Manufacturing of non-viral protein nanocages for biotechnological and biomedical applications
- 1iBB–Institute for Bioengineering and Biosciences, Department of Bioengineering, Instituto Superior Técnico, Universidade de Lisboa, Lisbon, Portugal
- 2Associate Laboratory i4HB–Institute for Health and Bioeconomy at Instituto Superior Técnico, Universidade de Lisboa, Lisbon, Portugal
Protein nanocages are highly ordered nanometer scale architectures, which are typically formed by homo- or hetero-self-assembly of multiple monomers into symmetric structures of different size and shape. The intrinsic characteristics of protein nanocages make them very attractive and promising as a biological nanomaterial. These include, among others, a high surface/volume ratio, multi-functionality, ease to modify or manipulate genetically or chemically, high stability, mono-dispersity, and biocompatibility. Since the beginning of the investigation into protein nanocages, several applications were conceived in a variety of areas such as drug delivery, vaccine development, bioimaging, biomineralization, nanomaterial synthesis and biocatalysis. The ability to generate large amounts of pure and well-folded protein assemblies is one of the keys to transform nanocages into clinically valuable products and move biomedical applications forward. This calls for the development of more efficient biomanufacturing processes and for the setting up of analytical techniques adequate for the quality control and characterization of the biological function and structure of nanocages. This review concisely covers and overviews the progress made since the emergence of protein nanocages as a new, next-generation class of biologics. A brief outline of non-viral protein nanocages is followed by a presentation of their main applications in the areas of bioengineering, biotechnology, and biomedicine. Afterwards, we focus on a description of the current processes used in the manufacturing of protein nanocages with particular emphasis on the most relevant aspects of production and purification. The state-of-the-art on current characterization techniques is then described and future alternative or complementary approaches in development are also discussed. Finally, a critical analysis of the limitations and drawbacks of the current manufacturing strategies is presented, alongside with the identification of the major challenges and bottlenecks.
1 Introduction
In recent years, nanoparticles have been explored for applications in several scientific areas from nanobiotechnology and biomedical sciences to materials science and synthetic biology. Examples of nanoparticles studied in the literature include protein-based nanoparticles, metal nanoparticles, polymer micelles, silica nanoparticles and quantum dots (Lee, 2018). Among these, protein-based nanoparticles spurred significant research interest given their enormous potential for biomedical purposes (Schreiber and Schiller, 2013; Bhaskar and Lim, 2017; Lee, 2018).
In Nature, the existence of cellular processes essential to life such as metabolic reactions and information exchange is dependent on biological compartmentalization. In addition to lipids, proteins are one of the main components of natural compartmentalization systems such as virus capsids. Nanoparticles based on functional proteins constitute an additional example of such bio-compartments. Several of these protein-based nano-compartments, with different structural and functional characteristics, are described in the literature, including toroid- and donut-shaped proteins, tubes and yoctowells, protein nanocages, bacterial microcompartments (BMCs), protein membrane-based organelles (PMBOs) and gas vesicle protein nanoparticles (GVNPs) (Diekmann and Pereira-Leal, 2013; Lim, 2013; Schreiber and Schiller, 2013). For example, GVNPs, which have membranes exclusively composed of proteins, are produced in a wide variety of prokaryotic microorganisms, from heterotrophic bacteria to halophilic Archaea (e.g., Halobacterium sp. NRC-1). These nanostructures are spindle- or cylinder-shaped with a hydrophobic interior, having dimensions from 30 to 250 nm in width and from 50 nm to 2 µm in length. The most interesting properties of GVNPs include structural stability, monodispersibility, non-toxicity, self-adjuvanticity and ease of engineering. Some studies in the literature describe applications of GVNPs in drug delivery, in antigen display for vaccines, as contrast agents for ultrasound imaging and as acoustic biosensors (DasSarma and DasSarma, 2015; Andar et al., 2017; Hill and Salmond, 2020; Kim et al., 2022; Pfeifer, 2022; Karan et al., 2023). Nevertheless, among all these protein-based nanoparticles, protein nanocages are one of the most relevant (Diekmann and Pereira-Leal, 2013; Lim, 2013; Schreiber and Schiller, 2013).
Protein nanocages can be defined as highly ordered, nano-scale architectures. In general, they are produced through the self-assembly of multiple monomers, which may be identical or distinct, into symmetric and homogeneous structures of different shape and size. These protein-based nanoparticles allow spatial control of biological processes and compartmentalization of toxic, unstable, and sensitive compounds (Lee et al., 2016; Pieters et al., 2016; Diaz et al., 2018; Lee, 2018).
The advantages of protein nanocages result from their distinctive intrinsic characteristics, including a high surface/volume ratio, multi-functionality and ease of modification or manipulation through genetic or chemical strategies. Additionally, high stability, monodispersibility, biocompatibility, low toxicity and biodegradability are very attractive properties in the context of biotechnological and biomedical applications. Other applications include biomineralization and nanomaterial synthesis (Bhaskar and Lim, 2017; Diaz et al., 2018; Lee, 2018).
Protein nanocages can be classified as virus-like particles (VLPs) or non-viral protein nanocages (NVPNs). VLPs, which constitute the major group of protein nanocages, present an extensive variability in terms of structures and dimensions (Roldão et al., 2010; Patterson et al., 2012; Smith et al., 2012; Wen et al., 2012; Hassani-Mehraban et al., 2015; Fu and Li, 2016; Li et al., 2017). While structurally similar to viruses, VLPs are not infectious since they lack genetic material (Diaz et al., 2018). NVPNs, on the other hand, are unrelated to viral particles. NVPNs are formed by the self-assembly of protein monomers. This process is critically determined by the nature of the interface between adjoining subunits (Lim, 2013; Bhaskar and Lim, 2017). A wide variety of NVPNs with different structural and functional characteristics have been described in the literature.
Key biomedical applications of NVPNs include drug delivery, vaccine development and diagnostic bioimaging (Lai et al., 2009; Suci et al., 2010; Ren et al., 2012; Jeon et al., 2013; Toita et al., 2013; Zhen et al., 2013; Moon et al., 2014; Ra et al., 2014; Lee E. J. et al., 2015; Lee W.et al., 2015). Non-natural, bioinspired NVPNs can also be designed de novo through the assembly of artificial, functional monomers (Vázquez and Villaverde, 2010; Schreiber and Schiller, 2013; Bhaskar and Lim, 2017; Diaz et al., 2018).
The development of biomedical applications of NVPNs requires large amounts of pure and well-folded nanoassemblies (Theil, 2012). Consequently, more efficient, flexible, and universal bioprocess technologies are needed to transform NVPNs into clinically valuable products. The development of such biomanufacturing processes should be accompanied by the setting up of adequate quality control strategies to characterize both biological function and structure of the obtained nanocages (Lee et al., 2016).
This review first overviews general and relevant concepts related to NVPNs, followed by a presentation of their main applications in bioengineering, biotechnology, and biomedicine. The processes currently used to manufacture protein nanocages are described, and the most important aspects of upstream and downstream processing are highlighted. State-of-the-art characterization techniques are then presented and future alternative or complementary approaches in development are also discussed. Finally, a brief critical analysis of the drawbacks of the current manufacturing strategies is presented, alongside the identification of the major challenges.
2 Non-viral protein nanocages
2.1 General aspects
Non-viral protein nanocages are formed by multiple protein monomers that self-assemble into precisely defined, symmetric, homogeneous and complex structures (Lim, 2013; Molino and Wang, 2014; Pieters et al., 2016; Lee, 2018). These nanometer size (10–100 nm) particles may originate from different prokaryotes and eukaryotes. Their structural characteristics are critical for important cellular functions, which include storage of minerals, regulation of iron homeostasis, chaperone activity for the protection of other proteins in response to high temperature, protection of DNA from oxidative damage, cargo transport of nucleic acids and catalytic support for enzymatic reactions (Lim, 2013; Molino and Wang, 2014; Diaz et al., 2018).
2.2 Structural and functional characteristics
NVPNs are robust, monodisperse and water soluble, and present high biocompatibility and biodegradability (Lim, 2013; Corsi and Mazzucchelli, 2016; Bhaskar and Lim, 2017; Diaz et al., 2018). Furthermore, they can be chemically or genetically modified to extend functions and applications beyond the natural ones. Such strategies rely on molecular, genetic, and crystal structure information available in the literature (Uchida et al., 2007; Moon et al., 2014; Giessen, 2016; Diaz et al., 2018). This versatility and ability to be used as a multipurpose platform constitutes one of their most interesting features. For example, since most NVPNs have intrinsic catalytic characteristics as well as the ability to carry different molecules in the inner core, it is possible to optimize them as reaction vessels and templates to synthesize metallic nanoparticles (Lim, 2013).
