Ferroptosis-associated circular RNAs: Opportunities and challenges in the diagnosis and treatment of cancer
- 1Department of Clinical Laboratory, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, China
- 2Key Clinical Laboratory of Henan Province, Zhengzhou, China
Ferroptosis is an emerging form of non-apoptotic regulated cell death which is different from cell death mechanisms such as autophagy, apoptosis and necrosis. It is characterized by iron-dependent lipid peroxide accumulation. Circular RNA (circRNA) is a newly studied evolutionarily conserved type of non-coding RNA with a covalent closed-loop structure. It exhibits universality, conservatism, stability and particularity. At present, the functions that have been studied and found include microRNA sponge, protein scaffold, transcription regulation, translation and production of peptides, etc. CircRNA can be used as a biomarker of tumors and is a hotspot in RNA biology research. Studies have shown that ferroptosis can participate in tumor regulation through the circRNA molecular pathway and then affect cancer progression, which may become a direction of cancer diagnosis and treatment in the future. This paper reviews the molecular biological mechanism of ferroptosis and the role of circular RNA in tumors and summarizes the circRNA related to ferroptosis in tumors, which may inspire research prospects for the precise prevention and treatment of cancer in the future.
1 Introduction
According to statistics, cancer mortality rates have been rising in the 20th century, and if cancer mortality rates continue to escalate, there will be an additional 3.2 million cancer deaths annually (Siegel et al., 2021). In 2020, breast cancer replaced lung cancer for the first time to become the world’s most diagnosed cancer. According to statistics, China accounts for 30% of all cancer deaths worldwide. Lung cancer remains the most common type of cancer in China and is the main cause of high cancer mortality. However, in terms of incidence based on gender, breast cancer is the most prevalent type of cancer among women. At present, China is undergoing a period of cancer transformation, and the burdens of lung cancer, gastrointestinal cancer and breast cancer are increasing (Cao et al., 2021).
Ferroptosis is an emerging form of cell death. The rapid development of cancer-related ferroptosis research provides new prospects for its application in cancer treatment. The main biochemical features of ferroptosis are metabolism of the xCT system, inhibition of L-Glutathione (GSH) and glutathione peroxidase 4 (GPX4), accumulation of iron ions and the occurrence of lipid peroxidation (Mou et al., 2019). Ferroptosis occurs through two main pathways: an exogenous or transporter-dependent pathway [(e.g., solute carrier family 7 member 11 (SLC7A11) and transferrin receptor (TRFC)] and an endogenous or enzyme-regulated pathway [(e.g., acyl-CoA synthetase long-chain family member (ACSL4) and apoptosis inducing factor mitochondria associated 2 (AIFM2)]. During the onset of ferroptosis, polyunsaturated fatty acids (PUFAs) are sensitive to both enzymatic and non-enzymatic oxidation (e.g., iron-dependent Fenton reaction) (Tang et al., 2021). In recent years, a growing number of studies have identified a key role for ferroptosis in cancer progression, such as in gastric cancer (Zhang et al., 2020), melanoma (Ubellacker et al., 2020), lung cancer (Lou et al., 2021; Wang et al., 2019), and pancreatic cancer (Ye et al., 2021).
A large number of studies have revealed the extent of systematic changes in RNA in cancer. Importantly, widespread changes in coding and non-coding RNAs (ncRNAs) [(e.g., microRNA (miRNA), long non-coding RNA (lncRNA), small nuclear RNA (snRNA) and circular RNA (circRNA)] in cancer affect many aspects of tumorigenesis (Goodall and Wickramasinghe, 2021). cCircRNAs have covalently closed structures and high stability and are involved in gene regulation. CircRNA exhibits a covalent closed-loop structure, which is produced by a process called reverse splicing in the pre-mRNA phase (Vo et al., 2019). CircRNA was initially considered splicing-related noise. In addition, circRNAs are tissue-specific and cell-specific, with cis-acting elements and trans-acting factors involved in the regulation of their production (Kristensen et al., 2019). Existing studies have found that circRNA regulates cancer progression by regulating miRNA expression or protein function in many cell processes by acting as a miRNA sponge (Yin et al., 2019), protein bait (Zang et al., 2020), or encoding peptide (Wu et al., 2020), or through another mode of action (Yin et al., 2019). CircRNAs are aberrantly expressed in cancer progression, and these aberrantly regulated circRNAs play key roles in promoting or suppressing tumors. In addition, due to their high stability, abundance and specific expression patterns, circRNAs may become biomarkers and therapeutic targets for cancer patients (Li et al., 2020a). Interestingly, ferroptosis is also involved in the regulation of cancer progression by circular RNA. Ferroptosis-related circRNAs may offer new directions for future cancer therapy (Xie and Guo, 2021). This review mainly discusses the mechanism of ferroptosis and circRNAs involved in cancer regulation, as well as the research prospects and biological significance of ferroptosis-associated circRNAs in future cancer early diagnosis and precision therapy.
2 Ferroptosis
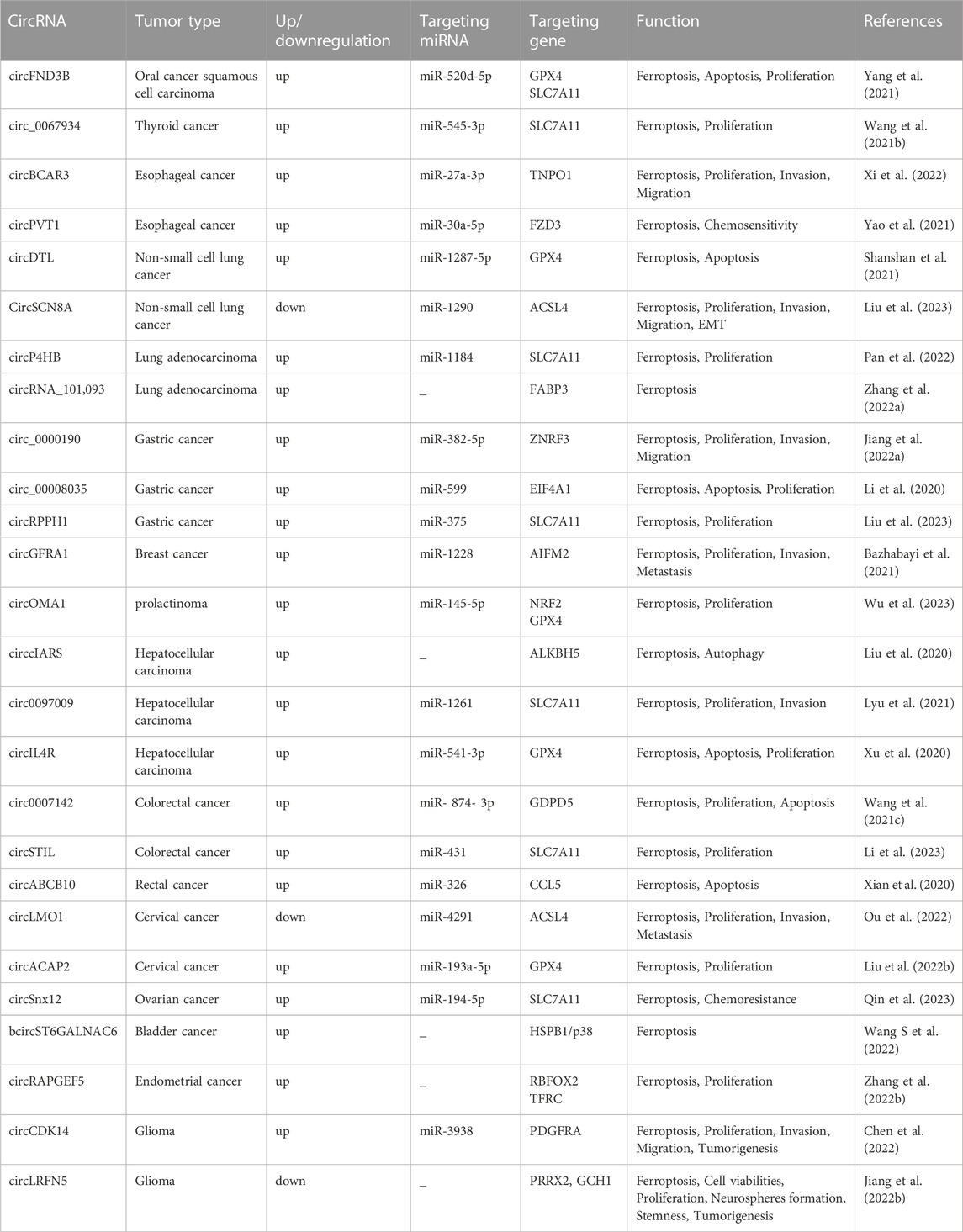
The definition of ferroptosis was first proposed by Dixon in 2012 and identifies it as an emerging mode of cell death (Dixon, 2017). Ferroptosis is mainly caused by high iron accumulation and lipid peroxidation. Iron-induced factors can affect glutathione peroxidase (GPx) levels through a variety of pathways, leading to elevated lipid reactive oxygen species (ROS) and a reduction in antioxidant capacity, ultimately leading to oxidative cell death (Li et al., 2020b). Ferroptosis has become a research hotspot in recent years. At present, the molecular mechanisms of ferroptosis are roughly divided into three pathways: deletion or activation of GPX4, iron metabolism and lipid peroxidation (Figure 1). Cysteine is introduced into the cell by the action of system xc-, while glutathione synthesis is supported by the action of glyoxylate carboligase (GCL) and glutathione synthetase (GSS) enzymes. At the same time, glutathione acts as a common substrate for GPX4, acting between the reduced and oxidized states. Inhibition of system xc- or GPX4 inactivation will result in lipid ROS accumulation and stimulate ferroptosis. The formation of ROS on lipids is mediated by hydrogen peroxide. PUFAs are incorporated into membrane PLs through the combined action of ACSL4 and lysophosphatidylcholine acyltransferase 3 (LPCAT3). Acyl-CoA synthetase long chain family member 3 (ACSL3) activates monounsaturated fatty acids (MUFAs), which compete with PUFAs for integration into PLs. Exogenous addition of MUFAs inhibits ferroptosis. In addition, lipophilic antioxidants and iron chelators may inhibit the occurrence of ferroptosis (Forcina and Dixon, 2019).
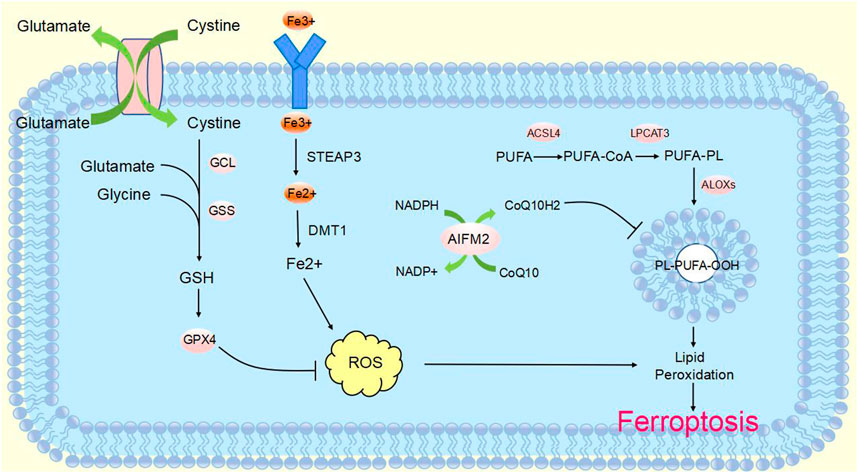
FIGURE 1. Molecular mechanisms of ferroptosisThe molecular mechanisms of ferroptosis can be roughly divided into three pathways. System xc-transports intracellular Glu to the extracellular space and extracellular Cys2 into the cell, which is then transformed into Cys for GSH synthesis. GPX4 ultimately reduces the accumulation of reactive oxygen species. The accumulation of lipid ROS leads to ferroptosis. Excess iron is the basis for ferroptosis execution. Circulated iron was combined with transferrin in the form of Fe3+, and then it entered into cells by TFR1. Iron in Fe3+ form was deoxidized to iron in Fe2+ by iron oxide reductase STEAP3. Ultimately, Fe2+ was released into a labile iron pool in the cytoplasm from the endosome mediated by DMT1, which led to ROS accumulation and eventual ferroptosis. ACSL4 and LPCAT3 promote the incorporation of PUFAs into phospholipids to form PUFA–PLs, which are vulnerable to free radical-initiated oxidation mediated by lipoxygenases (ALOXs). This ultimately causes lipid peroxidation and ferroptosis.
