020) 2020:3645157. doi: 10.1155/2020/3645157
192. de Vos L, Grünwald I, Bawden EG, Dietrich J, Scheckenbach K, Wiek C, et al. The Landscape of CD28, CD80, CD86, CTLA4, and ICOS DNA Methylation in Head and Neck Squamous Cell Carcinomas. Epigenetics (2020) 15(11):1195–212. doi: 10.1080/15592294.2020.1754675
193. Vicenzi E, Poli G. The Interferon-Stimulated Gene TRIM22: A Double-Edged Sword in HIV-1 Infection. Cytokine Growth Factor Rev (2018) 40:40–7. doi: 10.1016/j.cytogfr.2018.02.001
194. Lim K-H, Park E-S, Kim D-H, Cho Cho K, Kim KP, Park YK, et al. Suppression of Interferon-Mediated Anti-HBV Response by Single CpG Methylation in the 5′-UTR of TRIM22. Gut (2018) 67(1):166–78. doi: 10.1136/gutjnl-2016-312742
195. Coit P, Jeffries M, Altorok N, Dozmorov MG, Koelsch KA, Wren JD, et al. Genome-Wide DNA Methylation Study Suggests Epigenetic Accessibility and Transcriptional Poising of Interferon-Regulated Genes in Naive CD4+ T Cells From Lupus Patients. J Autoimmun (2013) 43:78–84. doi: 10.1016/j.jaut.2013.04.003
196. Geijtenbeek TB, van Kooyk Y. DC-SIGN: A Novel HIV Receptor on DCs That Mediates HIV-1 Transmission. Curr Top Microbiol Immunol (2003) 276:31–54. doi: 10.1007/978-3-662-06508-2_2
197. Geijtenbeek TB, Kwon DS, Torensma R, van Vliet SJ, van Duijnhoven GC, Middel J, et al. DC-SIGN, a Dendritic Cell–Specific HIV-1-Binding Protein That Enhances Trans-Infection of T Cells. Cell (2000) 100(5):587–97. doi: 10.1016/S0092-8674(00)80694-7
198. Bullwinkel J, Lüdemann A, Debarry J, Singh PB. Epigenotype Switching at the CD14 and CD209 Genes During Differentiation of Human Monocytes to Dendritic Cells. Epigenetics (2011) 6(1):45–51. doi: 10.4161/epi.6.1.13314
199. Brockman MA, Kwon DS, Tighe DP, Pavlik DF, Rosato PC, Sela J, et al. IL-10 Is Up-Regulated in Multiple Cell Types During Viremic HIV Infection and Reversibly Inhibits Virus-Specific T Cells. Blood (2009) 114(2):346–56. doi: 10.1182/blood-2008-12-191296
200. Fu L-H, Cong B, Zhen Y-F, Li S-J, Ma C-L, Ni Z-Y, et al. Methylation Status of the IL-10 Gene Promoter in the Peripheral Blood Mononuclear Cells of Rheumatoid Arthritis Patients. Yi Chuan (2007) 29(11):1357–61. doi: 10.1360/yc-007-1357
201. Alipour S, Nouri M, Khabbazi A, Samadi N, Babaloo Z, Abolhasani S, et al. Hypermethylation of IL-10 Gene Is Responsible for Its Low mRNA Expression in Behçet's Disease. J Cell Biochem (2018) 119(8):6614–22. doi: 10.1002/jcb.26809
202. Nold MF, Nold-Petry CA, Pott GB, Zepp JA, Saavedra MT, Kim S-H, et al. Endogenous IL-32 Controls Cytokine and HIV-1 Production. J Immunol (2008) 181(1):557–65. doi: 10.4049/jimmunol.181.1.557
203. Rasool ST, Tang H, Wu J, Li W, Mukhtar MM, Zhang J, et al. Increased Level of IL-32 During Human Immunodeficiency Virus Infection Suppresses HIV Replication. Immunol Lett (2008) 117(2):161–7. doi: 10.1016/j.imlet.2008.01.007