New functionalities can be engineered at the interface between monomers, and the external and internal surfaces of protein nanocages (Lim, 2013; Bhaskar and Lim, 2017; Diaz et al., 2018; Chen et al., 2021). The external surface can be conjugated with functional ligands to improve targeting of a therapeutic cargo, cell penetration and biodistribution. These ligands can be peptides, epitopes, or other small molecules. The possibility for multiple conjugation with different ligands is also attractive and limited only by steric hindrance (Domingo et al., 2001; Kickhoefer et al., 2009; Choi et al., 2011; Kang et al., 2012; Lim, 2013; Murata et al., 2015). The amino acids in the monomers that compose the inner surface of the NVPNs can be replaced by specific amino acids to enable the creation of anchors for the loading of molecules with different dimensions. Depending on the type of nanocage and on the dimensions of the molecule to be encapsulated, this loading process and the subsequent binding to the inner surface can be mediated by chemical interactions (covalent, ionic, hydrophobic) or through protein-protein interactions.
2.3 Self-assembly mechanism
Self-assembly is the key to nanocage architecture. If the underlying mechanism is known (Zhang and Orner, 2011; Doyle et al., 2013; Lv et al., 2021), self-assembly can be modulated by destabilizing interactions at the subunit interface. Therefore, nanocage disassembly and reassembly can be induced, which allows controlling both the molecular cargo release from the inner core and the encapsulation of payloads (e.g., therapeutic molecules or enzymes). Several reports study the conditions that permit disassembly without irreversibly damaging the protein nanocage and the subsequent reassembly into its original architecture (Kim et al., 2011; Lim, 2013; Ferrer-Miralles et al., 2015). Some of the most relevant factors are the pH (Dalmau et al., 2009; Kim et al., 2011; Peng and Lim, 2011), the ionic strength (Sánchez-Sánchez et al., 2014), the presence of reducing agents (Shen et al., 2015) and the presence of metals (Swift et al., 2009; Belval et al., 2016).
2.4 Designed NVPNs
Artificial nanocages can be designed and generated de novo by mimicking the intrinsic mechanisms of self-assembly of natural NVPNs. Starting from the structural characteristics (e.g., geometry, size) required for the target nanocage, functional monomers are selected and modified accordingly. The amino acid sequences in each monomer can be partially derived from natural nanocages or designed anew to promote self-assembly. Strategies and sub-methodologies used may include directed evolution, use of fusion proteins, redesign of key interfaces and the de novo design (Lv et al., 2021). Ultimately, monomer selection and design must guarantee that protein-protein interactions take place with minimal nonspecific aggregation (Vázquez and Villaverde, 2010; Lai et al., 2012b; Lim, 2013; Bhaskar and Lim, 2017; Diaz et al., 2018; Chen et al., 2021).
Hybrid protein nanocages, which are conjugated with components such as polymers, nucleotides, carbon hydrates or lipids, are yet another group of protein nanocages that can be considered. These nanocages may be of major importance for diagnosis and therapy applications, for example, in the context of targeting or modulation of immune response. However, hybrid NVPNs are not extensively described in the literature (Bhaskar and Lim, 2017).
2.5 Functionalization
Natural and artificial NVPNs can be functionalized to create nanoarchitectures more adjusted to the end applications (Moon et al., 2014; Giessen, 2016; Bhaskar and Lim, 2017). Two main strategies for the functionalization of NVPNs are described: genetic engineering and bioconjugation (Diaz et al., 2018). Genetic modifications allow a more precise control over the number, position and distribution of the introduced molecules (Lee et al., 2016). Specific techniques used include modular assembly (Kang et al., 2012; Guimaraes et al., 2013), improvement of payload encapsulation (Seebeck et al., 2006; Kar et al., 2011; Tamura et al., 2015), interface engineering (Peng et al., 2015; Chen et al., 2016), peptide display (Laplagne et al., 2004; Flenniken et al., 2006), and protein display (Phippen et al., 2016). Bioconjugation on the other hand consists in the attachment of molecules that cannot be introduced through genetic engineering. The conjugation can be performed through covalent (Flenniken et al., 2006; Falvo et al., 2013) or non-covalent bonds (Bhaskar and Lim, 2017; Diaz et al., 2018) (Bhaskar and Lim, 2017). The available literature indicates that genetic modification alone or in combination with bioconjugation is clearly the most efficient approach to modify NVPNs (Diaz et al., 2018).
2.6 Examples of natural and artificial NVPNs
A broad spectrum of NVPNs, including both natural and artificial variants, are documented in literature. Figure 1 presents a schematic representation of the three-dimensional (3D) structure of natural NVPNs that have been extensively studied. Similarly, Figure 2 shows a schematic representation of the 3D structure of some artificial NVPNs.
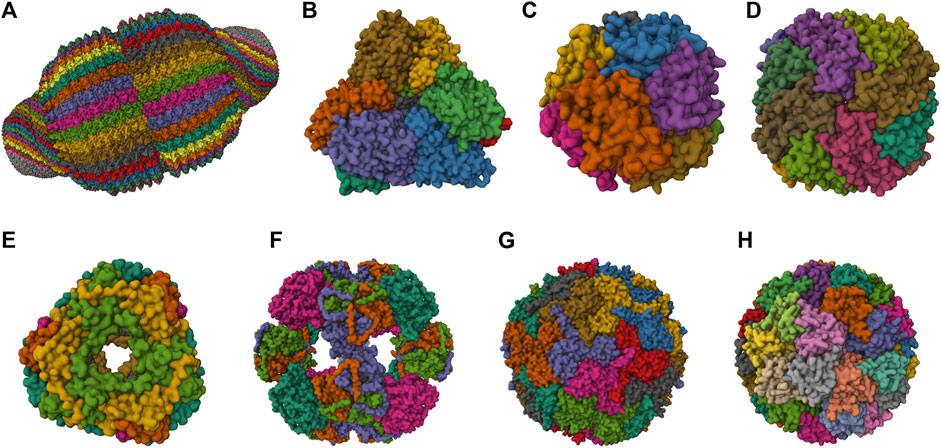
FIGURE 1. 3D structures of the most studied natural NVPNs. (A) Vault (PDB ID: 4V60) (Tanaka et al., 2009). (B) Aminopeptidase (PepA) (PDB ID: 3KL9) (Kim et al., 2010). (C) DNA-binding protein from starved cells (Dps) (PDB ID: 1QGH) (Ilari et al., 2000). (D) Ferritin (PDB ID: 2FHA) (Hempstead et al., 1997). (E) Heat shock protein (HSP) (PDB ID: 1SHS) (Kim et al., 1998a). (F) Dihydrolipoyl acetyltransferase (E2) (PDB ID: 1B5S) (Izard et al., 1999). (G) Encapsulin (PDB ID: 3DKT) (Sutter et al., 2008). (H) Lumazine synthase (PDB ID: 1RVV) (Ritsert et al., 1995). Representations created using the Mol* Viewer tool (Berman et al., 2000; Sehnal et al., 2021).
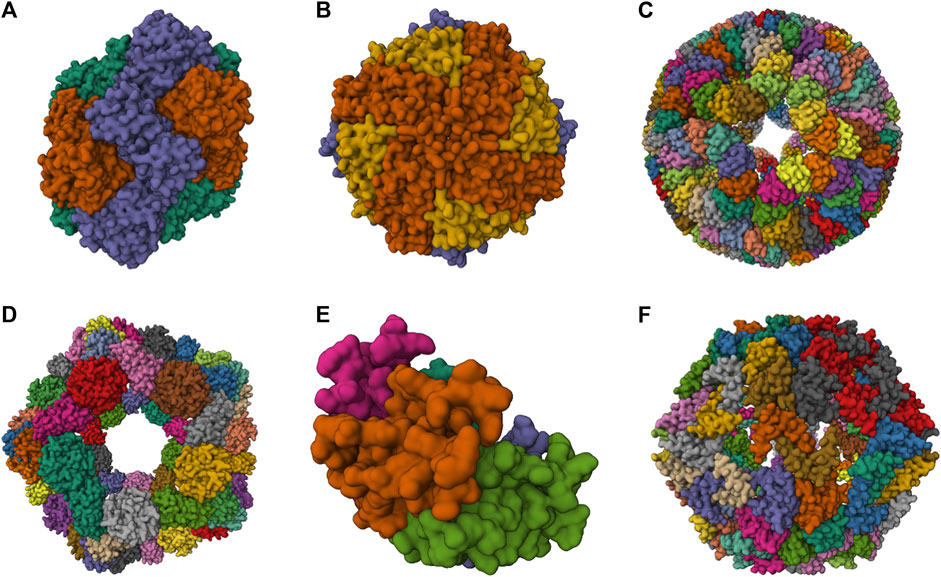
FIGURE 2. 3D structures of some artificial NVPNs. (A) 16-nm protein nanocages designed (PDB ID: 3VDX) (Lai et al., 2012a). (B) His6-HuHF and His6-SF nanocages (PDB ID: 7CPC) (Gu et al., 2020). (C) trp RNA-binding attenuation protein (TRAP) nanocages (PDB ID: 6RVW) (Malay et al., 2019). (D) I3-01 nanocages (PDB ID: 8ED3) (McCarthy and Gonen, 2022). (E) Prototype oxygen-impermeable protein nanocages (OIPNC) (PDB ID: 7WKC) (Gao et al., 2022). (F) TIP60 nanocages (PDB ID: 7EQ9) (Obata et al., 2021). Representations created using the Mol* Viewer tool (Berman et al., 2000; Sehnal et al., 2021).