2.1 Mechanism of ferroptosis
2.1.1 System xc- and GPX4 in ferroptosis
GPX4 is a selenoprotein originally discovered by Ursini that acts as a catalyst for the reduction of Phospholipid hydroperoxides (PLOOHs) in cells. GPX4 is a key molecule in the regulation of ferroptosis which functions mainly through the inhibition of lipid peroxidation. GPX4 converts GSH to oxidized glutathione (GSSG) and reduces toxic lipid peroxide (L-OOH) to alcohols (L-OH) (Li et al., 2020b). Direct inhibition of GPX4 or GSH synthesis can trigger ferroptosis, and glutathione is a key regulator of GPX4 (Wu et al., 2021). Inhibition of system xc-leads to depletion of GSH and inactivation of GPX4, which increases lipid peroxidation. The Fenton reaction of Fe2+ takes place, oxidizing lipids, producing large amounts of ROS and ultimately stimulating the onset of ferroptosis (Friedmann Angeli et al., 2014) (Figure 1). The level of cellular lipid peroxidation can be increased by lipoxygenase in addition to neutralizing GPX4-mediated lipid peroxidation. Lipoxygenase and GPX4 play opposite roles in ferroptosis. Lipid peroxidation is catalyzed by lipoxygenase, whereas GPX4 inhibits lipid peroxidation (Stockwell et al., 2020). Ding et al. (2021) found that the anti-TNBC effect of DMOCPTL mainly acts to induce GPX4 ubiquitination by directly binding to the GPX4 protein and then induce ferroptosis and apoptosis. This is the first report of ferroptosis induced by GPX4 ubiquitination. These findings reveal for the first time a new way to induce ferroptosis through GPX4 ubiquitination and provide ideas for us to further study the mechanism of ferroptosis.
2.1.2 FSP1 in ferroptosis
AIFM2 was originally defined as a proapoptotic gene that resists ferroptosis caused by GPX4 deficiency. It was later named ferroptosis suppressor protein 1 (FSP1) (Doll et al., 2019; Wu et al., 2002). FSP1 functions as an oxidoreductase at the plasma membrane, reducing ubiquinone (also known as coenzyme Q10) to the lipophilic free radical scavenger ubiquinone, which limits the accumulation of lipid ROS in the membrane under conditions independent of the GPX4 pathway (Bebber et al., 2020; Dixon et al., 2012). Extramitochondrial ubiquinone, as a lipophilic radical-trapping antioxidant (RTA), is produced by FSP1 from CoQ10 via a reduction reaction and reduces lipid radical levels through the recycling of a-tocopherol (a-TTH) (Zheng and Conrad, 2020) (Figure 1). FSP1 functions synergistically with GPX4 inhibitors and has been shown to trigger ferroptosis in many cancers. FSP1 plays a key role in the non-mitochondrial antioxidant system independently of the common GSH-GPX4 pathway. This suggests the existence of a novel ferroptosis inhibitory pathway and that inhibition of FSP1 may render cancer patients more sensitive to ferroptosis-inducing chemotherapeutic agents, thereby affecting cancer progression (Bersuker et al., 2019).
2.1.3 Iron in ferroptosis
Studies have found that excessive iron accumulation is a key cause of ferroptosis. Redox-active iron pools can catalyze the production of damaging free radicals from lipid peroxides via the Fenton reaction. Once lipid peroxide is not removed in time within the cell, increasing levels of lipid alkoxyl radicals will eventually trigger ferroptosis (Lu et al., 2017) (Figure 1). The name ferroptosis indicates that the process is iron dependent: intracellular iron levels are positively correlated with ferroptosis and can be inhibited by iron chelators. Transcriptional changes in key genes regulated by iron [(e.g., TFRC, iron responsive element binding protein 2 (IREB2) and ferritin heavy chain 1 (FTH1)] will affect the sensitivity to ferroptosis induced by erastin (Ma et al., 2016; Gao et al., 2015). Multiple metabolic pathways (e.g., enhanced iron uptake, reduced iron accumulation and inhibition of iron efflux) will result in increased iron levels and promote iron uptake through integrated signaling pathways in animal models (Chen et al., 2020). Iron produces a large amount of ROS, which may promote tumor development. However, iron-dependent accumulation of reactive oxygen species can also lead to ferroptosis, which can inhibit tumor cells. Some researchers have proposed a hypothesis to explain this contradiction, namely, that redox signaling of iron and thiols is in dynamic equilibrium in cancer cells to help cells resist ferroptosis, but this idea requires more evidence and further research (Toyokuni et al., 2017). To date, there are three known pathways involved in the iron-dependent accumulation of lipid ROS in iron-induced blepharoptosis. First, ROS are produced by iron through the Fenton reaction, which is a non-enzymatic inorganic chemical reaction. Second, the reactive oxygen species produced by lipid self-oxidation are controlled by iron-catalyzed enzymes. Third, reactive oxygen species are produced by the oxidation of arachidonic acid (AA) by iron-containing oxygenase. Iron metabolism plays an important role in ferroptosis, and its regulatory mechanism still needs to be further studied (Lei et al., 2019). Sun et al. (2015) found that heat shock protein beta-1 (HSPB1) could further downregulate intracellular iron content by reducing the level of telomeric DNA-binding factor (TRF1). Thus, high levels of HSPB1, a key gene in ferroptosis, could hinder the development of ferroptosis. In addition, the expression of IREB2, the main transcription factor that inhibits iron metabolism, can significantly increase the expression of ferritin light chain (FTL) and FTH1, thereby inhibiting erastin-induced ferroptosis (Gammella et al., 2015). In addition, Fang et al. found that iron released from heme degradation, which is regulated by the involvement of heme oxygenase-1 (HO-1), is closely associated with ferroptosis. However, there are conflicting data showing that HO-1 can promote and inhibit ferroptosis (Fang et al., 2020). In conclusion, iron metabolism regulates a variety of genes and molecular pathways, affects sensitivity to ferroptosis, and plays an important role in ferroptosis.
2.1.4 Lipid peroxidation in ferroptosis
Lipid peroxidation also plays an important role in the progression of ferroptosis. Ferroptosis is triggered when an imbalance in the endogenous antioxidant system of the cell leads to massive accumulation of lipid ROS, causing toxicity and disruption of membrane structure. Thus, oxidative stress and cellular antioxidant levels are important regulators of lipid peroxidation that can induce ferroptosis (Kajarabille and Latunde-Dada, 2019). Oxidation and depletion of PUFAs may cause changes in membrane structure and fluidity, increasing membrane permeability and reducing membrane integrity. It is hypothesized that the thinning of the membrane and changes in curvature when ferroptosis occurs will reduce the stability of the membrane and ultimately lead to the formation of pores and micelles (Yang and Stockwell, 2016; Agmon et al., 2018; Snaebjornsson et al., 2020). Polyunsaturated fatty acid-containing phospholipid (PUFA-PL) oxidation has been demonstrated to stimulate ferroptosis. Genetic disruption of ACSL4 and LPCAT3 confirmed resistance to ferroptosis (Dixon et al., 2015). ACSL4 preferentially activates polyunsaturated fatty acids (such as AA) to incorporate lipids, while LPCAT3 preferentially incorporates polyunsaturated fatty acid coenzyme A into membrane phospholipids. Thus, deletion of ACSL4 and LPCAT3 can reduce the membrane burden of oxidizable PUFA-PLs and modulate ferroptosis (Shindou and Shimizu, 2009) (Figure 1). In addition to its typical function in the electron transport chain, CoQ10 can also “moonlight” with respect to lipophilic antioxidants. The loss of CoQ10 may induce or sensitize ferroptosis and participate in the regulation of ferroptosis to a certain extent (Shimada et al., 2016). Lysyl oxidase (LOX) also plays a role in the process of lipid peroxidation. In the absence of LOX enzyme activity, free intracellular iron is sufficient to catalyze Fenton chemistry on lipid peroxides to generate toxic hydroxyl or lipid alkoxy radicals to promote lipid peroxidation and ferroptosis (Kim et al., 2016). In addition, Sun et al. (2015) found that protein kinase C-mediated phosphorylation of HSPB1 could hinder lipid reactive oxygen species accumulation to counteract ferroptosis. Inhibition of the HSF1-HSPB1 pathway and phosphorylation of HSPB1 resulted in enhanced anticancer effects in xenograft mouse tumors. Phosphorylation of HSPB1 acts as an important inhibitor of ferroptosis, mainly by inhibiting iron uptake and lipid ROS accumulation. NFE2 like bZIP transcription factor 2 (NRF2) is also a key molecule in ferroptosis, and in tumor cells, NRF2 prevents the accumulation of lipid peroxides and lipid oxidation of target proteins for survival purposes. Although kelch like ECH associated protein 1 (KEAP1) can produce an anti-ferroptosis effect by activating NRF2, many active lipids are in turn involved in inhibiting the function of NRF2 target genes, which in turn cannot be ignored in the ferroptosis cascade response (Dodson et al., 2019). In addition, GPX4 acts as an antioxidant enzyme, and its stimulation or inactivation can also modulate ferroptosis by affecting lipid peroxidation. Glutathione, a coregulator of GPX4, prevents peroxidation of cells and membranes, thereby neutralizing lipid peroxidation and ensuring membrane fluidity. Inhibition of GPX4 leads to increased levels of ROS, while overexpression of GPX4 downregulates ROS, thereby inhibiting the onset of ferroptosis (Ribas et al., 2014; Yang et al., 2014; Su et al., 2019). The key mechanisms of ferroptosis are iron accumulation, lipid peroxidation, and GPX4 regulation. The mechanisms of ferroptosis are very complex, and more details need to be explored in depth.
3 Function of CircRNA in cancer
CircRNA is a non-coding RNA that is widely found in eukaryotes. It was originally defined as a product of abnormal RNA splicing and was also referred to as “transcriptional noise”. However, with the development of high-throughput genomic technologies, circular RNA can be studied in depth (Nisar et al., 2021). CircRNAs are made from pre-mRNA by back-splicing, where the downstream 3′splice site is linked to the upstream 5′splice site, which in turn is cyclized to form a covalent closed loop. They can be formed from introns or one or more exons. In addition, they are not susceptible to degradation by exonucleases, as they lack 3′and 5′ends (Zhang et al., 2014; Fontemaggi et al., 2021) (Figure 2). CircRNA biogenesis is mediated by four main mechanisms: intron base-pairing-driven cyclization (Figure 2A), RBP-driven cyclization (Figure 2B), lariat-driven cyclization (Figure 2C), and Pre-tRNAs generate tricRNAs (Figure 2D) (Wang et al., 2021a; Hu et al., 2023; Harper et al., 2021). Circular RNAs are classified into three main types according to their constituents: ciRNA, EIciRNA and exonic circRNA, which account for 85% of circular RNAs (Fontemaggi et al., 2021; Eger et al., 2018).