204. Zhao Z, Lan M, Li J, Dong Q, Li X, Liu B, et al. The Proinflammatory Cytokine Tnfα Induces DNA Demethylation–Dependent and–Independent Activation of Interleukin-32 Expression. J Biol Chem (2019) 294(17):6785–95. doi: 10.1074/jbc.RA118.006255
205. Meyer B, Chavez RA, Munro JE, Chiaroni-Clarke RC, Akikusa JD, Allen RC, et al. DNA Methylation at IL32 in Juvenile Idiopathic Arthritis. Sci Rep (2015) 5(1):1–12. doi: 10.1038/srep11063
206. Li W, Sun W, Liu L, Yang F, Li Y, Chen Y, et al. IL-32: A Host Proinflammatory Factor Against Influenza Viral Replication Is Upregulated by Aberrant Epigenetic Modifications During Influenza A Virus Infection. J Immunol (2010) 185(9):5056–65. doi: 10.4049/jimmunol.0902667
207. Battistini A, Marsili G, Sgarbanti M, Ensoli B, Hiscott J. IRF Regulation of HIV-1 Long Terminal Repeat Activity. J Interferon Cytokine Res (2002) 22(1):27–37. doi: 10.1089/107999002753452638
208. Sgarbanti M, Remoli AL, Marsili G, Ridolfi B, Borsetti A, Perrotti E, et al. IRF-1 Is Required for Full NF-κB Transcriptional Activity at the Human Immunodeficiency Virus Type 1 Long Terminal Repeat Enhancer. J Virol (2008) 82(7):3632–41. doi: 10.1128/JVI.00599-07
209. Kim J, Bhattacharjee R, Khalyfa A, Kheirandish-Gozal L, Capdevila OS, Wang Y, et al. DNA Methylation in Inflammatory Genes Among Children With Obstructive Sleep Apnea. Am J Respir Crit Care Med (2012) 185(3):330–8. doi: 10.1164/rccm.201106-1026OC
210. Hersperger AR, Pereyra F, Nason M, Demers K, Sheth P, Shin LY, et al. Perforin Expression Directly Ex Vivo by HIV-Specific CD8+ T-Cells Is a Correlate of HIV Elite Control. PloS Pathog (2010) 6(5):e1000917. doi: 10.1371/journal.ppat.1000917
211. Maltby VE, Lea RA, Sanders KA, White N, Benton MC, Scott RJ, et al. Differential Methylation at MHC in CD4+ T Cells Is Associated With Multiple Sclerosis Independently of HLA-DRB1. Clin Epigenet (2017) 9(1):71. doi: 10.1186/s13148-017-0371-1
212. Lu Q, Wu A, Ray D, Deng C, Attwood J, Hanash S, et al. DNA Methylation and Chromatin Structure Regulate T Cell Perforin Gene Expression. J Immunol (2003) 170(10):5124–32. doi: 10.4049/jimmunol.170.10.5124
213. Renauer P, Coit P, Jeffries MA, Merrill JT, McCune WJ, Maksimowicz-McKinnon K, et al. DNA Methylation Patterns in Naïve CD4+ T Cells Identify Epigenetic Susceptibility Loci for Malar Rash and Discoid Rash in Systemic Lupus Erythematosus. Lupus Sci Med (2015) 2(1):e000101. doi: 10.1136/lupus-2015-000101
214. Kulakova O, Kabilov MR, Danilova LV, Popova EV, Baturina OA, Tsareva EY, et al. Whole-Genome DNA Methylation Analysis of Peripheral Blood Mononuclear Cells in Multiple Sclerosis Patients With Different Disease Courses. Acta Naturae (англоязычная версия) (2016) 8(3):103–110.