2.6.1 Natural NVPNs
The most relevant structural and functional characteristics of representative natural NVPNs are summarized in Table 1. Vault nanocages (Figure 1A) have extending caps at their ends, a hinged waist region, 8 small pores (2 nm in diameter) and a significant internal volume. Each vault nanocage is composed of several protein and non-protein elements, with the major vault protein (MJV) representing 70% of the overall mass. An interesting property is the dynamic ability of the vault nanocages, since they can open and close transiently, allowing the incorporation of small molecules and macromolecules within the inner core (Kickhoefer et al., 1996; Kickhoefer et al., 2005; Kong et al., 1999; Poderycki et al., 2006; Anderson et al., 2007; Ryu et al., 2008; Goldsmith et al., 2009; Tanaka et al., 2009; Yu et al., 2017).
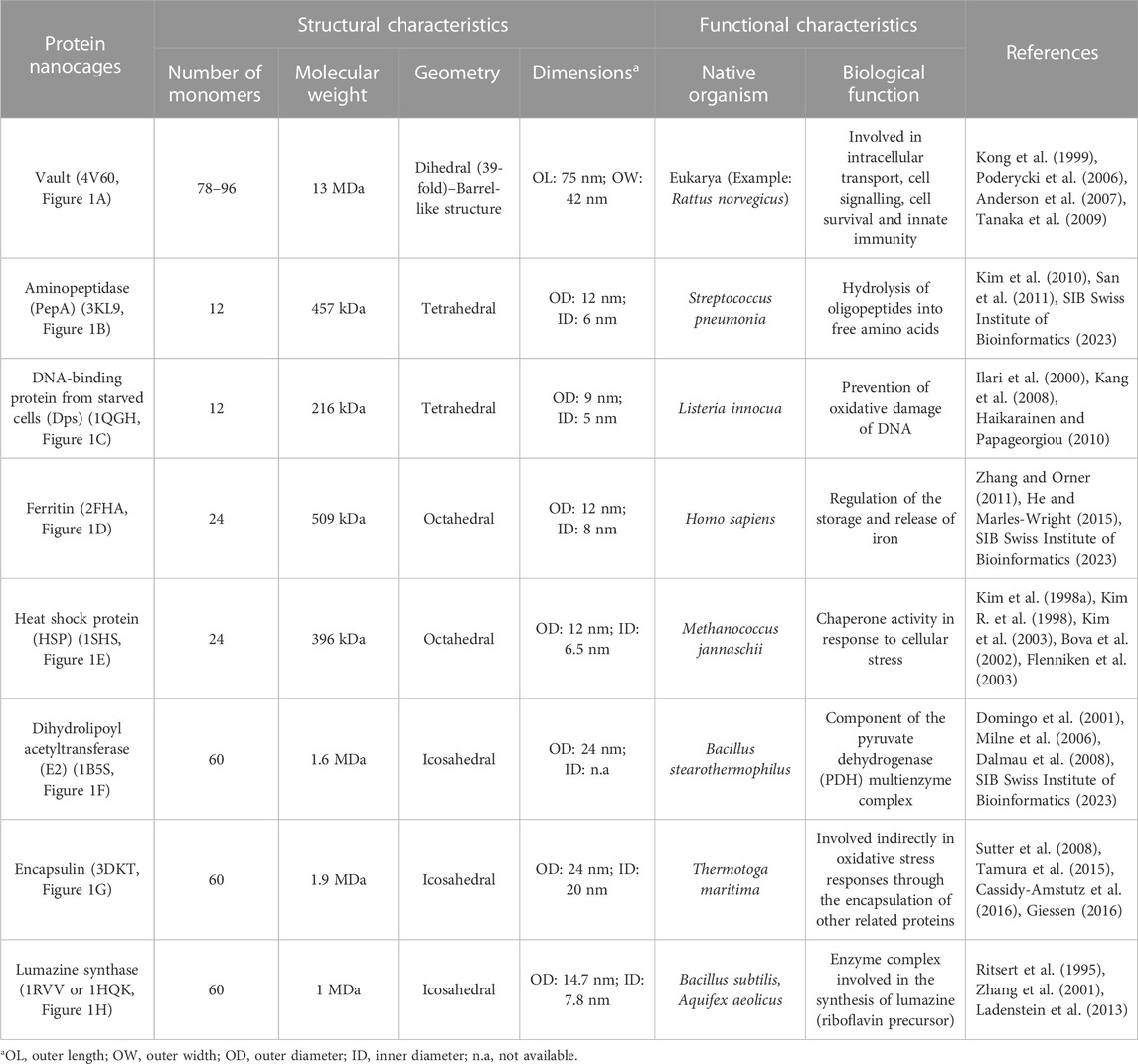
TABLE 1. A summary of the structural and functional characteristics of the most studied natural NVPNs. The indicated code corresponds to the respective PDB ID.
PepA nanocages are formed by the self-assembly of aminopeptidase (PepA), a zinc-dependent metallopeptidase (Figure 1B). The resulting nanoparticles have an inner cavity with negative charge and 8 pores on the edges and faces with diameters of 1 and 3 nm. The channels are useful for the traffic of molecules. PepA nanocages were used as templates for the size-controlled synthesis of ultrasmall platinum nanoparticles, with the formation of a multifunctional biohybrid catalyst (Kim et al., 2010; San et al., 2011).
DNA-binding protein from starved cells (Dps) belongs to the ferritin subfamily. Dps nanocages (Figure 1C) have small pores (0.8 nm) through which small molecules can diffuse. Dps nanocages were used as templates for the formation of metallic-protein nanoparticles to improve the endogenous catalytic activity of ferritins (Ilari et al., 2000; Kang et al., 2008; Haikarainen and Papageorgiou, 2010).
Ferritin nanocages (Figure 1D) have an internal cavity that matches the size of the iron core. In this core, iron is stored in an insoluble non-toxic state. The cages have 8 hydrophilic pores (4 nm) that facilitate the movement of iron atoms and other small molecules. Ferritin nanocages are quite stable at high temperature and in a wide range of pH values. Self-assembly, disassembly and reassembly can be controlled by metal ions. The nanocages were applied in biomineralization and as nanocarrier of biological and non-biological molecules (Lawson et al., 1991; Hempstead et al., 1997; Zhang and Orner, 2011; He and Marles-Wright, 2015; Kuruppu et al., 2015; Chen et al., 2016; Wang et al., 2017; Palombarini et al., 2020; Zhang et al., 2020).
Small heat shock protein (sHSP) nanocages (Figure 1E) have 8 pores (3 nm and 17 nm) that allow molecular trafficking. Advantages of the cages include high stability at high temperatures (up to 70°C) and in a broad range of pH values (5–11). sHSP nanocages were used as nanoreactors, for biomineralization, for drug delivery and for bioimaging. The functionalization reported includes the introduction of modifications with organic molecules outside and inside of the nanocages (Kim et al., 1998a; Kim et al., 1998b; Kim R. et al., 1998; Bova et al., 2002; Flenniken et al., 2003; Kim et al., 2003; Varpness et al., 2005; Flenniken et al., 2006; Abedin et al., 2009).
Dihydrolipoyl acetyltransferase (E2) nanocages from Bacillus stearothermophilus (Figure 1F) have 12 pores (5 nm) and high stability at extreme temperatures due to the thermophilic nature of the native organism. E2 proteins can be modified simultaneously at the outer and inner surfaces to allow loading of drugs inside and to display functional epitopes outside (Izard et al., 1999; Domingo et al., 2001; Jung et al., 2002; Milne et al., 2006; Dalmau et al., 2008).
Encapsulin nanocages (Figure 1G) have a characteristic large central cavity (20 nm) that makes them interesting as a cargo delivery nanoplatform and as a nanoreactor (Sutter et al., 2008; Rahmanpour and Bugg, 2013; Moon et al., 2014; Tamura et al., 2015; Cassidy-Amstutz et al., 2016; Giessen, 2016). Lumazine synthase nanocages (Figure 1H), which have a 7.8 nm inner cavity with negative charge, were used as molecular carriers through encapsulation. Other applications include the biomineralization of iron. A key characteristic of these nanocages is the stability at high temperatures (up to 95°C) (Schott et al., 1990; Ritsert et al., 1995; Shenton et al., 2001; Zhang et al., 2001; Seebeck et al., 2006; Ladenstein et al., 2013; Ra et al., 2014).
2.6.2 Artificial NVPNs
Table 2 summarizes structural characteristics of artificial NVPNs reported in the literature. The formation of symmetric and homogenous 16-nm protein nanocages (Figure 2A) results from the self-assembly of monomers obtained by a fusion process. Each monomer is a geometrically controlled fusion of two natural protein oligomers, which are connected by an α-helical linker. One of those oligomers is the trimeric bromoperoxidase and the other is a dimeric M1 virus matrix protein (Lai et al., 2012a).