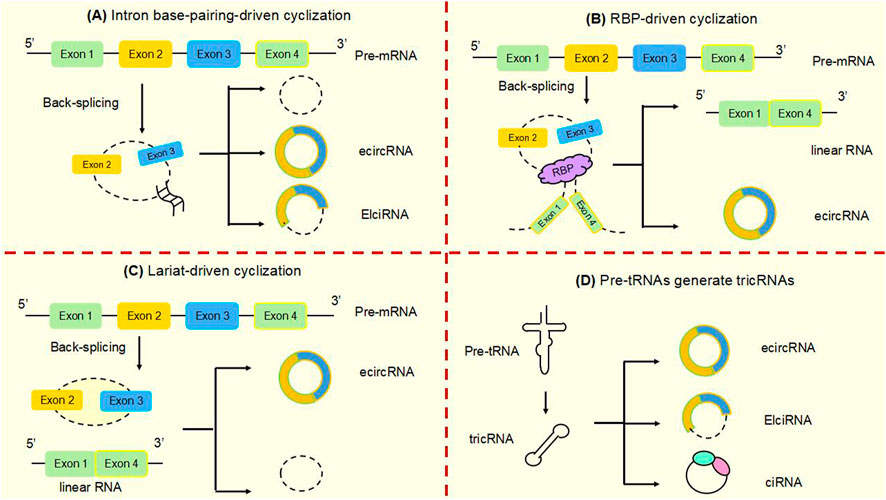
FIGURE 2. Biogenesis of circular RNAsCircRNAs are mainly produced from primary transcripts by back-splicing. The biogenesis of cyclic RNA is generated by four main mechanisms: (A) Intron base-pairing driven cyclization. Flanking intron complementation occurs by base pairing, allowing splice sites to join and drive cyclization. (B) RBP-driven cyclization. RBPs drive the cyclization of pre-mRNAs by binding to introns flanking introns in linear transcripts, leading to closer proximity of the splice site. (C) Lariat-driven cyclization. Back-splicing forms a lasso containing introns and skipped exons. Introns are spliced and subsequently form ecircRNA. (D) Pre-tRNA produces tricRNA. Pre-tRNA containing introns is sheared to produce triple RNA, then intron ends join and form triple RNA.
An increasing number of studies have shown that specific circular RNAs have the characteristics of carcinogenesis or tumor inhibition in the context of human cancers including gastric cancer (Zhang et al., 2019), non-small cell lung cancer (Chen et al., 2019a), breast cancer (Wang et al., 2018), bladder cancer (Cheng et al., 2021), and esophageal cancer (Meng et al., 2021). The specific expression and high stability of circRNAs in cells makes them a potential new target in cancer therapy (Fontemaggi et al., 2021). Therefore, further understanding of the biological functions and roles of circRNAs in different cancers, as well as the detection of signaling pathways, is important for the early diagnosis and precise treatment of cancer.
3.1 Role of CircRNA as MiRNA sponge in cancer
Studies have shown that circular RNAs can act as miRNA sponges and gene transcripts and that interactions between circRNAs and proteins regulate gene expression. Moreover, circRNA can be translated into protein. Many previous studies have found that circRNA disorders manifest different functions and roles in cancer (Figure 3). However, the most widely studied is the role of miRNA sponge. CircRNA ciRS-7 was the first circRNA identified as a miR sponge, containing more than 70 miR-7 binding sites, ciRS-7 expression significantly repressed miR-7, resulting in reduced miR-7 activity and increased levels of miR-7-targeted transcripts, acting as a miR-7 inhibitor (Hansen et al., 2013a; Hansen et al., 2013b) (Figure 3A). found that circNRIP1, a sponge for miR-595, inhibited the oncogenic effects of esophageal cancer cell growth, migration and invasion through competitive binding of circNRIP1 and miR-595 to target the semaphorin 4D (SEMA4D) and PI3K/Akt signaling pathways in esophageal cancer cells (Zhou et al., 2021). Similarly, in hepatocellular carcinoma, circFBLIM1 can act as a competitive endogenous RNA (ceRNA) to regulate filamin binding LIM protein 1 (FBLIM1) expression levels through the sponge miR-346. CircFBLIM1 may become a biomarker and therapeutic target for hepatocellular carcinoma (Bai et al., 2018). Chen et al. also observed this function of circular RNA in bladder urothelial carcinoma. CircPRMT5 can absorb miR-30c by acting as a sponge and promote EMT in Urinary bladder tumor cells. CircPRMT5 is a key regulator in promoting EMT and invasiveness in UCB cells. Taken together, a wealth of experiments validate that circRNAs can act as miRNA sponges in competitively binding downstream target genes to influence cancer progression: this is one of the most common functions of circRNAs.
FIGURE 3. The main functions of circular RNAsAt present, several well-known functions of circular RNAs are as follows. (A) MiRNA sponges. CircRNAs can act as miRNA molecule sponges to prevent their binding to mRNA targets, which is the most widely studied function of circRNAs. (B) Protein decoys. CircRNAs can regulate the function of proteins by working as protein decoys or scaffolds. (C) Interact with mRNAs. CircRNA can directly bind mRNA, enhance mRNA stability and regulate gene expression. (D) Form functional circRNP complexes. CircRNA can bind proteins to form circRNP complexes that regulate downstream signaling pathways. (E) Regulating translation. CircRNAs can regulate translation, and m6A-mediated and internal ribosome entry sites (IRES) are the main mechanisms by which circRNAs initiate translation. (F) CircRNAs in exosomes. CircRNAs can be loaded into exosomes, resulting in exo-circRNAs that function as messenger RNAs for cellular communication by transferring them to recipient cells or neighboring cells. (G) Modulating transcription and splicing. CircRNA interacts with Pol II and snRNP, thereby regulating the transcription and splicing of parental genes. (H) Protein Recruitment. CircRNA can recruit proteins to specific motifs, promote protein assembly, and regulate gene expression.
3.2 Interaction between CircRNA and RBP regulates cancer development
RNA binding proteins (RBPs) are a class of proteins that specifically recognize RNA binding regions and bind to each other (Tang and Hann, 2020). RBPs are widely involved in gene transcription and translation and control the production, maturation, localization, translation and degradation of cellular RNA (Okholm et al., 2020; Dominguez et al., 2018). As circular RNAs have been studied in depth, an increasing number of studies have confirmed that circRNAs can interact with RBPs to regulate cancer progression.
Circ-cux1 can promote aerobic glycolysis and increase the invasiveness of neuroblastoma cells by interacting with EWS RNA binding proteins. It also promotes association with myc-associated zinc finger protein (MAS) and stimulates enhanced transcriptional activity of MAS, suggesting that circ-cux1 is involved in regulating neuroblastoma progression by targeting myc associated zinc finger protein (MAZ) through binding to EWS RNA binding protein 1 (EWSR1) (Li et al., 2019). Furthermore, in lung adenocarcinoma, circdcun1d4 can bind the HuR protein, which elevates HuR activity, and acts as a scaffold to interact with thioredoxin interacting protein (TXNIP) mRNA, increasing the stability of TXNIP mRNA, which then plays a protective role in lung adenocarcinoma (Liang et al., 2021). reported that circnsun2 binds to insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) to target high mobility group AT-hook 2 (HMGA2), forming the circnsun2/IGF2BP2/HMGA2 RNA protein ternary complex, which is involved in promoting colorectal carcinoma formation (Chen et al., 2019b). Lu et al. (2020) found that heterogeneous ribonucleoprotein L (HnRNP-L) is a multifunctional RBP and that overexpression of HnRNP-L is involved in upregulating circCSPP1 in combination with miR-520h to target early growth response 1 (EGR1), promoting proliferation and apoptosis of prostate tumor cells and leading to increased progression of prostate cancer (Lu et al., 2021). HuR is a widely studied RBP. found that cIARS can interact with alkB homolog 5, RNA demethylase (ALKBH5) to act as a negative regulator in hepatocellular carcinoma and that downregulation of ALKBH5 significantly inhibited si-CIARS-induced iron deposition events, autophagic flux and ferritin phagocytosis (Liu et al., 2020) (Figure 3B). In conclusion, these findings suggest that circRNAs, as RBP sponges, interact with specific protein subunits to affect cancer progression, and more mechanisms need to be found.
3.3 Regulation of mRNA stability
CircRNAs have variety of biological functions and their functions as miRNA sponges and protein scaffolds interacting with RBPs have been extensively studied. In addition, circRNAs can directly bind mRNAs to affect gene expression levels, and regulate mRNA stability. CircRNA zinc finger protein 609 (circZNF609) has a role in regulating mRNA stability. Previous studies have shown that circZNF609 is a circRNA capable of promoting myogenic cell proliferation and regulating the malignant progression of rhabdomyosarcoma (RMS), a childhood skeletal muscle malignancy (Rossi et al., 2019). CircZNF609 was found by Rossi et al. to regulate cytoskeleton-associated protein 5 (CKAP5) mRNA stability and mediate tumor progression. Specifically, circZNF609 has multiple binding sites in ELAV-like RNA binding protein 1 (ELAVL1) (Beltran et al., 2022). CircZNF609 interacts with CKAP5 transcripts to promote ELAVL1 binding to mRNA, stimulate mRNA levels and translation, enhance mRNA stability and lead to changes in microtubule (MT) dynamics, thereby induce tumor cell proliferation.
Similarly, in cervical cancer cells, hsa circ0032434 (circPCNX) also plays a role in regulating mRNA stability. CircPCNX is an abundant target of F-box family protein (AUF1). Overexpression of circPCNX specifically blocks the association of AUF1 with p21 (CDKN1A) mRNA, thereby enhancing the stability of p21 mRNA and increasing the level of p21, which acts as a major inhibitor of cell proliferation, inhibiting the proliferation of cervical cancer cells and impeding the malignant development of cervical cancer (Tsitsipatis et al., 2021) (Figure 3C).
3.4 Role of functional circRNP complexes
CircRNA can also form functional circRNP complexes that regulate downstream signaling pathways and influence tumorigenesis. CircFoxo3 can affect tumor proliferation by regulating cyclins. CircFoxo3 forms the circFoxo3-p21-CDK2 ternary complex by binding to the cell cycle protein cell cycle protein-dependent kinase 2 (CDK2) and cell cycle protein kinase inhibitor 1 (p21), which cuts off the transition from G1 to S phase and eliminates the effect of p21 on the inhibitory effect of p21 on the cyclin A/CDK2 complex causes cells to stagnate in G1 phase and fail to transition to S phase, preventing normal cell cycle progression. CircFoxo3 interacts with p21 and CDK2 to form a functional circRNP complex that affects tumor cell proliferation and may be a direction for cancer therapy (Du et al., 2016) (Figure 3D).