215. Yu X, Shang H, Jiang Y. ICAM-1 in HIV Infection and Underlying Mechanisms. Cytokine (2020) 125:154830. doi: 10.1016/j.cyto.2019.154830
216. Shalaby SM, Mackawy AMH, Atef DM, Atef RM, Saeed J. Promoter Methylation and Expression of Intercellular Adhesion Molecule 1 Gene in Blood of Autoimmune Thyroiditis Patients. Mol Biol Rep (2019) 46(5):5345–53. doi: 10.1007/s11033-019-04990-6
217. Friedrich MG, Chandrasoma S, Siegmund KD, Weisenberger DJ, Cheng JC, Toma MI, et al. Prognostic Relevance of Methylation Markers in Patients With Non-Muscle Invasive Bladder Carcinoma. Eur J Cancer (2005) 41(17):2769–78. doi: 10.1016/j.ejca.2005.07.019
218. Shyamala N, Gundapaneni KK, Galimudi RK, Tupurani MA, Padala C, Puranam K, et al. PCSK9 Genetic (Rs11591147) and Epigenetic (DNA Methylation) Modifications Associated With PCSK9 Expression and Serum Proteins in CAD Patients. J Gene Med (2021) 23:e3346. doi: 10.1002/jgm.3346
219. Radhakrishna U, Albayrak S, Alpay-Savasan Z, Zeb A, Turkoglu O, Sobolewski P, et al. Genome-Wide DNA Methylation Analysis and Epigenetic Variations Associated With Congenital Aortic Valve Stenosis (AVS). PloS One (2016) 11(5):e0154010. doi: 10.1371/journal.pone.0154010
220. Guardiola M, Oliva I, Sanchez M, Plana N, Masana L, Monk D, et al. Pcsk9 Promoter Methylation Is Associated With Small Ldl Particles In Patients With Type 2 Diabetes And Metabolic Syndrome. Atherosclerosis (2019) 287:e43. doi: 10.1016/j.atherosclerosis.2019.06.123
221. Brown TR. I am the Berlin Patient: A Personal Reflection. AIDS Res Hum Retroviruses (2015) 31(1):2–3. doi: 10.1089/aid.2014.0224
222. Peterson CW, Kiem H-P. Lessons From London and Berlin: Designing a Scalable Gene Therapy Approach for HIV Cure. Cell Stem Cell (2019) 24(5):685–7. doi: 10.1016/j.stem.2019.04.010
223. Corley MJ, Sacdalan C, Pang APS, Chomchey N, Ratnaratorn N, Valcour V, et al. Abrupt and Altered Cell-Type-Specific DNA Methylation Profiles in Blood During Acute HIV Infection Persists Despite Prompt Initiation of ART. PloS Pathog (2021) 17(8):e1009785. doi: 10.1371/journal.ppat.1009785
224. Shrivastava S, Ray RM, Holguin L, Echavarria L, Grepo N, Scott TA, et al. Exosome-Mediated Stable Epigenetic Repression of HIV-1. Nat Commun (2021) 12(1):1–14. doi: 10.1038/s41467-021-25839-2
225. Deeks SG, Lewin SR, Ross AL, Ananworanich J, Benkirane M, Cannon P, et al. International AIDS Society Global Scientific Strategy: Towards an HIV Cure 2016. Nat Med (2016) 22(8):839–50. doi: 10.1038/nm.4108
226. Kumar A, Darcis G, Van Lint C, Herbein G. Epigenetic Control of HIV-1 Post Integration Latency: Implications for Therapy. Clin Epigenet (2015) 7(1):1–12. doi: 10.1186/s13148-015-0137-6
227. Bouchat S, Delacourt N, Kula A, Darcis G, Van Driessche B, Corazza F, et al. Sequential Treatment With 5-Aza-2'-Deoxycytidine and Deacetylase Inhibitors Reactivates HIV-1. EMBO Mol Med (2016) 8(2):117–38. doi: 10.15252/emmm.201505557