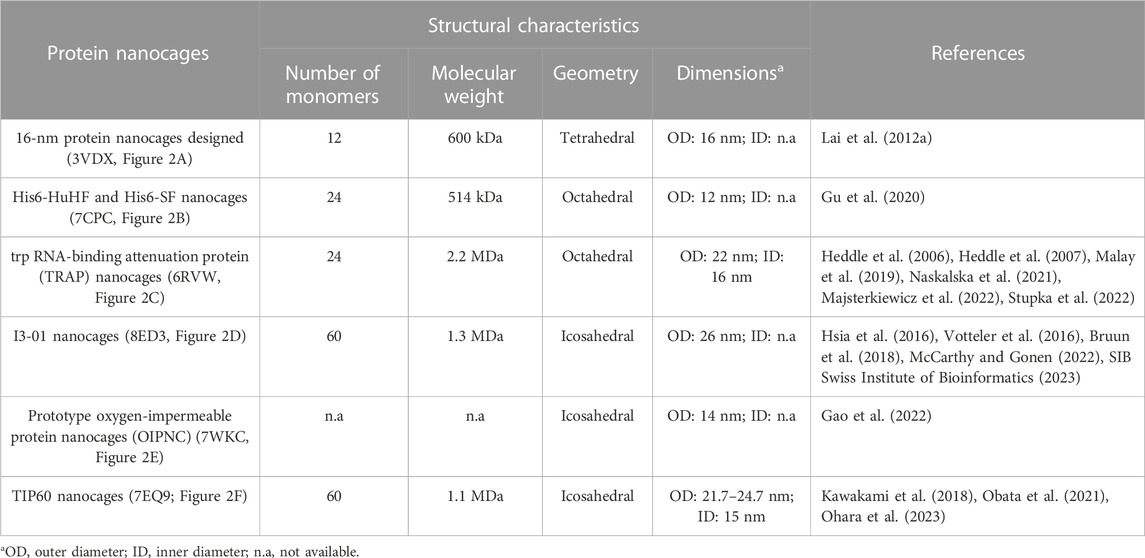
TABLE 2. A summary of the structural characteristics for some artificial NVPNs. The indicated code corresponds to the respective PDB ID.
His6-HuHF and His6-SF nanocages (Figure 2B) with regulatable self-assembly were created based on two recombinant ferritins (rHuHF and rSF). Histidine motifs were incorporated in one of their subunit interfaces. Two different switches (metal- and pH-based) were developed to control the assembly-disassembly of the nanocages. This may lead to more efficient encapsulation of molecules within the nanocages compared to the traditional methods reported (Gu et al., 2020).
The trp RNA-binding attenuation protein (TRAP) nanocages (Figure 2C) result from the assembly of 24 ring-shaped proteins derived from the natural TRAP from B. stearothermophilus. Each constituent TRAP ring is formed by 11 monomers that were engineered to include a cysteine residue. Contrary to the usual situation, the complete nanocage is formed not through a network of protein-protein interactions but through the bridging of opposing thiols of the cysteine residues between TRAP rings via single gold (I) ions. The fully assembled TRAP nanocages present six square apertures (4 nm). TRAP nanocages are stable up to 95°C and at high concentration of denaturing agents (e.g., 7 M urea). Although they are susceptible to reducing agents, this could be a promising characteristic for delivery to targets that contain this type of agents. The fact that the assembly relies on a metal-induced process is very useful since it provides a more rigorous control of assembly-disassembly and, eventually, a programmable mechanism. The nanocages can also be labelled with a dye in each ring-shaped monomer, which could be useful for bioimaging applications (Heddle et al., 2006; Heddle et al., 2007; Malay et al., 2019; Naskalska et al., 2021; Majsterkiewicz et al., 2022; Stupka et al., 2022).
I3-01 nanocages (Figure 2D) are hollow architectures that result from the self-assembly of multiple monomers corresponding to a trimeric 2-keto-3-deoxy-6-phosphogluconate (KDPG) aldolase from Thermotoga maritima. These aldolase monomers were engineered to contain complementary hydrophobic interfaces. The nanocages have several large pores (9 nm). I3-01 nanocages are stable up to 80°C and in the presence of high concentration of denaturing agents (e.g., 6.7 M guanidine hydrochloride). Applications include synthetic biology, targeted drug delivery, and vaccine development (Hsia et al., 2016; Votteler et al., 2016; Bruun et al., 2018; McCarthy and Gonen, 2022).
Prototype oxygen-impermeable protein nanocages (OIPNC) (Figure 2E) are derived from the pentameric β-carboxysome from Thermosynechococcus elongatus BP-1 (CcmL). Self-assembly of the proteins into nanocages occurs in the presence of quantum dots as templates by protein-quantum dots interfacial engineering. An advantage of these nanocages is the permeability to O2 in a switchable process controlled by molecular patches. Future interesting applications include their use as nanocarriers and nanoreactors (Gao et al., 2022).
TIP60 nanocages (Figure 2F) are hollow spheres with 20 triangular pores that were created using a fusion protein design approach. Each monomer is a genetic fusion of two proteins (one pentameric- LSm and one dimeric- MyoX-coil) with a three-residue linker. A reversible assembly and disassembly mechanism based on metal ions and chelators was developed for TIP60 nanocages (Kawakami et al., 2018; Obata et al., 2021; Ohara et al., 2023).
3 Applications of non-viral protein nanocages
Applications of NVPNs can be found in a wide variety of areas and fields of study, with particular emphasis on those related to bioengineering, biotechnology, and biomedicine. Representative applications in drug delivery, vaccine development, bioimaging and diagnostic imaging, biomineralization and nanomaterials synthesis, and biocatalysis are briefly discussed in the following sections and presented with more details in the Supplementary Table S1.
3.1 Drug delivery
NVPNs constitute an excellent vehicle for the encapsulation, targeted delivery, and controlled release of drugs, which can range from small molecules to larger biomolecules like nucleic acids or proteins. Several NVPNs (e.g., Dps, encapsulin, ferritin, sHSP, and TRAP nanocages) were functionalized through genetic or chemical modifications to contain targeting molecules (e.g., biotin, hepatocellular carcinoma cell binding peptides, neuropilin 1-binding peptides and PTD4 cell-penetrating peptides) and to allow the incorporation of different cargos (e.g., SnCe6 photosensitizer, aldoxorubicin, doxorubicin, OSU03012 anticancer drug, small interfering RNA, curcumin). In general, results show that NVPN-mediated delivery can be performed successfully and that the desired effect is achieved efficiently (Suci et al., 2010; Bellini et al., 2014; Moon et al., 2014; Murata et al., 2015; Guan et al., 2018; Naskalska et al., 2021; Ji et al., 2022).
3.2 Vaccine development
NVPNs can be used as platforms for antigen display, offering the possibility of co-delivery of adjuvants, targeted delivery, immune modulation, and antigen stabilisation. Engineered protein nanocages including E2, ferritin, I3-01, lumazine synthase, sHSP and vault nanocages were demonstrated as a potential vaccine platform, with the triggering of strong immune responses (namely, CD8+ and CD4+ T-cell responses). Different types of antigens and other molecules were displayed on the outer and/or inner surfaces of the nanocages, such as human melanoma-associated antigen gp100, MHC I-restricted SIINFEKL peptide epitopes, SIINFEKL and ISQAVHAAHAEINEAGR peptides, and transmission-blocking and blood-stage malaria antigens (Wang et al., 2003; Kar et al., 2012; Molino et al., 2013; Han et al., 2014; Ra et al., 2014; Bruun et al., 2018).
3.3 Bioimaging and diagnostic imaging
The incorporation of contrast agents into NVPNs offers the possibility of extending their applications to the visualization of biological processes, detection of diseases, and monitoring of therapies. Ferritin and sHSP nanocages were tested in the context of bioimaging and diagnostic imaging applications. For example, the conjugation of NVPNs with targeting peptides (e.g., RGD and DEVD) and a fluorescent molecule (e.g., Cy5.5) allowed the imaging of caspase activity inside tumor cells (Choi et al., 2011). In another study, labelled protein nanocages loaded with an iron oxide nanoparticle catalyzed the oxidation of peroxidase substrates, which allowed the subsequent visualization of tumor tissue (Fan et al., 2012). Protein nanocages combined with metallic nanoparticles also showed promise in real-time in vivo photoacoustic imaging of tumor cells, and in positron emission tomography imaging when combined with a copper radionuclide (Wang et al., 2016). Finally, molecular magnetic resonance imaging was possible with a protein nanocage engineered with a targeting molecule (e.g., neuropilin 1-binding peptide) and combined with gadolinium (III)-chelated contrast agents (Kawano et al., 2018).
3.4 Biomineralization and nanomaterials synthesis
NVPNs can be used to control and direct the formation of nanomaterials with specific properties, for example, by serving as templates for the growth of inorganic minerals, by encapsulating metal nanoparticles, quantum dots, or magnetic nanoparticles, or by acting as microreactors for the controlled synthesis of nanomaterials. For example, natural and engineered Dps and sHSP nanocages were used as a nanoscale platform for the synthesis of monodisperse and homogeneous iron oxide nanoparticles (Allen et al., 2002; Flenniken et al., 2003). In another application, a synthetic polymer with modifiable groups was successfully incorporated into HspG41C protein nanocages (Abedin et al., 2009).