CircRNA SCAR forms a SCAR-ATP synthase, H+ transporting mitochondrial F1 complex, beta subunit (ATP5B) scaffold in the mPTP complex, and ATP5B is a regulator of the mitochondrial permeability transition pore (mPTP) complex to block mPTP opening and inhibit mROS export to maintain mROS homeostasis in vivo. CircSCAR overexpression inhibits mROS in non-alcoholic steatohepatitis (NASH) generation and eliminates mROS export after lipid processing. This mitochondrial circRNA forms a functional circRNP complex that may be a therapeutic target for NASH (Zhao et al., 2020).
3.5 Role of circRNA in regulating translation
In addition to the functions of circRNAs described above, circRNAs can also regulate the translation of proteins or polypeptides. N6-methyladenosine (m6A)-mediated and internal ribosome entry sites (IRES) are the main mechanisms by which circRNAs initiate translation. The most abundant base modification of m6A RNA promotes efficient initiation of circRNA protein translation in human cells. M6A-driven translation requires the initiation factor eukaryotic translation initiation factor 4 gamma 2 (EIF4G2) and the m6A reader YTH N6-methyladenosine RNA binding protein F3 (YTHDF3) and is enhanced by the methyltransferase methyltransferase 3, N6-adenosine-methyltransferase complex catalytic subunit (METTL3), inhibited by the demethylase alpha-ketoglutarate dependent dioxygenase (FTO), and upregulated in response to heat shock (Hu et al., 2023; Yang et al., 2017; Lei et al., 2020) (Figure 3E). In the cytoplasm, the m6A-binding protein YTHDF1 promotes the translation of m6A-modified mRNA, and YTHDF2 accelerates the decay of m6A-modified transcripts (Shi et al., 2017). Song et al. (2022) found that eukaryotic translation initiation factor 3 subunit j (eIF3j) inhibits the translation process by inducing the dissociation of the eIF3 complex from circSfl. The C-terminal of eIF3j and the RNA regulators in the circS fluoride translation region (UTR) are key factors in the inhibition of eIF3j, and reveal the physiological relevance of eIF3j mediated circSfl translation inhibition to heat shock response. This provides a new direction for the cap less translation regulation of eIF3 and the involvement of circRNA in regulating translation.
3.6 Exosome-circRNAs (exo-circRNAs)
Exosomes play the role of messengers for intercellular communication. CircRNAs can be wrapped into exosomes to form exocytotic circRNAs that communicate by transferring to recipient cells or adjacent cells (Bai et al., 2019). The exosome circRNA_101093 (cir93) maintains elevated levels of intracellular cir93 in lung adenocarcinoma to regulate AA and inhibit the onset of ferroptosis. Exosomes and cir93 play an important role in the development of ferroptosis in lung adenocarcinoma, and blocking exosomes may be an approach to lung adenocarcinoma treatment (Zhang et al., 2022a) (Figure 3F). In colorectal cancer, circLPAR1 was detected as loaded in exosomes and its expression was reduced in plasma exosomes but recovered after surgical treatment. Exosomal circLPAR1 was internalized and direct binding of eIF3h by exosomal circLPAR1 blocked METTL3-eIF3h binding, inhibited translation of oncogene bromodomain containing 4 (BRD4), and reduced malignant progression of colorectal cancer (Zheng and Conrad, 2020). Exosomal circRNA has been shown to have important functions in cancer, and further mechanisms await our discovery.
3.7 Modulating transcription and splicing
Besides to their roles in the cytoplasm, circRNAs are also key factors in the regulation of transcription and splicing in the nucleus. Some circRNAs are now known to interact with RNA polymerase II (Pol II), thereby regulating the transcription and splicing of parental genes. Li et al. (2015) demonstrated that in the nucleolus, circRNAs can regulate the expression of nuclear genes, where EIciRNAs promote the transcription of parental genes in a cis manner by interacting with u U1 small nuclear ribonucleoprotein (snRNP) or binding to the Pol II promoter (Figure 3G). Ciankrd52, as a ciRNA, has a stronger R-loop formation capacity than its homologous pre-mRNA. Pre-mRNA is released from the R-loop by ciankrd52 substitution and ciRNA is removed by RNase H1 to achieve significant transcriptional elongation. This R-loop-dependent ciRNA degradation represses ciRNA levels by recruiting RNase H1 on the one hand, and resolves R-loop transcriptional elongation at some GC-rich ciRNA-producing motifs on the other (Li et al., 2021a).
CircRNA can also regulate gene shearing. SEPALATA3 (SEP3) is a homologous MADS box TF required for the homozygous heterozygous phenotype of Arabidopsis flowers. Exon 6 of SEP3 generates circSEP3 by reverse splicing (Conn et al., 2017). loop, whereas the linear RNA equivalent binds weakly to DNA in comparison. The production of R-loop represses transcription, splicing factor recruitment and AS2-4 are consistent. Taken together, this suggests that circRNAs may form R-loops more efficiently than linear RNAs to regulate gene transcription and splicing (Liu et al., 2022a).
3.8 Protein Recruitment
CircRNA interacts with proteins in a variety of ways. On the one hand, circRNAs act as protein scaffolds and facilitate the interactions between enzymes and substrates. On the other hand, circRNAs can also recruit proteins to specific motifs and stimulate protein assembly. CircMbl, a circular RNA with the first demonstrated protein sponge function, is involved in regulating the feedback pathway between the multifunctional proteotoxic mushroom (MBL) and circMb1. MBL promotes reverse shearing of circMbl by binding to a circMbl flanking intron containing a conserved MBL binding site. This suggests that the level of MBL can regulate the production of circMbl (Ashwal-Fluss et al., 2014).
Moreover, circNDUFB2 also functions as a protein scaffold, facilitating the interaction of tripartite motif containing 25 (TRIM25) and IGF2BPs to form a TRIM25/circNDUFB2/IGF2BPs ternary complex that induces ubiquitination and degradation of IGF2BPs, an effect enhanced by m6A modification of circNDUFB2. CircNDUFB2 regulates the development of non-small cell lung cancer by regulating the ubiquitination and degradation of IGF2BP2, confirming its role as a protein scaffold (Li et al., 2021b) (Figure 3H).
4 Ferroptosis related CircRNA involvement in regulating cancer progression
In addition to the fact that circRNAs can influence cancer progression by regulating cell death (e.g., cell autophagy, apoptosis), recent studies have confirmed that circular RNAs can promote or inhibit ferroptosis by regulating key ferroptosis proteins after transcription and regulate tumor progression in various cancers through different molecular pathways. Thus, ferroptosis involving circRNAs may become a novel approach for cancer therapy (Xie and Guo, 2021) (Table 1) (Figure 4).
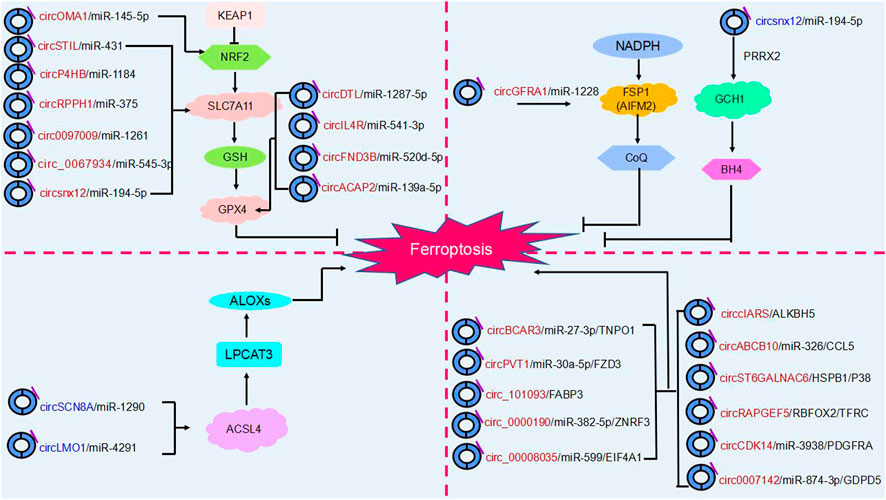
FIGURE 4. The regulatory role of circRNAs in ferroptosis. Ferroptosis is mainly regulated by four molecular pathways: the glutathione pathway, the ubiquinone pathway (NADPH- FSP1-CoQ10]], the tetrahydrobiopterin (GCH1- BH4] pathway and (ACSL4- LPCAT3- PUFA-PLs). As shown in the figure, circRNAs are involved in cancer progression by participating in four major molecular pathways regulating ferroptosis. Red circRNAs are upregulated in cancers, and blue circRNAs are downregulated in cancers.
4.1 Oral squamous cell carcinoma
The role of circFNDC3B in oral squamous cell carcinoma has been demonstrated. CircFNDC3B acts as a pro-oncogene in oral squamous cell carcinoma. It was found that knockdown of circFNDC3B resulted in elevated Fe2+, ROS accumulation, and downregulation of GPX4 and SLC7A11 expression levels, which stimulated ferroptosis and inhibited oral squamous cell carcinoma development. CircFNDC3B acts as a ceRNA binding miR-520d-5p to regulate SLC7A11. Overexpression of SLC7A11 reversed the ferroptosis caused by circFNDC3B depletion. CircFNDC3B may be a potential therapeutic target for oral squamous cell carcinoma (Yang et al., 2021).
4.2 Thyroid cancer
Circ_0067934 has been found to play a pro-carcinogenic role in a variety of cancers, and it was found that in thyroid cancer, knockdown of circ_0067934 enhanced Fe2+, intracellular iron and ROS levels and promoted the development of ferroptosis, while inhibition of miR-545-3p and overexpression of SLC7A11 reversed this effect. circ_0067934 competitively binding to miR-545-3p upregulates SLC7A11 to inhibit ferroptosis, promotes thyroid cancer development and advances thyroid cancer treatment (Wang et al., 2021b).
4.3 Esophageal cancer
CircBCAR3 was found to interact with miR-27a-3p to upregulate transportin 1 (TNPO1) expression, promote esophageal cancer cell proliferation, migration, invasion and ferroptosis, and play an oncogenic role in esophageal cancer by promoting esophageal tumorigenesis and metastasis in mice (Xi et al., 2022).
CircPVT1 also plays a role in esophageal cancer as a circular RNA. CircPVT1 sponge miR-30a-5p targets and binds frizzled class receptor 3 (FZD3), affects the expression levels of p-β-catenin, GPX4 and SLC7A11, inhibits cellular ferroptosis, affects esophageal cancer 5-FU chemotherapy sensitivity and promotes the development of esophageal cancer. This could be a potential target for esophageal cancer therapy (Yao et al., 2021).
4.4 Lung cancer
In China, where the incidence and mortality of lung cancer are high (Wang et al., 2020). Showed that downregulation of circDTL promotes ferroptosis in non-small cell lung cancer cells. Mechanistically, circDTL targets GPX4 via the circDTL sponge miR1287-5p to inhibit the onset of cellular ferroptosis, which in turn exerts its tumorigenic effects (Shanshan et al., 2021).
CircP4HB was found to be a novel ferritinase inhibitor in lung adenocarcinoma. CircP4HB affects GSH synthesis by targeting SLC7A11 through the sponge miR-1184, inhibiting ferroptosis and promoting tumor cell growth. circP4HB may be used as a molecular target for the treatment of lung cancer in the future (Pan et al., 2022).