228. Fenaux P. Inhibitors of DNA Methylation: Beyond Myelodysplastic Syndromes. Nat Clin Pract Oncol (2005) 2(1):S36–44. doi: 10.1038/ncponc0351
229. Kaminskas E, Farrell AT, Wang Y-C, Sridhara R, Pazdur R. FDA Drug Approval Summary: Azacitidine (5-Azacytidine, Vidaza™) for Injectable Suspension. Oncologist (2005) 10(3):176–82. doi: 10.1634/theoncologist.10-3-176
230. Fenaux P, Mufti GJ, Hellstrom-Lindberg E, Santini V, Finelli C, Giagounidis A, et al. Efficacy of Azacitidine Compared With That of Conventional Care Regimens in the Treatment of Higher-Risk Myelodysplastic Syndromes: A Randomised, Open-Label, Phase III Study. Lancet Oncol (2009) 10(3):223–32. doi: 10.1016/S1470-2045(09)70003-8
231. Fenaux P, Mufti GJ, Santini V, Finelli C, Giagounidis A, Schoch R, et al. Azacitidine (AZA) Treatment Prolongs Overall Survival (OS) in Higher-Risk MDS Patients Compared With Conventional Care Regimens (CCR): Results of the AZA-001 Phase III Study. Blood (2007) 110(11):817–7. doi: 10.1182/blood.V110.11.817.817
232. Kantarjian H, Issa J-PJ, Rosenfeld CS, Bennett JM, Albitar M, DiPersio J, et al. Decitabine Improves Patient Outcomes in Myelodysplastic Syndromes. Cancer (2006) 106(8):1794–803. doi: 10.1002/cncr.21792
233. Ebina H, Misawa N, Kanemura Y, Koyanagi Y. Harnessing the CRISPR/Cas9 System to Disrupt Latent HIV-1 Provirus. Sci Rep (2013) 3(1):2510. doi: 10.1038/srep02510
234. Liu Z, Chen S, Jin X, Wang Q, Yang K, Li C, et al. Genome Editing of the HIV Co-Receptors CCR5 and CXCR4 by CRISPR-Cas9 Protects CD4+ T Cells From HIV-1 Infection. Cell Biosci (2017) 7(1):1–15. doi: 10.1186/s13578-017-0174-2
235. Kang H, Minder P, Park MA, Mesquitta W-T, Torbett BE, Slukvin II, et al. CCR5 Disruption in Induced Pluripotent Stem Cells Using CRISPR/Cas9 Provides Selective Resistance of Immune Cells to CCR5-Tropic HIV-1 Virus. Mol Ther Nucleic Acids (2015) 4:e268. doi: 10.1038/mtna.2015.42
236. Hou P, Chen S, Wang S, Yu X, Yu C. Genome Editing of CXCR4 by CRISPR/cas9 Confers Cells Resistant to HIV-1 Infection. Sci Rep (2015) 5(1):1–12. doi: 10.1038/srep15577
237. Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, et al. Analysis of Off-Target Effects of CRISPR/Cas-Derived RNA-Guided Endonucleases and Nickases. Genome Res (2014) 24(1):132–41. doi: 10.1101/gr.162339.113
238. Ledford H. CRISPR Gene Editing in Human Embryos Wreaks Chromosomal Mayhem. Nature (2020) 583(7814):17–8. doi: 10.1038/d41586-020-01906-4
239. McDonald JI, Celik H, Rois LE, Fishberger G, Fowler T, Rees R, et al. Reprogrammable CRISPR/Cas9-Based System for Inducing Site-Specific DNA Methylation. Biol Open (2016) 5(6):866–74. doi: 10.1242/bio.019067
240. Vojta A, Dobrinić P, Tadić V, Bočkor L, Korać P, Julg B, et al. Repurposing the CRISPR-Cas9 System for Targeted DNA Methylation. Nucleic Acids Res (2016) 44(12):5615–28. doi: 10.1093/nar/gkw159