3.5 Biocatalysis
The use of NVPNs for enzyme encapsulation and immobilization, substrate channelling and modulation/tuning of enzyme properties offers an opportunity to improve catalytic efficiency, alter selectivity and specificity, and enhance stability and recyclability. For example, bioinorganic hybrid catalysts with interesting characteristics (namely, greater stability and prevention of agglomeration) were created by incorporating an enzyme (e.g., manganese peroxidase) inside a NVPN (such as ferritin, PepA, sHSP and vault nanocages) and using the biomineralization capacities (e.g., iron oxide and platinum) of these nanostructures (Ensign et al., 2004; Varpness et al., 2005).
4 Non-viral protein nanocages manufacturing
4.1 Overview
Efficient manufacturing of NVPNs nanostructures, all the way from the laboratory to the industrial scale, is crucial for the development of applications. However, few scientific studies available in the literature have dealt with the biomanufacturing of natural and artificial NVPNs. In addition to basic and applied research, it is essential to focus on bioprocess development, as this will play a pivotal role in bringing protein nanocages closer to the market.
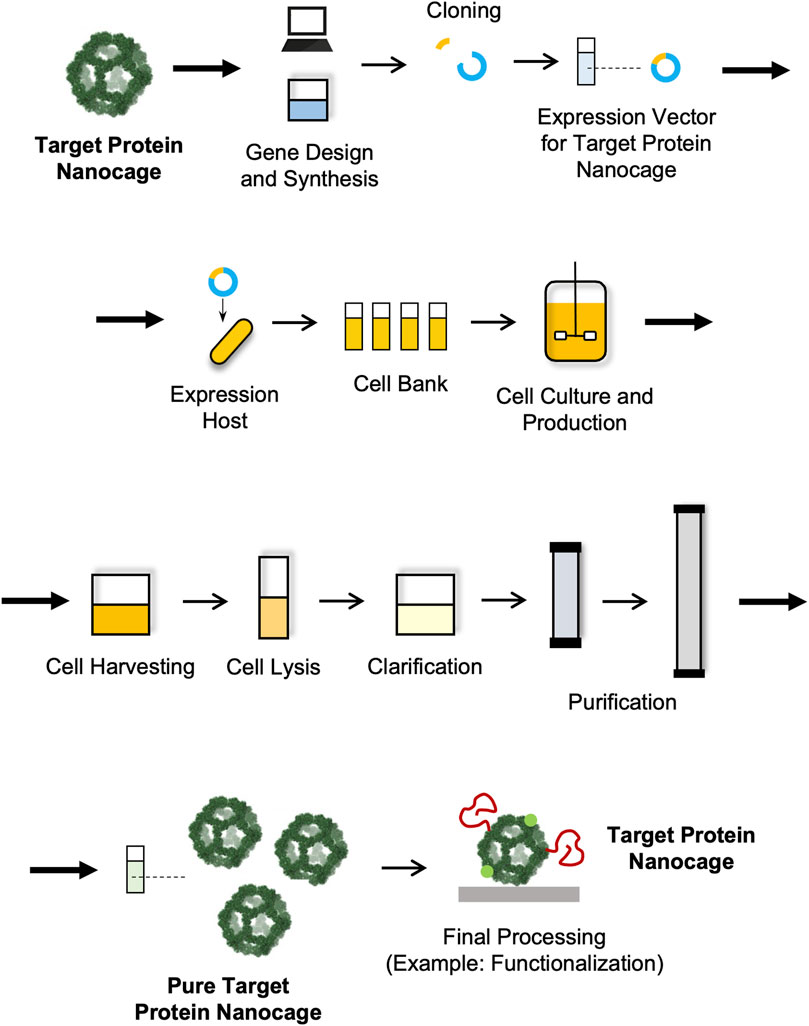
Like in the case of other biological and biopharmaceutical products, the manufacturing of NVPNs involves a sequence of actions that are designed with the objective of producing a certain amount of product with specific quality features (João et al., 2021). These actions can be categorized into the upstream and the downstream processing sections (Section 4.2 Upstream Processing and Section 4.3 Downstream Processing) (Figure 3). The upstream processing involves the generation of the producer host cells (including host selection and cloning), cell banks implementation, inoculum preparation, cell cultivation and protein nanocage expression (Owczarek et al., 2019; Puetz and Wurm, 2019). The downstream processing includes all the unit operations required to purify the NVPNs to a point where final product specifications are met. Lastly, the manufacturing will require a final processing stage, which may comprise formulation, functionalization and sterilization steps to yield the desired final nanocages (Diaz et al., 2018; João et al., 2021).
Since NVPNs are complex biologicals with an intrinsic variability in terms of composition, stability and biological activity, another important aspect to consider during process development is the implementation of analytical and characterization approaches (Section 4.4 Analytical and Characterization Technologies). Clearly, the set-up of such methodologies is essential to determine and evaluate the structural and functional features of the final nanocages, but also to adequately monitor the performance of the biomanufacturing process throughout all its steps (João et al., 2021).
4.2 Upstream processing
Cultivation of producer host cells and protein expression is at the core of a NVPN manufacturing process. While the ultimate purpose is to produce large amounts of protein nanocages, scientific studies focused on application development require amounts that can be generated easily with simple lab-based protocols. In general, these protocols are directed at low-volume batches and do not rely on a strict control of operating conditions. This results in low host cell densities and in reduced nanocage yields. New and optimized strategies are clearly required that can be used in large-scale settings in order to increase volumetric productivity and consequently decrease production costs (Diaz et al., 2018; Tripathi and Shrivastava, 2019; Chen et al., 2021; João et al., 2021).
4.2.1 Selection of host cells
The upstream processing comprises the selection of the host cells, the preparation and optimization of the expression vector and its subsequent transformation/transfection into the host. All these aspects are crucial for obtaining a quality bioproduct and a high productivity (Puetz and Wurm, 2019; Tripathi and Shrivastava, 2019). While the natural host can be selected if the desired NVPN is expressed naturally (Schott et al., 1990; Sutter et al., 2008), nanocage production uses mainly expression systems/host dedicated to recombinant protein production (Diaz et al., 2018). Here, a variety of expression hosts are available that include bacteria, yeasts and multicellular fungi, insect, mammalian and plant cells (Diaz et al., 2018; Puetz and Wurm, 2019; Tripathi and Shrivastava, 2019). NVPNs are mostly produced recombinantly in bacterial cells, and in particular in Escherichia coli, a very-well studied host that grows fast, is easy to cultivate and propagate at low cost, and displays high productivity. However, E. coli is also associated with disadvantages such as the lack of proper post-translational modifications (PTMs), the formation of inclusion bodies and the propensity to generate endotoxin contamination due to its Gram-negative nature (Owczarek et al., 2019; Puetz and Wurm, 2019; Tripathi and Shrivastava, 2019). These can be circumvented using yeasts such as Saccharomyces cerevisiae and Pichia pastoris, which grow fast, are easy to manipulate genetically and can perform PTMs. Mammalian cells are more suitable for production of larger and more complex nanocages that might also require PTMs. The most common and used cell lines are Chinese hamster ovary (CHO), Sp2/0 and NS0. Insect cells like Sf9 can also be used for nanocage production through a baculovirus expression vector system (Owczarek et al., 2019; Puetz and Wurm, 2019; Tripathi and Shrivastava, 2019).
4.2.2 Design of expression vector and preparation of recombinant host cells
The selection of host cells is accompanied by the design and construction of an expression vector/system that can drive a highly efficient nanocage expression (Puetz and Wurm, 2019). The expression vectors will comprise the gene that codes for the NVPN but also any additional genetic elements required to improve stability or enable functionalization. These can be point mutations to change or remove standard reactive residues or to introduce unnatural amino acids, or sequences of functional peptides or proteins that are fused to the N- or C-terminus or internal loops of the monomers (Diaz et al., 2018).
Most types of NVPNs reported in the literature (PepA, Dps, E2 nanocages, encapsulin, ferritin, sHSP, lumazine synthase, I3-01 nanocages, Tet8-M nanocages, TIP60 nanocages and TRAP nanocages) were produced in E. coli (Table 3) using pET-based expression vectors. Table 3 lists the most used key strains of E. coli as well as alternatives in terms of producer host organisms. Contrary to most NVPNs, vault nanocages (Figure 1A) cannot be produced in E. coli due to their eukaryotic origin, which implies complex post-translational modifications and protein folding (Diaz et al., 2018). In this specific case, yeast, insect, or mammalian cells must be used as producer hosts (Table 3). In yeast, the expression vector comprises a glyceraldehyde 3-phosphate dehydrogenase promoter (Wang et al., 2018) and in insect cells a Bac-to-Bac method based on infection with a recombinant baculovirus is necessary (Stephen et al., 2001; Kickhoefer et al., 2005; Poderycki et al., 2006; Champion et al., 2009; Kar et al., 2011; Kar et al., 2012; Wang et al., 2015).