Similarly, the exosome circRNA_101093 (cir93) plays an important role in ferroptosis and regulates the development of lung adenocarcinoma. Overexpression of cir93 inhibited erastin-induced ferroptosis and reduced lipid peroxidation to inhibit ferroptosis occurrence by upregulating fatty acid binding protein 3 (FABP3) expression. In addition, cir93 interacted with FABP3 thereby regulating NAT (a product of taurine and AA) to inhibit ACSL4, LPCAT3 and phospholipid transfer protein (PLTP) to reduce AA impeding ferroptosis progression. This may hold promise for future lung adenocarcinoma treatment (Zhang et al., 2022a).
In lung cancer, circSCN8A overexpression increased Fe2+, ROS and MDA levels and decreased GSH levels. Western blotting demonstrated that circSCN8A overexpression downregulated SLC7A11 and GPX4 protein levels and promoted ferroptosis, which was reversed by the addition of Ferrostatin-1 (Fer-1), an ferroptosis inhibitor. In addition, circSCN8A bound miR-1290 to enhance ACSL4 expression to promote lung cancer proliferation and metastasis and inhibit ferroptosis. Thus, circSCN8A may be a promising therapeutic target against lung cancer (Liu et al., 2023).
4.5 Gastric cancer
Jiang et al. (2022a) found that circ_ 0000190 targeted the tumor suppressor zinc and ring finger 3 (ZNRF3) by competitively binding to miR-382-5p, inhibited gastric cancer cell proliferation, migration and invasion, and stimulated ferroptosis to occur, ultimately leading to inhibition of gastric cancer progression. Upregulation of circ_0000199 or ZNRF3 expression may be an effective strategy for the treatment of GC.
The role of Circ_0008035 in gastric cancer has been confirmed by several studies. However, how Circ_0008035 regulates ferroptosis has not been reported yet. Li et al. (2020) found knockdown of circ_0008035 could promote ferroptosis, and circ_0008035 acted as a sponge for miR-599 and suppressed the expression of its target gene eukaryotic translation initiation factor 4A1 (EIF4A1). Overexpression of EIF4A1 reversed the effects of knockdown of circ_0008035 on iron accumulation, lipid peroxidation and mitochondrial membrane potential in gastric cancer cells. In conclusion, the circ_0008035/miR-599/EIF4A1 axis could regulate the occurrence of ferroptosis in gastric cancer cells and thus affect gastric cancer progression. Moreover, Circ0008035 was found to be another pathway regulating cellular ferroptosis affecting gastric cancer progression. Gao et al. found that circ0008035 increased E2F transcription factor 2 (E2F2) expression by acting as a ceRNA for miR-302a. Circ0008035 inhibited dextromethorphan-induced ferroptosis, and overexpression of miR-302a or knockdown of E2F transcription factor 7 (E2F7) effectively reversed this effect (Gao and Wang, 2023). In other words, circ0008035 can affect ferroptosis regulation of tumorigenesis through two independent parallel pathways, which provides a viable therapeutic option for gastric cancer.
CircRPPH1 also plays a regulatory role in gastric cancer. CircRPPH1 as a miR-375 sponge positively regulates SLC7A11 expression and has been shown to be a direct target of miR-377 in cancer, thereby regulating cellular ferroptosis. Knockdown of circRPPH1 promoted GSH synthesis, caused lipid peroxidation, induced cellular ferroptosis, and inhibited gastric cancer progression. And inhibition of miR-375 or overexpression of SLC7A11 partially reversed this effect. This may be a future research direction for gastric cancer treatment (Liu et al., 2022b).
4.6 Breast cancer
Breast cancer is the most prevalent cancer in women. In breast cancer, circGFRA1b enhances the activity of AIFM2 by binding to miR-1228, and the expression of GPX4 and the GSH/GSSG ratio are upregulated. In other words, HER2-positive breast cancer cells can inhibit ferroptosis by activating two pathways. Therapies targeting these two pathways may be able to inhibit breast cancer progression (Bazhabayi et al., 2021).
4.7 Prolactinoma
CircOMA1 is a pro-oncogene. CircOMA1 was first identified to regulate ferroptosis to affect proliferation of prolactinomas. CircOMA1 acts as a miR-145-5p sponge to regulate the expression level of glutamate-cysteine ligase, modifier subunit (GCLM), inhibit ferroptosis and promote tumor development. Moreover, circOMA1 also impedes ferroptosis by upregulating the expression of NRF2, GPX4 and xCT, key genes of ferroptosis. In other words, circOMA1 inhibits ferroptosis and promotes the development of prolactinoma through two parallel and independent pathways, which provides a new pathway for the treatment of prolactinoma (Wu et al., 2023).
4.8 Hepatocellular carcinoma
In hepatocellular carcinoma, the involvement of the circRNA cIARS in the regulation of ferroptosis through the inhibition of ALKBH5 was demonstrated by rescue experiments to influence cancer progression and represents a key point in the treatment of hepatocellular carcinoma (Liu et al., 2020).
SLC7A11 is a key regulator of iron uptake by cancer cells. Downregulation of the circ0097009 gene promotes iron uptake by hepatocellular carcinoma cells through two independent pathways: system xc- and GPX4. This study confirms that circ0097009 and miR-1261 competitively bind to target SLC7A11 to mediate hepatocellular carcinoma progression through regulation of ferroptosis (Lyu et al., 2021).
In addition, knockdown of circIL4R in hepatocellular carcinoma upregulated cellular iron level and oxidative stress and promoted erastin-induced ferroptosis. CircIL4R competitively bound to miR-541-3p, and inhibition of miR-541-3p replied to the tumor suppression caused by circIL4R knockdown and activated ferroptosis. GPX4 plays an important role in regulating ferroptosis as a target gene of miR-541-3p. In conclusion, circIL4R inhibits ferroptosis through miR-541-3p/GPX4 axis and promotes hepatocellular carcinogenesis (Xu et al., 2020).
4.9 Colorectal cancer
Circ_0007142 is overexpressed in colorectal cancer and targets glycerophosphodiester phosphodiesterase domain containing 5 (GDPD5) through binding to miR-874-3p, triggering the onset of ferroptosis and thus inhibiting the progression of colorectal cancer. In conclusion, demonstrated that circ_0007142 can be involved in the regulation of ferroptosis and colorectal cancer progression. Circ_0007142 may be a ferritinase marker for the treatment of colorectal cancer (Wang et al., 2021b).
Similarly, circSTIL plays an important role in colorectal cancer. Knockdown of circSTIL induces cellular Fe2+ and lipid ROS accumulation, activates cellular ferroptosis, and inhibits colorectal cancer cell proliferation. In addition, circSTIL targets SLC7A11 by competitively binding to miR-431 to regulate ferroptosis, providing a new target for the treatment of colorectal cancer (Li et al., 2023).
4.10 Rectal cancer
Numerous studies have shown that circABCB10 plays an important role in a variety of cancers. However, the reports of circABCB10 on ferroptosis are still relatively few and deserve further consideration and research. Xian et al. (2020) found that knockdown of circABCB10 in vitro stimulated ferroptosis in colorectal carcinoma cells. CircABCB10 acts as a miR-326 sponge competing for binding to C-C motif chemokine ligand 5 (CCL5), regulating the expression level of CCL5, and inhibition of miR-326 partially reversed circABCB10-induced ferroptosis. In conclusion, knockdown of circABCB10 promotes ferroptosis in colorectal carcinoma cells through miR-326/CCL5 axis and inhibits the further development of colorectal cancer, providing a new perspective on the progression of colorectal cancer.
4.11 Cervical cancer
In addition, circLMO1 inhibits the growth of cervical cancer cells by adsorbing miR-4192 to repress the target gene ACSL4, which promotes ferroptosis in cervical cancer cells. In conclusion, circLMO1 is expressed at low levels in cervical cancer, and overexpression of circLMO1 may be useful in cervical cancer treatment (Ou et al., 2022).
Similarly, circACAP2 plays a ceRNA role in cervical cancer cells, acting directly with miR-193a-5p to target GPX4, a key protein for ferroptosis, to inhibit ROS accumulation and intracellular iron levels. This suggests that circACAP2 inhibits ferroptosis and promotes the malignant progression of cervical cancer (Liu and Chen, 2022). In conclusion, circRNA related ferroptosis is involved in the regulation of cancer progression, which provides a new idea for cancer targeted therapy. However, the current understanding of its mechanism is not sufficiently comprehensive and thorough. Therefore, further study is urgently required.
4.12 Ovarian cancer
Cisplatin chemotherapy resistance (DDP) is one of the modalities of ovarian cancer treatment, however, it was found that knockdown of circSnx12 stimulated ferroptosis production and enhanced cisplatin sensitivity. In addition, circSnx12 inhibits ferroptosis and promotes ovarian cancer progression by targeting SLC7A11 through the sponge miR-194-5p. In conclusion, circSnx12 can regulate ferroptosis and chemoresistance, which are crucial for the treatment of ovarian cancer (Qin et al., 2023).
4.13 Bladder cancer
CircST6GALNAC6 plays an oncogene in bladder cancer, however, whether circST6GALNAC6 has a regulatory role on ferroptosis is currently unknown. Wang et al. first found that circST6GALNAC6 could directly bind to HSPB1, blocking erastin-induced phosphorylation of HSPB1 and promoting cellular ferroptosis. CircST6GALNAC6 could also inhibit HSPB1 to suppress bladder cancer cell proliferation and promote cellular ferroptosis by activating the P38 MAPK signaling pathway (Wang L et al., 2022; Wang S et al., 2022b).
4.14 Endometrial cancer
It was found that in endometrial cancer, circRAPGEF5 acts with RNA binding fox-1 homolog 2 (RBFOX2) to block the binding of RBFOX2 to pre-mRNA and inhibit TFRC expression. Overexpression of circRAPGEF5 leads to a reduction in the pool of unstable iron and a reduction in lipid peroxidation in endometrial cancer cells, resulting in resistance to ferroptosis. These findings may validate the emerging mechanism by which circRNA mediates ferroptosis through regulation of selective splicing (Zhang X. et al, 2022).
4.15 Glioma
In glioma, circCDK14 was found to reduce the likelihood of ferroptosis occurring in glioma cells by regulating the expression levels of platelet derived growth factor receptor alpha (PDGFRA), which is closely associated with poor prognosis in glioma patients and promotes the malignant progression of gliomas (Chen et al., 2022). CircLRFN5 can mediate paired related homeobox 2 (PRRX2) via the ubiquitin proteasome pathway, and in gliomas, PRRX2 activates transcriptional upregulation of GTP cyclohydrolase 1 (GCH1) expression, a substance that inhibits ferroptosis by producing the antioxidant tetrahydrobiopterin (BH4). This implies that circLRFN5 can mediate ferroptosis in glioma (Jiang et al., 2022b).
5 Conclusion and future perspective
Tumors are a disease type commanding substantial interest worldwide. The diagnostic and therapeutic targets of tumors have been a hot topic of research in this field. In recent years, various studies have confirmed that ferroptosis can bind non-coding RNAs to regulate tumors. MiRNAs and lncRNAs have been extensively studied. As an emerging ncRNA, circRNAs have been shown to be associated with ferroptosis.
Overall, ferroptosis is mainly regulated through four independent parallel molecular pathways (Figure 4). The first is the glutathione pathway. For example, the circRNA circACAP2 promotes the malignant progression of cervical cancer by competitively binding to and targeting the GPX4 ferroptosis key protein and inhibiting the onset of ferroptosis (Liu J. et al, 2022). The second pathway is the ubiquinone pathway (NADPH-FSP1-CoQ10). CircGFRA1 binding to miR-1228 enhances FSP1 expression to inhibit ferroptosis and promote the development of HER2-positive breast cancer (Bazhabayi et al., 2021). Third, in the GCH1-BH4 pathway, circLRFN5 regulates GCH1-activated ferroptosis via PRRX2 and inhibits glioblastoma progression (Jiang et al., 2022b). Moreover, ACSL4 and LPCAT3 mediate the depletion of PUFA-PLs. The circular RNA circLMO1 regulates ACSL4 by triggering miR-4291, which induces ferroptosis and suppresses cervical cancer malignancy (Ou et al., 2022).