241. Choudhury SR, Cui Y, Lubecka K, Stefanska B, Irudayaraj J. CRISPR-Dcas9 Mediated TET1 Targeting for Selective DNA Demethylation at BRCA1 Promoter. Oncotarget (2016) 7(29):46545. doi: 10.18632/oncotarget.10234
242. Beltrán-García J, Osca-Verdegal R, Mena-Mollá S, García-Giménez JL. Epigenetic IVD Tests for Personalized Precision Medicine in Cancer. Front Genet (2019) 10:621. doi: 10.3389/fgene.2019.00621
243. Rahat B, Ali T, Sapehia D, Mahajan A, Kaur J. Circulating Cell-Free Nucleic Acids as Epigenetic Biomarkers in Precision Medicine. Front Genet (2020) 11:844–4. doi: 10.3389/fgene.2020.00844
244. Lamb YN, Dhillon S. Epi proColon® 2.0 CE: A Blood-Based Screening Test for Colorectal Cancer. Mol Diagn Ther (2017) 21(2):225–32. doi: 10.1007/s40291-017-0259-y
245. Trenti E, D'Elia C, Mian C, Schwienbacher C, Hanspeter E, Pycha A, et al. Diagnostic Predictive Value of the Bladder EpiCheck Test in the Follow-Up of Patients With non–Muscle-Invasive Bladder Cancer. Cancer Cytopathol (2019) 127(7):465–9. doi: 10.1002/cncy.22152
246. Aref-Eshghi E, Kerkhof J, Pedro VP, France GDI, Barat-Houari M, Ruiz-Pallares N, et al. Evaluation of DNA Methylation Episignatures for Diagnosis and Phenotype Correlations in 42 Mendelian Neurodevelopmental Disorders. Am J Hum Genet (2020) 106(3):356–70. doi: 10.1016/j.ajhg.2020.01.019
247. Sadikovic B, Levy MA, Aref-Eshghi E. Functional Annotation of Genomic Variation: DNA Methylation Episignatures in Neurodevelopmental Mendelian Disorders. Hum Mol Genet (2020) 29(R1):R27–32. doi: 10.1093/hmg/ddaa144
248. Matsunaga A, Hishima T, Tanaka N, Yamasaki M, Yoshida L, Mochizuki M, et al. DNA Methylation Profiling can Classify HIV-Associated Lymphomas. AIDS (2014) 28(4):503–10. doi: 10.1097/QAD.0000000000000120
249. Mays VM, So BT, Cochran SD, Detels R, Benjamin R, Allen E, et al. HIV Disease in Ethnic Minorities: Implications of Racial/Ethnic Differences in Disease Susceptibility and Drug Dosage Response for HIV Infection and Treatment. Handb Health Psychol (2001) 801–16.
Keywords: HIV, epigenetic regulation, DNA methylation, epigenome-wide methylation, epi-therapeutics
Citation: Arumugam T, Ramphal U, Adimulam T, Chinniah R and Ramsuran V (2021) Deciphering DNA Methylation in HIV Infection. Front. Immunol. 12:795121. doi: 10.3389/fimmu.2021.795121
Received: 14 October 2021; Accepted: 17 November 2021;
Published: 02 December 2021.
Edited by:
Esaki M. Shankar, Central University of Tamil Nadu, IndiaReviewed by:
Arumugam Rajavelu, Indian Institute of Technology Madras, IndiaThangavel Samikkannu, Texas A&M University Kingsville, United States
Copyright © 2021 Arumugam, Ramphal, Adimulam, Chinniah and Ramsuran. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Veron Ramsuran, RamsuranV@ukzn.ac.za
 Thilona Arumugam
Thilona Arumugam Upasana Ramphal
Upasana Ramphal Theolan Adimulam
Theolan Adimulam Romona Chinniah1
Romona Chinniah1 Veron Ramsuran
Veron Ramsuran