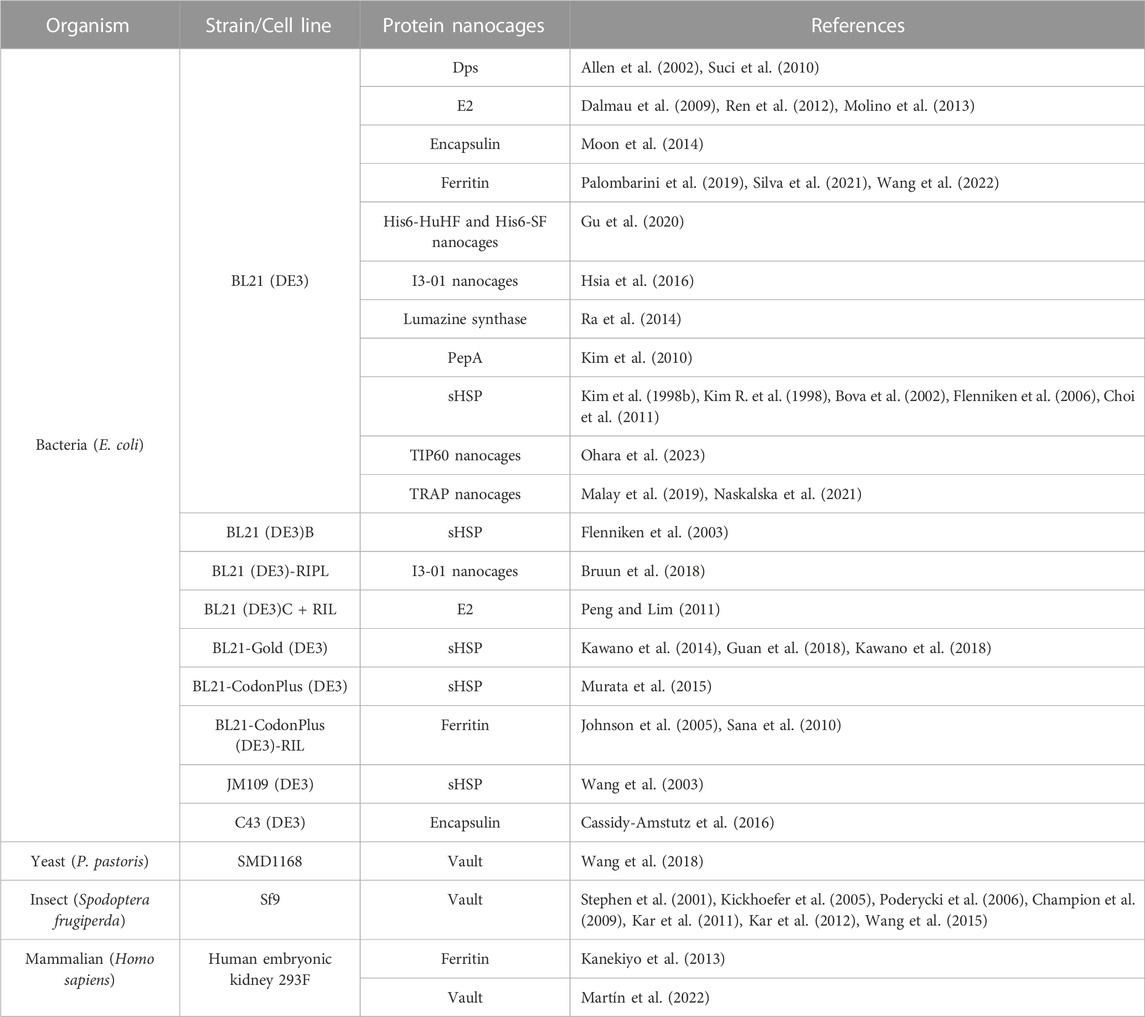
TABLE 3. A summary of organisms and respective strains or cell lines used in the production of NVPNs.
Once recombinant host cells with the expression vector are established, host cell banks are prepared that must be properly characterized and stored. This allows the maintenance of the reproducibility and consistency of the process, since each batch of protein nanocages will be manufactured using the same cell source (Puetz and Wurm, 2019; João et al., 2021).
4.2.3 Cultivation of host cells and NVPNs expression
Following bank preparation, host cells are cultivated and the NVPN is expressed. Initially, a screening should be performed using small-scale cultures to identify and evaluate the impact of cultivation and operation parameters on protein expression levels. These parameters include media composition, temperature, agitation, aeration, cell density, pH, inducer concentration and induction time, among others (Tripathi, 2016; Tripathi and Shrivastava, 2019). After the process is defined at a small scale, scale-up and pilot studies are performed in highly controlled bioreactors as a steppingstone to industrial scale implementation. Bioreactors can be operated in batch, semi-batch, and continuous/perfusion modes, depending on the method used to supply nutrients, circulate the culture medium or recover the target NVPN. Large scale manufacturing needs to be run using optimal operation conditions that maximize expression and yield of NVPNs. Specific parameters of the bioreactor (aeration, dissolved oxygen, CO2, and hydrodynamic shear) also need to be tested and optimized to guarantee high specific and volumetric productivities. A Design of Experiments (DOE) approach may be used as a more efficient strategy to optimize these parameters and understand the interaction effects between them (Tripathi, 2016; Tripathi and Shrivastava, 2019; João et al., 2021).
NVPNs studied in the literature are produced mostly in shake flask cultures with volumes up to 4 L (Zou et al., 2016a; 2016b), mainly using Luria-Bertani (LB) (Allen et al., 2002; Bellini et al., 2014; Zou et al., 2016b) or 2× YT growth media (Murata et al., 2015; Guan et al., 2018; Kawano et al., 2018; Cristie-David and Marsh, 2019). When under the control of a T7 promoter, expression is induced with the addition of 0.1 mM (Cristie-David and Marsh, 2019; Gu et al., 2020; Ohara et al., 2023) to 1 mM (Santambrogio et al., 2000; Dalmau et al., 2008; 2009; Jeon et al., 2013; Kawano et al., 2014; Zou et al., 2016a; 2016b) of isopropyl β-D-1-thiogalactopyranoside (IPTG) to the culture during the exponential growth phase. Other parameters that may vary are the induction time, between 2 h (Kim R. et al., 1998; Kim et al., 1998a; Santambrogio et al., 2000; Allen et al., 2002; Choi et al., 2011) and 30 h (Jeon et al., 2013; Zou et al., 2016a; Zou et al., 2016b; Cristie-David and Marsh, 2019; Palombarini et al., 2019), and the temperature at and after induction, which is frequently 20°C–25°C (Jeon et al., 2013; Zou et al., 2016a; Zou et al., 2016b; Palombarini et al., 2019) or 37°C (Zou et al., 2016a; Zou et al., 2016b). Overall, data regarding selection and optimization of process and operating conditions, as well as potential scale-up approaches, is very limited.
A key aspect to consider when using prokaryotic expression systems is whether protein overexpression leads to the accumulation of NVPNs as intracellular inclusion bodies. To minimize this, conditions should be optimized to increase nanocage solubility. For example, Zou and co-workers evaluated the effect of temperature (20°C, 15 h and 37°C, 5 h) on the expression of the heavy and light chains of human ferritin and on the co-expression of molecular chaperones. The authors concluded that the amount of soluble protein nanocages increased with the lower temperature and with the presence of chaperones to help in the folding process (Zou et al., 2016a; Zou et al., 2016b). If formation of inclusion bodies cannot be avoided altogether, solubilization and refolding steps must be considered in the downstream processing (Tripathi, 2016; Tripathi and Shrivastava, 2019). A study by Palombarini et al. (2019) on the expression of ferritin monomers optimized the concentration of IPTG (0.1, 0.5, and 1 mM), the induction time (4, 8, and 16 h) and the induction temperature (25°C and 37°C). The addition of 0.5 mM IPTG at 25°C for 16 h was identified as optimal and subsequently implemented at a large-scale production of ferritin nanocages by the biotechnology company GeneScript (Palombarini et al., 2019).
Martín and co-workers developed a strategy for the production of vault nanocages in mammalian cells as a faster and more efficient alternative to the traditional expression in yeast or insect cells. An engineered vault nanocage (His-tagged major vault protein) was successfully produced in the human embryonic kidney 293F cell line through transient gene expression (Martín et al., 2022).
4.2.4 Alternative NVPNs expression strategy
Cell-free protein synthesis has been widely explored in recent years as an alternative to cell-based expression systems but reports of its use in NVPNs production are scarce. In one example, Mrazek described the obtention of engineered vaults using a cell-free wheat germ expression system and either a DNA vector or an mRNA encoding the major vault protein. Notably, the author was able to simultaneously package passenger molecules in the internal cavity of the formed vaults by adding them to the synthesis mixture (Mrazek, 2016). However, due to its current limitations and challenges, cell-free protein synthesis does not seem to be the most suitable strategy for consistent production of NVPNs at large-scale (Kwon and Jewett, 2015; Des Soye et al., 2018; Failmezger et al., 2018; Tran et al., 2018; Gregorio et al., 2019; Colant et al., 2021).