However, research into the involvement of circRNAs in the regulation of ferroptosis in cancer development is relatively sparse and still in its infancy. Ferroptosis in tumor biology is complex and highly context-dependent with respect to treatment. Exploring the pathogenesis of ferroptosis and binding to circRNAs in various tumors is a direction for our future research.
Author contributions
LM conceived the structure of manuscript and revised the manuscript. RY and LM consulted the related paper and wrote the manuscript. JW drafted the figures. ZL proofread this manuscript. ZZ and ZY are responsible for editing the article format.
Funding
This study was supported by grants from the National Natural Science Foundation of China (82172351), Medical Science and Technology Research Project of Henan Province (SBGJ202102131 and SBGJ202102139), Key scientific research project plan of colleges and universities in Henan Province (22A320074) and Natural Science Foundation of Henan Province (212300410395).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Agmon, E., Solon, J., Bassereau, P., and Stockwell, B. R. (2018). Modeling the effects of lipid peroxidation during ferroptosis on membrane properties. Sci. Rep. 8, 5155. doi:10.1038/s41598-018-23408-0
Ashwal-Fluss, R., Meyer, M., Pamudurti, N. R., Ivanov, A., Bartok, O., Hanan, M., et al. (2014). circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 56, 55–66. doi:10.1016/j.molcel.2014.08.019
Bai, H., Lei, K., Huang, F., Jiang, Z., and Zhou, X. (2019). Exo-circRNAs: A new paradigm for anticancer therapy. Mol. Cancer 18, 56. doi:10.1186/s12943-019-0986-2
Bai, N., Peng, E., Qiu, X., Lyu, N., Zhang, Z., Tao, Y., et al. (2018). circFBLIM1 act as a ceRNA to promote hepatocellular cancer progression by sponging miR-346. J. Exp. Clin. Cancer Res. 37, 172. doi:10.1186/s13046-018-0838-8
Bazhabayi, M., Qiu, X., Li, X., Yang, A., Wen, W., Zhang, X., et al. (2021). CircGFRA1 facilitates the malignant progression of HER-2-positive breast cancer via acting as a sponge of miR-1228 and enhancing AIFM2 expression. J. Cell Mol. Med. 25, 10248–10256. doi:10.1111/jcmm.16963
Bebber, C. M., Muller, F., Prieto Clemente, L., Weber, J., and von Karstedt, S. (2020). Ferroptosis in Cancer Cell Biology, 12. doi:10.3390/cancers12010164Cancers (Basel)
Beltran, M., Rossi, F., and Bozzoni, I. (2022). CircZNF609 as a prototype to elucidate the biological function of circRNA-mRNA interactions. Mol. Cell Oncol. 9, 2055939. doi:10.1080/23723556.2022.2055939
Bersuker, K., Hendricks, J. M., Li, Z., Magtanong, L., Ford, B., Tang, P. H., et al. (2019). The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature 575, 688–692. doi:10.1038/s41586-019-1705-2
Cao, W., Chen, H. D., Yu, Y. W., Li, N., and Chen, W. Q. (2021). Changing profiles of cancer burden worldwide and in China: A secondary analysis of the global cancer statistics 2020. Chin. Med. J. Engl. 134, 783–791. doi:10.1097/CM9.0000000000001474
Chen, L., Nan, A., Zhang, N., Jia, Y., Li, X., Ling, Y., et al. (2019a). Circular RNA 100146 functions as an oncogene through direct binding to miR-361-3p and miR-615-5p in non-small cell lung cancer. Mol. Cancer 18, 13. doi:10.1186/s12943-019-0943-0
Chen, R. X., Chen, X., Xia, L. P., Zhang, J. X., Pan, Z. Z., Ma, X. D., et al. (2019b). N(6)-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat. Commun. 10, 4695. doi:10.1038/s41467-019-12651-2
Chen, S., Zhang, Z., Zhang, B., Huang, Q., Liu, Y., Qiu, Y., et al. (2022). CircCDK14 promotes tumor progression and resists ferroptosis in glioma by regulating PDGFRA. Int. J. Biol. Sci. 18, 841–857. doi:10.7150/ijbs.66114
Chen, X., Yu, C., Kang, R., and Tang, D. (2020). Iron metabolism in ferroptosis. Front. Cell Dev. Biol. 8, 590226. doi:10.3389/fcell.2020.590226
Cheng, F., Zheng, B., Si, S., Wang, J., Zhao, G., Yao, Z., et al. (2021). The roles of CircRNAs in bladder cancer: Biomarkers, tumorigenesis drivers, and therapeutic targets. Front. Cell Dev. Biol. 9, 666863. doi:10.3389/fcell.2021.666863
Conn, V. M., Hugouvieux, V., Nayak, A., Conos, S. A., Capovilla, G., Cildir, G., et al. (2017). A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat. Plants 3, 17053. doi:10.1038/nplants.2017.53
Ding, Y., Chen, X., Liu, C., Ge, W., Wang, Q., Hao, X., et al. (2021). Identification of a small molecule as inducer of ferroptosis and apoptosis through ubiquitination of GPX4 in triple negative breast cancer cells. J. Hematol. Oncol. 14, 19. doi:10.1186/s13045-020-01016-8
Dixon, S. J., Lemberg, K. M., Lamprecht, M. R., Skouta, R., Zaitsev, E. M., Gleason, C. E., et al. (2012). Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 149, 1060–1072. doi:10.1016/j.cell.2012.03.042
Dixon, S. J., Winter, G. E., Musavi, L. S., Lee, E. D., Snijder, B., Rebsamen, M., et al. (2015). Human haploid cell genetics reveals roles for lipid metabolism genes in nonapoptotic cell death. ACS Chem. Biol. 10, 1604–1609. doi:10.1021/acschembio.5b00245
Dodson, M., Castro-Portuguez, R., and Zhang, D. D. (2019). NRF2 plays a critical role in mitigating lipid peroxidation and ferroptosis. Redox Biol. 23, 101107. doi:10.1016/j.redox.2019.101107
Doll, S., Freitas, F. P., Shah, R., Aldrovandi, M., da Silva, M. C., Ingold, I., et al. (2019). FSP1 is a glutathione-independent ferroptosis suppressor. Nature 575, 693–698. doi:10.1038/s41586-019-1707-0
Dominguez, D., Freese, P., Alexis, M. S., Su, A., Hochman, M., Palden, T., et al. (2018). Sequence, structure, and context preferences of human RNA binding proteins. Mol. Cell 70, 854–867. doi:10.1016/j.molcel.2018.05.001
Du, W. W., Yang, W., Liu, E., Yang, Z., Dhaliwal, P., and Yang, B. B. (2016). Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 44, 2846–2858. doi:10.1093/nar/gkw027
Eger, N., Schoppe, L., Schuster, S., Laufs, U., and Boeckel, J. N. (2018). Circular RNA splicing. Adv. Exp. Med. Biol. 1087, 41–52. doi:10.1007/978-981-13-1426-1_4
Fang, X., Cai, Z., Wang, H., Han, D., Cheng, Q., Zhang, P., et al. (2020). Loss of cardiac ferritin H facilitates cardiomyopathy via slc7a11-mediated ferroptosis. Circ. Res. 127, 486–501. doi:10.1161/CIRCRESAHA.120.316509
Fontemaggi, G., Turco, C., Esposito, G., and Di Agostino, S. (2021). New molecular mechanisms and clinical impact of circRNAs in human cancer. Cancers (Basel) 13, 3154. doi:10.3390/cancers13133154
Forcina, G. C., and Dixon, S. J. (2019). GPX4 at the crossroads of lipid homeostasis and ferroptosis. Proteomics 19, e1800311. doi:10.1002/pmic.201800311
Friedmann Angeli, J. P., Schneider, M., Proneth, B., Tyurina, Y. Y., Tyurin, V. A., Hammond, V. J., et al. (2014). Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat. Cell Biol. 16, 1180–1191. doi:10.1038/ncb3064
Gammella, E., Recalcati, S., Rybinska, I., Buratti, P., and Cairo, G. (2015), Iron-induced damage in cardiomyopathy: Oxidative-dependent and independent mechanisms. Oxid. Med. Cell Longev. 2015, 230182. doi:10.1155/2015/230182
Gao, M., Monian, P., Quadri, N., Ramasamy, R., and Jiang, X. (2015). Glutaminolysis and transferrin regulate ferroptosis. Mol. Cell 59, 298–308. doi:10.1016/j.molcel.2015.06.011
Gao, X., and Wang, X. L. (2023). Dexmedetomidine promotes ferroptotic cell death in gastric cancer via hsa_circ_0008035/miR-302a/E2F7 axis. Kaohsiung J. Med. Sci. 39, 390–403. doi:10.1002/kjm2.12650
Goodall, G. J., and Wickramasinghe, V. O. (2021). RNA in cancer. Nat. Rev. Cancer 21, 22–36. doi:10.1038/s41568-020-00306-0
Hansen, T. B., Jensen, T. I., Clausen, B. H., Bramsen, J. B., Finsen, B., Damgaard, C. K., et al. (2013a). Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388. doi:10.1038/nature11993
Hansen, T. B., Kjems, J., and Damgaard, C. K. (2013b). Circular RNA and miR-7 in cancer. Cancer Res. 73, 5609–5612. doi:10.1158/0008-5472.CAN-13-1568
Harper, K. L., Mottram, T. J., and Whitehouse, A. (2021). Insights into the evolving roles of circular RNAs in cancer. Cancers (Basel) 13. doi:10.3390/cancers13164180
Hu, F., Peng, Y., Fan, X., Zhang, X., and Jin, Z. (2023). Circular RNAs: Implications of signaling pathways and bioinformatics in human cancer. Cancer Biol. Med. 20, 104–128. doi:10.20892/j.issn.2095-3941.2022.0466
Jiang, M., Mo, R., Liu, C., and Wu, H. (2022a). Circ_0000190 sponges miR-382-5p to suppress cell proliferation and motility and promote cell death by targeting ZNRF3 in gastric cancer. J. Biochem., mvac003. doi:10.1093/jb/mvac003
Jiang, Y., Zhao, J., Li, R., Liu, Y., Zhou, L., Wang, C., et al. (2022b). CircLRFN5 inhibits the progression of glioblastoma via PRRX2/GCH1 mediated ferroptosis. J. Exp. Clin. Cancer Res. 41, 307. doi:10.1186/s13046-022-02518-8
Kajarabille, N., and Latunde-Dada, G. O. (2019). Programmed cell-death by ferroptosis: Antioxidants as mitigators. Int. J. Mol. Sci. 20, 4968. doi:10.3390/ijms20194968
Kim, S. E., Zhang, L., Ma, K., Riegman, M., Chen, F., Ingold, I., et al. (2016). Ultrasmall nanoparticles induce ferroptosis in nutrient-deprived cancer cells and suppress tumour growth. Nat. Nanotechnol. 11, 977–985. doi:10.1038/nnano.2016.164
Kristensen, L. S., Andersen, M. S., Stagsted, L. V. W., Ebbesen, K. K., Hansen, T. B., and Kjems, J. (2019). The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 20, 675–691. doi:10.1038/s41576-019-0158-7
Lei, M., Zheng, G., Ning, Q., Zheng, J., and Dong, D. (2020). Translation and functional roles of circular RNAs in human cancer. Mol. Cancer 19, 30. doi:10.1186/s12943-020-1135-7
Lei, P., Bai, T., and Sun, Y. (2019). Mechanisms of ferroptosis and relations with regulated cell death: A review. Front. Physiol. 10, 139. doi:10.3389/fphys.2019.00139
Li, B., Zhu, L., Lu, C., Wang, C., Wang, H., Jin, H., et al. (2021b). circNDUFB2 inhibits non-small cell lung cancer progression via destabilizing IGF2BPs and activating anti-tumor immunity. Nat. Commun. 12, 295. doi:10.1038/s41467-020-20527-z
Li, C., Tian, Y., Liang, Y., and Li, Q. (2020). Circ_0008035 contributes to cell proliferation and inhibits apoptosis and ferroptosis in gastric cancer via miR-599/EIF4A1 axis. Cancer Cell Int. 20, 84. doi:10.1186/s12935-020-01168-0
Li, H., Yang, F., Hu, A., Wang, X., Fang, E., Chen, Y., et al. (2019). Therapeutic targeting of circ-CUX1/EWSR1/MAZ axis inhibits glycolysis and neuroblastoma progression. EMBO Mol. Med. 11, e10835. doi:10.15252/emmm.201910835
Li, J., Cao, F., Yin, H. L., Huang, Z. J., Lin, Z. T., Mao, N., et al. (2020b). Ferroptosis: Past, present and future. Cell Death Dis. 11, 88. doi:10.1038/s41419-020-2298-2
Li, J., Sun, D., Pu, W., Wang, J., and Peng, Y. (2020a). Circular RNAs in cancer: Biogenesis, function, and clinical significance. Trends Cancer 6, 319–336. doi:10.1016/j.trecan.2020.01.012
Li, Q., Li, K., Guo, Q., and Yang, T. (2023). CircRNA circSTIL inhibits ferroptosis in colorectal cancer via miR-431/SLC7A11 axis. Environ. Toxicol. doi:10.1002/tox.23670
Li, X., Zhang, J. L., Lei, Y. N., Liu, X. Q., Xue, W., Zhang, Y., et al. (2021a). Linking circular intronic RNA degradation and function in transcription by RNase H1. Sci. China Life Sci. 64, 1795–1809. doi:10.1007/s11427-021-1993-6
Li, Z., Huang, C., Bao, C., Chen, L., Lin, M., Wang, X., et al. (2015). Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 22, 256–264. doi:10.1038/nsmb.2959
Liang, Y., Wang, H., Chen, B., Mao, Q., Xia, W., Zhang, T., et al. (2021). circDCUN1D4 suppresses tumor metastasis and glycolysis in lung adenocarcinoma by stabilizing TXNIP expression. Mol. Ther. Nucleic Acids 23, 355–368. doi:10.1016/j.omtn.2020.11.012
Liu, B., Ma, H., Liu, X., and Xing, W. (2023). CircSCN8A suppresses malignant progression and induces ferroptosis in non-small cell lung cancer by regulating miR-1290/ACSL4 axis. Cell Cycle 22, 758–776. doi:10.1080/15384101.2022.2154543
Liu, C. X., and Chen, L. L. (2022a). Circular RNAs: Characterization, cellular roles, and applications. Cell 185, 2016–2034. doi:10.1016/j.cell.2022.04.021
Liu, J., Yang, H., Deng, J., Jiang, R., Meng, E., and Wu, H. (2022b). CircRPPH1 promotes the stemness of gastric cancer cells by targeting miR-375/SLC7A11 axis. Environ. Toxicol. 38, 115–125. doi:10.1002/tox.23668
Liu, Y., Li, L., Yang, Z., Wen, D., and Hu, Z. (2022a). Circular RNA circACAP2 suppresses ferroptosis of cervical cancer during malignant progression by miR-193a-5p/GPX4. J. Oncol. 2022, 5228874. doi:10.1155/2022/5228874
Liu, Z., Wang, Q., Wang, X., Xu, Z., Wei, X., and Li, J. (2020). Circular RNA cIARS regulates ferroptosis in HCC cells through interacting with RNA binding protein ALKBH5. Cell Death Discov. 6, 72. doi:10.1038/s41420-020-00306-x
Lou, J. S., Zhao, L. P., Huang, Z. H., Chen, X. Y., Xu, J. T., Tai, W. C., et al. (2021). Ginkgetin derived from Ginkgo biloba leaves enhances the therapeutic effect of cisplatin via ferroptosis-mediated disruption of the Nrf2/HO-1 axis in EGFR wild-type non-small-cell lung cancer. Phytomedicine 80, 153370. doi:10.1016/j.phymed.2020.153370
Lu, B., Chen, X. B., Ying, M. D., He, Q. J., Cao, J., and Yang, B. (2017). The role of ferroptosis in cancer development and treatment response. Front. Pharmacol. 8, 992. doi:10.3389/fphar.2017.00992
Lu, J., Wang, Y. H., Yoon, C., Huang, X. Y., Xu, Y., Xie, J. W., et al. (2020). Circular RNA circ-RanGAP1 regulates VEGFA expression by targeting miR-877-3p to facilitate gastric cancer invasion and metastasis. Cancer Lett. 471, 38–48. doi:10.1016/j.canlet.2019.11.038
Lu, J., Zhong, C., Luo, J., Shu, F., Lv, D., Liu, Z., et al. (2021). HnRNP-L-regulated circCSPP1/miR-520h/EGR1 axis modulates autophagy and promotes progression in prostate cancer. Mol. Ther. Nucleic Acids 26, 927–944. doi:10.1016/j.omtn.2021.10.006
Lyu, N., Zeng, Y., Kong, Y., Chen, Q., Deng, H., Ou, S., et al. (2021). Ferroptosis is involved in the progression of hepatocellular carcinoma through the circ0097009/miR-1261/SLC7A11 axis. Ann. Transl. Med. 9, 675. doi:10.21037/atm-21-997
Ma, S., Henson, E. S., Chen, Y., and Gibson, S. B. (2016). Ferroptosis is induced following siramesine and lapatinib treatment of breast cancer cells. Cell Death Dis. 7, e2307. doi:10.1038/cddis.2016.208
Meng, L., Zheng, Y., Liu, S., Ju, Y., Ren, S., Sang, Y., et al. (2021). ZEB1 represses biogenesis of circ-DOCK5 to facilitate metastasis in esophageal squamous cell carcinoma via a positive feedback loop with TGF-β. Cancer Lett. 519, 117–129. doi:10.1016/j.canlet.2021.06.026
Mou, Y., Wang, J., Wu, J., He, D., Zhang, C., Duan, C., et al. (2019). Ferroptosis, a new form of cell death: Opportunities and challenges in cancer. J. Hematol. Oncol. 12, 34. doi:10.1186/s13045-019-0720-y
Nisar, S., Bhat, A. A., Singh, M., Karedath, T., Rizwan, A., Hashem, S., et al. (2021). Insights into the role of CircRNAs: Biogenesis, characterization, functional, and clinical impact in human malignancies. Front. Cell Dev. Biol. 9, 617281. doi:10.3389/fcell.2021.617281
Okholm, T. L. H., Sathe, S., Park, S. S., Kamstrup, A. B., Rasmussen, A. M., Shankar, A., et al. (2020). Transcriptome-wide profiles of circular RNA and RNA-binding protein interactions reveal effects on circular RNA biogenesis and cancer pathway expression. Genome Med. 12, 112. doi:10.1186/s13073-020-00812-8
Ou, R., Lu, S., Wang, L., Wang, Y., Lv, M., Li, T., et al. (2022). Circular RNA circLMO1 suppresses cervical cancer growth and metastasis by triggering miR-4291/ACSL4-mediated ferroptosis. Front. Oncol. 12, 858598. doi:10.3389/fonc.2022.858598
Pan, C.-F., Wei, K., Ma, Z.-J., He, Y.-Z., Huang, J.-J., Guo, Z.-Z., et al. (2022). CircP4HB regulates ferroptosis via SLC7A11-mediated glutathione synthesis in lung adenocarcinoma. Transl. Lung Cancer Res. 11, 366–380. doi:10.21037/tlcr-22-138
Qin, K., Zhang, F., Wang, H., Wang, N., Qiu, H., Jia, X., et al. (2023). circRNA circSnx12 confers Cisplatin chemoresistance to ovarian cancer by inhibiting ferroptosis through a miR-194-5p/SLC7A11 axis. BMB Rep. 56, 184–189. doi:10.5483/BMBRep.2022-0175
Ribas, V., Garcia-Ruiz, C., and Fernandez-Checa, J. C. (2014). Glutathione and mitochondria. Front. Pharmacol. 5, 151. doi:10.3389/fphar.2014.00151
Rossi, F., Legnini, I., Megiorni, F., Colantoni, A., Santini, T., Morlando, M., et al. (2019). Circ-ZNF609 regulates G1-S progression in rhabdomyosarcoma. Oncogene 38, 3843–3854. doi:10.1038/s41388-019-0699-4
Shanshan, W., Hongying, M., Jingjing, F., Yiming, Y., Yu, R., and Rui, Y. (2021). CircDTL functions as an oncogene and regulates both apoptosis and ferroptosis in non-small cell lung cancer cells. Front. Genet. 12, 743505. doi:10.3389/fgene.2021.743505
Shi, H., Wang, X., Lu, Z., Zhao, B. S., Ma, H., Hsu, P. J., et al. (2017). YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 27, 315–328. doi:10.1038/cr.2017.15
Shimada, K., Skouta, R., Kaplan, A., Yang, W. S., Hayano, M., Dixon, S. J., et al. (2016). Global survey of cell death mechanisms reveals metabolic regulation of ferroptosis. Nat. Chem. Biol. 12, 497–503. doi:10.1038/nchembio.2079
Shindou, H., and Shimizu, T. (2009). Acyl-CoA:lysophospholipid acyltransferases. J. Biol. Chem. 284, 1–5. doi:10.1074/jbc.R800046200
Siegel, R. L., Miller, K. D., Fuchs, H. E., and Jemal, A. (2021). Cancer statistics, 2021. CA Cancer J. Clin. 71, 7–33. doi:10.3322/caac.21654
Snaebjornsson, M. T., Janaki-Raman, S., and Schulze, A. (2020). Greasing the wheels of the cancer machine: The role of lipid metabolism in cancer. Cell Metab. 31, 62–76. doi:10.1016/j.cmet.2019.11.010
Song, Z., Lin, J., Su, R., Ji, Y., Jia, R., Li, S., et al. (2022). eIF3j inhibits translation of a subset of circular RNAs in eukaryotic cells. Nucleic Acids Res. 50, 11529–11549. doi:10.1093/nar/gkac980
Stockwell, B. R., Jiang, X., and Gu, W. (2020). Emerging mechanisms and disease relevance of ferroptosis. Trends Cell Biol. 30, 478–490. doi:10.1016/j.tcb.2020.02.009
Su, L. J., Zhang, J. H., Gomez, H., Murugan, R., Hong, X., Xu, D., et al. (2019). Reactive oxygen species-induced lipid peroxidation in apoptosis, autophagy, and ferroptosis. Oxid. Med. Cell Longev., 5080843. doi:10.1155/2019/5080843