4.3 Downstream Processing
4.3.1 Overview
The downstream processing encompasses the extraction, isolation, and purification of the target NVPNs from the broth culture obtained in the upstream stage. The final end-product must conform to a predetermined set of specifications that are established with the final intended use in consideration. Ideally, the overall downstream process will comprise a small number of high-yield unit operations so as to minimize complexity, residence time and processing costs (João et al., 2021). Typically, unit operations are selected and implemented that explore different physical-chemical properties of the specific protein nanocages and of the associated impurities (genomic DNA, RNA, host proteins, and endotoxins). In general, the downstream processing of NVPNs will include primary isolation and recovery steps such as cell harvesting, cell lysis and clarification, and then a number of purification steps (Tripathi, 2016; Tripathi and Shrivastava, 2019; João et al., 2021). An overall description of this downstream processing is illustrated in Figure 4.
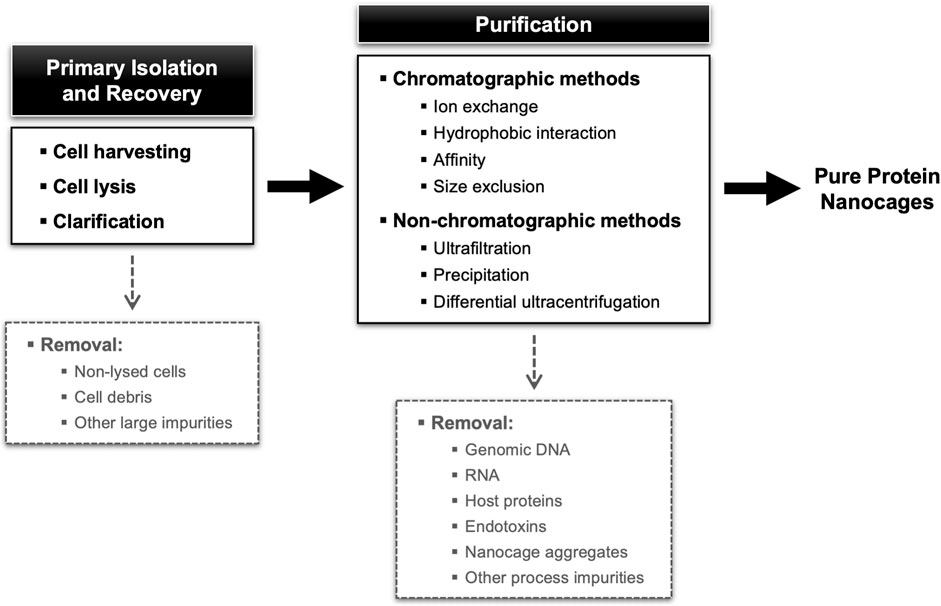
FIGURE 4. Block diagram for a standard NVPNs downstream processing showing different possibilities of unit operations.
4.3.2 Primary isolation and recovery
In the first step of the downstream processing, NVPN producer cells are harvested from the culture broth, for example, by centrifugation at moderate speeds (e.g., 5,000 × g) (Kim et al., 1998b; Zou et al., 2016a; 2016b; Martín et al., 2022). Next, cells are resuspended in an appropriate buffer, and the intracellular nanocages are released via a specific cell lysis step. The most common options at lab scale are ultrasonication (Allen et al., 2002; Suci et al., 2010; Choi et al., 2011; Jeon et al., 2013; Zhen et al., 2013; Bellini et al., 2014; Cassidy-Amstutz et al., 2016; Martín et al., 2022), French press (Jung et al., 2002; Suci et al., 2010; Peng and Lim, 2011) and Dounce homogenization cell lysis (Stephen et al., 2001; Poderycki et al., 2006). In some instances, lysozyme is added to the lysis buffer to weaken/break down bacterial cell walls and hence improve lysis (Allen et al., 2002; Suci et al., 2010; Bellini et al., 2014). Nucleases (e.g., DNase, RNase) are also added often to lysis buffers to promote degradation of nucleic acids as they are released (Allen et al., 2002; Suci et al., 2010; Bellini et al., 2014). Protease inhibitors are also an option in some cases (Stephen et al., 2001; Poderycki et al., 2006; Bruun et al., 2018; Martín et al., 2022). For large-scale operation, high-pressure homogenizers and bead milling will certainly be more suitable than sonication, French press or Dounce homogenization cell lysis (Tripathi and Shrivastava, 2019).
After cell lysis, a clarification step based on centrifugation or filtration is normally implemented to remove cell debris and particulate matter. If a situation exists where nanocage inclusion bodies are formed, these must be separated from cell debris and recovered, solubilized and then adequately refolded (Dalmau et al., 2008; Dalmau et al., 2009; Tripathi, 2016; Tripathi and Shrivastava, 2019). For example, Jeon and co-workers have shown that inclusion bodies of ferritin nanocages can be solubilized with 8 M urea and then refolded on a nickel ion chelate affinity column with a downward gradient of urea from 8 M to 0 M (Jeon et al., 2013).
Clarified lysates containing the NVPNs are sometimes subjected to a heat treatment prior to purification. This approach explores the fact that protein nanocages are in general thermostable at high temperatures. Thus, heating clarified E. coli cell lysates at 65°C–90°C will promote the denaturation and precipitation of host proteins, which are subsequently removed by centrifugation (e.g., 12,000 × g), without affecting the structure of the thermostable nanocages (Diaz et al., 2018). This strategy was applied successfully to lysates containing a range of different nanocages, including Dps (Allen et al., 2002; Suci et al., 2010), E2 nanocages (Dalmau et al., 2008; 2009), encapsulin (Moon et al., 2014; Cassidy-Amstutz et al., 2016), ferritin (Santambrogio et al., 2000; Sana et al., 2010; Falvo et al., 2013; Zhen et al., 2013; Bellini et al., 2014; He and Marles-Wright, 2015), sHSP (Flenniken et al., 2006) and I3-01 nanocages (Hsia et al., 2016).
Several studies also report the enzymatic treatment of nanocage-containing clarified E. coli cell lysates. In the majority of cases, a DNase or an RNase is used directly in the clarified lysates (Santambrogio et al., 2000; Murata et al., 2015) or previously added to the lysis buffer (Allen et al., 2002; Flenniken et al., 2003; Flenniken et al., 2006; Suci et al., 2010; Bellini et al., 2014; Ohara et al., 2023) to degrade host cell nucleic acid impurities such as genomic DNA and RNA that are co-released from cells during lysis. The resulting products of degradation (e.g., short oligonucleotides) are easier to remove in subsequent purification steps. Protease inhibitors may also be added at this stage to minimize enzymatic degradation of nanocages (Bruun et al., 2018).
4.3.3 Purification
The NVPNs-containing clarified lysates generated in the intermediate recovery stage and entering the final purification stage will often contain host derived impurities that must be removed to obtain a bulk product complying with final-specifications (Figure 4). Apart from genomic DNA, RNA, host proteins and other macromolecules (e.g., lipopolysaccharides in the case of E. coli), it is also important to remove misfolded nanocages or aggregates. Given this range of impurities, the purification stage is likely to comprise more than one step. Data on the downstream processing available in the literature primarily comes from laboratory protocols. This limited information makes it difficult to compare and weigh the merits of the different methodologies used.
4.3.3.1 Chromatographic methods
Column chromatography is the preferred method for obtaining highly pure nanocages. The interaction modes explored include ion exchange (IEX), hydrophobic interaction (HIC), affinity (AC) and size exclusion (SEC). In IEX, HIC, and AC, nanocages are retained by interacting with the stationary phase, whereas impurities flowthrough and/or elute under suitable buffer conditions. Nanocages are then eluted using an adequate buffer. In general, the nanocage-containing fractions obtained are concentrated relatively to the feed (Owczarek et al., 2019; João et al., 2021). In spite of its known shortcomings, SEC is an attractive purification option because nanocages are in general larger than most host-derived impurities.
The large size of nanocages (10–100 nm) is likely to impact the performance of IEX, HIC, and AC. For once, and on account of their size, the diffusion coefficients of protein nanocages are inherently small. Furthermore, since most chromatographic matrices feature pores with diameters that seldom exceed the 30 nm (Tiainen et al., 2007), intrapore diffusion will be hindered. This may translate into internal mass transfer limitations, which can result in broad peaks, low recovery, and the need to use small flow rates. This proximity of nanocage and pore size is also likely to translate into poor binding capacities. One possible way to overcome these capacity limitations, is to use stationary phases like chromatographic membranes and monoliths that are engineered to accommodate very large pores (>200 nm). This strategy is in line with what is used for the purification of other very large biologicals such as plasmids, bacteriophages or VLPs (Prazeres, 2009; João et al., 2021).
4.3.3.1.1 Ion exchange and hydrophobic interaction chromatography
In IEX, anion-exchange (AEX) resins are used almost exclusively due to the anionic nature of the outer surface of most NVPNs. However, negatively charged impurities such as nucleic acids and lipopolysaccharides can also bind to anion exchangers and thus affect binding capacity and performance. The feed to AEX columns is usually a clarified lysate of the producer host cells (microbial or mammalian). Examples of resins used include strong anion-exchangers with quaternary amine functional groups such as Uno-Q, Q Sepharose, HiTrap Q and HiPrep Q (Kim et al., 1998b; Kim R. et al., 1998; Allen et al., 2002; Dalmau et al., 2008; 2009; Choi et al., 2011; Ren et al., 2012; Kanekiyo et al., 2013; Molino et al., 2013; Kawano et al., 2014; Kawano et al., 2018; Murata et al., 2015; Peng et al., 2015) and weak anion-exchangers such as DEAE Sepharose (Gu et al., 2020). In most cases, sodium chloride gradients (up to 1 M) are employed to increase the ionic strength and elute the nanocages. If an adequate combination of stationary phase, operating conditions and elution scheme is used, a substantial amount of host impurities can be removed by AEX. HIC has also been used to purify ferritin nanocages using a Phenyl Sepharose resin (Johnson et al., 2005; Sana et al., 2010). The key disadvantage of HIC, however, is the need to use large amounts of salts to promote binding.