Sun, X., Ou, Z., Xie, M., Kang, R., Fan, Y., Niu, X., et al. (2015). HSPB1 as a novel regulator of ferroptotic cancer cell death. Oncogene 34, 5617–5625. doi:10.1038/onc.2015.32
Tang, D., Chen, X., Kang, R., and Kroemer, G. (2021). Ferroptosis: Molecular mechanisms and health implications. Cell Res. 31, 107–125. doi:10.1038/s41422-020-00441-1
Tang, Q., and Hann, S. S. (2020). Biological roles and mechanisms of circular RNA in human cancers. Onco Targets Ther. 13, 2067–2092. doi:10.2147/OTT.S233672
Toyokuni, S., Ito, F., Yamashita, K., Okazaki, Y., and Akatsuka, S. (2017). Iron and thiol redox signaling in cancer: An exquisite balance to escape ferroptosis. Free Radic. Biol. Med. 108, 610–626. doi:10.1016/j.freeradbiomed.2017.04.024
Tsitsipatis, D., Grammatikakis, I., Driscoll, R. K., Yang, X., Abdelmohsen, K., Harris, S. C., et al. (2021). AUF1 ligand circPCNX reduces cell proliferation by competing with p21 mRNA to increase p21 production. Nucleic Acids Res. 49, 1631–1646. doi:10.1093/nar/gkaa1246
Ubellacker, J. M., Tasdogan, A., Ramesh, V., Shen, B., Mitchell, E. C., Martin-Sandoval, M. S., et al. (2020). Lymph protects metastasizing melanoma cells from ferroptosis. Nature 585, 113–118. doi:10.1038/s41586-020-2623-z
Vo, J. N., Cieslik, M., Zhang, Y., Shukla, S., Xiao, L., Zhang, Y., et al. (2019). The landscape of circular RNA in cancer. Cell 176, 869–881. doi:10.1016/j.cell.2018.12.021
Wang, G., Sun, D., Li, W., and Xin, Y. (2021b). CircRNA_100290 promotes GC cell proliferation and invasion via the miR-29b-3p/ITGA11 axis and is regulated by EIF4A3. Cancer Cell Int. 21, 324. doi:10.1186/s12935-021-01964-2
Wang, H. H., Ma, J. N., and Zhan, X. R. (2021c). Circular RNA Circ_0067934 attenuates ferroptosis of thyroid cancer cells by miR-545-3p/slc7a11 signaling. Front. Endocrinol. (Lausanne) 12, 670031. doi:10.3389/fendo.2021.670031
Wang, H., Xiao, Y., Wu, L., and Ma, D. (2018). Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-000911/miR-449a pathway in breast carcinogenesis. Int. J. Oncol. 52, 743–754. doi:10.3892/ijo.2018.4265
Wang, L., Wu, S., He, H., Ai, K., Xu, R., Zhang, L., et al. (2022). CircRNA-ST6GALNAC6 increases the sensitivity of bladder cancer cells to erastin-induced ferroptosis by regulating the HSPB1/P38 axis. Lab. Investig. 102, 1323–1334. doi:10.1038/s41374-022-00826-3
Wang, M., Mao, C., Ouyang, L., Liu, Y., Lai, W., Liu, N., et al. (2019). Long noncoding RNA LINC00336 inhibits ferroptosis in lung cancer by functioning as a competing endogenous RNA. Cell Death Differ. 26, 2329–2343. doi:10.1038/s41418-019-0304-y
Wang, S., Wang, Y., Li, Q., Li, X., and Feng, X. (2022). A novel circular RNA confers trastuzumab resistance in human epidermal growth factor receptor 2-positive breast cancer through regulating ferroptosis. Environ. Toxicol. 37, 1597–1607. doi:10.1002/tox.23509
Wang, X., Zhang, H., Yang, H., Bai, M., Ning, T., Deng, T., et al. (2020). Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol. Oncol. 14, 539–555. doi:10.1002/1878-0261.12629
Wang, C. C., Han, C. D., Zhao, Q., and Chen, X. (2021a). Circular RNAs and complex diseases: From experimental results to computational models. Brief. Bioinform 22. doi:10.1093/bib/bbab286
Wu, M., Xu, L. G., Li, X., Zhai, Z., and Shu, H. B. (2002). AMID, an apoptosis-inducing factor-homologous mitochondrion-associated protein, induces caspase-independent apoptosis. J. Biol. Chem. 277, 25617–25623. doi:10.1074/jbc.M202285200
Wu, N., Zhu, D., Li, J., Li, X., Zhu, Z., Rao, Q., et al. (2023). CircOMA1 modulates cabergoline resistance by downregulating ferroptosis in prolactinoma. J. Endocrinol. doi:10.1007/s40618-023-02010-w
Wu, P., Mo, Y., Peng, M., Tang, T., Zhong, Y., Deng, X., et al. (2020). Emerging role of tumor-related functional peptides encoded by lncRNA and circRNA. Mol. Cancer 19, 22. doi:10.1186/s12943-020-1147-3
Wu, X., Li, Y., Zhang, S., and Zhou, X. (2021). Ferroptosis as a novel therapeutic target for cardiovascular disease. Theranostics 11, 3052–3059. doi:10.7150/thno.54113
Xi, Y., Shen, Y., Wu, D., Zhang, J., Lin, C., Wang, L., et al. (2022). CircBCAR3 accelerates esophageal cancer tumorigenesis and metastasis via sponging miR-27a-3p. Mol. Cancer 21, 145. doi:10.1186/s12943-022-01615-8
Xian, Z. Y., Hu, B., Wang, T., Cai, J. L., Zeng, J. Y., Zou, Q., et al. (2020). CircABCB10 silencing inhibits the cell ferroptosis and apoptosis by regulating the miR-326/CCL5 axis in rectal cancer. Neoplasma 67, 1063–1073. doi:10.4149/neo_2020_191024N1084
Xie, B., and Guo, Y. (2021). Molecular mechanism of cell ferroptosis and research progress in regulation of ferroptosis by noncoding RNAs in tumor cells. Cell Death Discov. 7, 101. doi:10.1038/s41420-021-00483-3
Xu, Q., Zhou, L., Yang, G., Meng, F., Wan, Y., Wang, L., et al. (2020). CircIL4R facilitates the tumorigenesis and inhibits ferroptosis in hepatocellular carcinoma by regulating the miR-541-3p/GPX4 axis. Cell Biol. Int. 44, 2344–2356. doi:10.1002/cbin.11444
Yang, J., Cao, X. H., Luan, K. F., and Huang, Y. D. (2021). Circular RNA FNDC3B protects oral squamous cell carcinoma cells from ferroptosis and contributes to the malignant progression by regulating miR-520d-5p/slc7a11 Axis. Front. Oncol. 11, 672724. doi:10.3389/fonc.2021.672724
Yang, W. S., SriRamaratnam, R., Welsch, M. E., Shimada, K., Skouta, R., Viswanathan, V. S., et al. (2014). Regulation of ferroptotic cancer cell death by GPX4. Cell 156, 317–331. doi:10.1016/j.cell.2013.12.010
Yang, W. S., and Stockwell, B. R. (2016). Ferroptosis: Death by lipid peroxidation. Trends Cell Biol. 26, 165–176. doi:10.1016/j.tcb.2015.10.014
Yang, Y., Fan, X., Mao, M., Song, X., Wu, P., Zhang, Y., et al. (2017). Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 27, 626–641. doi:10.1038/cr.2017.31
Yao, W., Wang, J., Meng, F., Zhu, Z., Jia, X., Xu, L., et al. (2021). Circular RNA CircPVT1 inhibits 5-fluorouracil chemosensitivity by regulating ferroptosis through MiR-30a-5p/FZD3 Axis in esophageal cancer cells. Front. Oncol. 11, 780938. doi:10.3389/fonc.2021.780938
Ye, Z., Zhuo, Q., Hu, Q., Xu, X., Mengqi, L., Zhang, Z., et al. (2021). FBW7-NRA41-SCD1 axis synchronously regulates apoptosis and ferroptosis in pancreatic cancer cells. Redox Biol. 38, 101807. doi:10.1016/j.redox.2020.101807
Yin, Y., Long, J., He, Q., Li, Y., Liao, Y., He, P., et al. (2019). Emerging roles of circRNA in formation and progression of cancer. J. Cancer 10, 5015–5021. doi:10.7150/jca.30828
Zang, J., Lu, D., and Xu, A. (2020). The interaction of circRNAs and RNA binding proteins: An important part of circRNA maintenance and function. J. Neurosci. Res. 98, 87–97. doi:10.1002/jnr.24356
Zhang, H., Deng, T., Liu, R., Ning, T., Yang, H., Liu, D., et al. (2020). CAF secreted miR-522 suppresses ferroptosis and promotes acquired chemo-resistance in gastric cancer. Mol. Cancer 19, 43. doi:10.1186/s12943-020-01168-8
Zhang, J., Chen, S., Wei, S., Cheng, S., Shi, R., Zhao, R., et al. (2022b). CircRAPGEF5 interacts with RBFOX2 to confer ferroptosis resistance by modulating alternative splicing of TFRC in endometrial cancer. Redox Biol. 57, 102493. doi:10.1016/j.redox.2022.102493
Zhang, X. O., Wang, H. B., Zhang, Y., Lu, X., Chen, L. L., and Yang, L. (2014). Complementary sequence-mediated exon circularization. Cell 159, 134–147. doi:10.1016/j.cell.2014.09.001
Zhang, X., Wang, S., Wang, H., Cao, J., Huang, X., Chen, Z., et al. (2019). Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol. Cancer 18, 20. doi:10.1186/s12943-018-0935-5
Zhang, X., Xu, Y., Ma, L., Yu, K., Niu, Y., Xu, X., et al. (2022a). Essential roles of exosome and circRNA_101093 on ferroptosis desensitization in lung adenocarcinoma. Cancer Commun. (Lond) 42, 287–313. doi:10.1002/cac2.12275
Zhao, Q., Liu, J., Deng, H., Ma, R., Liao, J. Y., Liang, H., et al. (2020). Targeting mitochondria-located circRNA SCAR alleviates NASH via reducing mROS output. Cell 183, 76–93. doi:10.1016/j.cell.2020.08.009
Zheng, J., and Conrad, M. (2020). The metabolic underpinnings of ferroptosis. Cell Metab. 32, 920–937. doi:10.1016/j.cmet.2020.10.011
Keywords: ferroptosis, circular RNA, cancer, biomarker, signaling pathway
Citation: Yang R, Ma L, Wan J, Li Z, Yang Z, Zhao Z and Ming L (2023) Ferroptosis-associated circular RNAs: Opportunities and challenges in the diagnosis and treatment of cancer. Front. Cell Dev. Biol. 11:1160381. doi: 10.3389/fcell.2023.1160381
Received: 07 February 2023; Accepted: 11 April 2023;
Published: 20 April 2023.
Edited by:
Zhe-Sheng Chen, St. John’s University, United StatesReviewed by:
Weipu Mao, Southeast University, ChinaQingbin Cui, University of Toledo College of Medicine and Life Sciences, United States
Copyright © 2023 Yang, Ma, Wan, Li, Yang, Zhao and Ming. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liang Ming, mingliang@zzu.edu.cn
†These authors have contributed equally to this work
 Ruotong Yang
Ruotong Yang Liwei Ma1,2†
Liwei Ma1,2†  Junhu Wan
Junhu Wan