4.3.3.1.2 Size exclusion chromatography
The removal of most impurities by AEX is usually followed by a SEC polishing step. Examples of SEC resins used here include Superose 6 (Allen et al., 2002; Dalmau et al., 2008; 2009; Kanekiyo et al., 2013; Molino et al., 2013), Superdex 200 (Choi et al., 2011; Peng et al., 2015; Gu et al., 2020), TSKgel G3000SW (Kawano et al., 2014; Kawano et al., 2018; Murata et al., 2015) and Sephacryl S-200 (Kim R. et al., 1998; Kim et al., 1998b). The key goal when using SEC for polishing is to separate nanocages from similarly sized impurities such as nanocage aggregates and misassembled variants. Baseline peak separation of the later impurities will in general be difficult to achieve due to the limitations of resolution inherent to SEC. Removal of traces of host impurities and buffer exchange are also afforded by SEC.
4.3.3.1.3 Affinity chromatography
AC is often used for the purification of engineered variants of natural and artificial NVPNs. The method requires the incorporation of an affinity tag in the primary sequence of monomers, to enable the capture of assembled nanocages by an affinity resin modified with the appropriate ligand. For example, poly-histidine tags in combination with nickel ions chelate affinity resins (e.g., Ni-NTA, Ni Sepharose 6, Ni-NTA-Sefinose, HiTrap Chelating) are widely used to purify a range of protein nanocages (Kim et al., 2010; Kim et al., 2017; Jeon et al., 2013; Zou et al., 2016a; Zou et al., 2016b; Guan et al., 2018; Ohara et al., 2023). Similarly, vault nanocages produced in 293F mammalian cells were purified using commercial magnetic particles in a platform based on immobilized metal affinity chromatography (IMAC) chemistry (Martín et al., 2022). In another instance, a maltose-binding protein (MBP) domain was fused to the N-terminus of TriEst, an esterase monomer used to assemble artificial protein nanocages Tet8-M. Purification was then performed with a maltose affinity resin (MBP-Trap) (Cristie-David and Marsh, 2019). While AC provides high selectivity and specificity, leading to high purity levels, in some cases a second purification step is performed, for example, by SEC (Hsia et al., 2016; Kim et al., 2017; Bruun et al., 2018; Cristie-David and Marsh, 2019) or an AEX (Kim et al., 2010). Possible downsides of AC are related to the requirement to add affinity tags to monomers. Apart from the extra effort involved, the presence of tags can eventually compromise the self-assemble process or alter the properties of the original nanocages.
4.3.3.1.4 Combinations of chromatographic steps
Although the AEX-SEC combination is widely used, Santambrogio and co-workers proposed a scheme for the purification of the heavy chain of mouse ferritin nanocages that combines a first SEC step (Sepharose 6B resin) followed by an AEX step (HiTrap Q resin) (Santambrogio et al., 2000). A similar approach was used by Jung et al. (2002) when purifying E2 nanocages (SEC with a Superdex 200 resin and AEX with a Mono-Q resin). Finally, Bova and co-workers established a three-step chromatographic purification of sHSP nanocages, which involved AEX (Mono-Q resin), HIC (Phenyl Sepharose resin) and SEC (Superose 6 resin) (Bova et al., 2002). While combinations of chromatographic steps are the norm when purifying nanocages, some reports describe the purification of NVPNs with either a single AEX (DEAE Sepharose and HiPrep Q) (Peng and Lim, 2011; Bellini et al., 2014) or a single SEC step (Superose 6, Superdex 200 and Sephacryl S-400) (Flenniken et al., 2003; Suci et al., 2010; Zhen et al., 2013; Moon et al., 2014; Wang et al., 2016; Ohara et al., 2023).
One downstream processing that is particularly interesting and unique among NVPNs involves artificial TRAP nanocages. Unlike other protein nanocages that undergo purification after self-assembly, TRAP nanocages are assembled in vitro in the presence of gold ions. This means that the individual sub-units of TRAP nanocages, which are composed of 11 monomeric TRAP proteins, must be purified in advance. Heddle and his group elaborated a downstream strategy that starts with a heat treatment of the clarified E. coli lysate containing the TRAP rings followed by AEX (Q Sepharose or HiTrap Q resins) and SEC (Superdex 200 resin) steps (Heddle et al., 2006; Heddle et al., 2007; Malay et al., 2019; Naskalska et al., 2021; Stupka et al., 2022).
4.3.3.1.5 Pre-chromatography processing
In some studies the target protein nanocages are precipitated with ammonium sulphate prior to AEX or SEC chromatography to remove nucleic acid impurities (Kim R. et al., 1998; Kim et al., 1998b; Santambrogio et al., 2000; Jung et al., 2002; Hsia et al., 2016; de Turris et al., 2017; Gu et al., 2020). Differential ultracentrifugation is also described as pre-purification step before chromatography (AEX or SEC) (He and Marles-Wright, 2015). For example, encapsulins were subjected to ultracentrifugation with sucrose gradient [e.g., 10%–50% (w/v)]. However, this approach has clear drawbacks such as the need for high centrifugation speed (e.g., 100,000 × g) and extensive centrifugation time (e.g., 18 h), and the lack of scalability (Moon et al., 2014; Cassidy-Amstutz et al., 2016).
4.3.3.1.6 Chromatographic process yields
Despite limited, some process yield data for purified NVPNs can be summarized. For example, yields of 56 mg and 25 mg per liter of cell culture were obtained for ferritin nanocages assembled from heavy chain sub-units and purified with a single AEX step (Bellini et al., 2014) and for ferritin nanocages purified with a single SEC step (Zhen et al., 2013), respectively. On the other hand, yields of 15 mg (purity >90%) and 10 mg (purity of 96%) per liter of cell culture were obtained when using Ni-affinity chromatography for the purification of the heavy and the light chains of ferritin nanocages, respectively (Zou et al., 2016a; Zou et al., 2016b). For an IMAC-based platform, Martín and co-workers verified a recovery of 90.4% in terms of the vault nanocages in the soluble fraction, obtaining a protein concentration of 20 μg mL−1. These authors concluded that 5 mg of the magnetic nanoparticles can capture 30 μg of vault nanocages, with a purity greater than 85% (Martín et al., 2022). A two-step chromatography purification (SEC + AEX) yielded 15 mg and 7 mg per liter of cell culture of the heavy and the light chains of ferritin, respectively (Santambrogio et al., 2000). Other reported values include 20 mg L−1 of cell culture for E2 nanocages purified by AEX + SEC (Dalmau et al., 2008; Dalmau et al., 2009), 400 mg L−1 of cell culture for artificial protein nanocages Tet8-M purified with AC (Cristie-David and Marsh, 2019) and 50 mg L−1 of cell culture and 40 mg L−1 of cell culture for encapsulin and ferritin nanocages, respectively, when purified by tandem ammonium sulphate precipitation and SEC (Cassidy-Amstutz et al., 2016; de Turris et al., 2017). A final yield of purified TRAP rings of 1–2 mg L−1 of cell culture was obtained with a combination of AEX and SEC steps (Heddle et al., 2006; Heddle et al., 2007; Malay et al., 2019; Naskalska et al., 2021; Stupka et al., 2022).
4.3.3.2 Non-chromatographic methods
Palombarini and co-workers devised and suggested an alternative methodology for large-scale NVPNs purification that eliminates chromatographic steps while maintaining high efficiency and potentially reducing costs (Palombarini et al., 2019). Specifically, a ferritin nanocage-containing lysate obtained by sonication was subjected to an initial heat treatment and then clarified by vacuum filtration aided by diatomaceous earth. Next, the clarified lysate was purified by crossflow ultrafiltration using a 100 kDa cut-off membrane module. The system was operated in concentration and diafiltration modes, producing a stream with a final concentration of 20 g L−1. The process was able to eliminate critical impurities, including genomic DNA and non-targeted proteins. The methodology offers the advantage of regenerating and reusing the filtration modules multiple times, depending on the sample source and quality.
The immunogenic and pyrogenic nature of endotoxins, which mainly include lipopolysaccharides (LPS) from Gram-negative bacteria, is a significant concern when purifying biological products. Regulatory agencies such as the Food and Drug Administration (FDA) and the European Medicines Agency (EMA) establish maximum allowed limits for the quantity of endotoxins that must be met when manufacturing nanocages for biomedical applications (Schwarz et al., 2014; Franco et al., 2018). When exploring methods to remove endotoxins, it is crucial to consider the substantial size of nanocages, which makes them more prone to interact with endotoxins, as well as the requirement to preserve their architecture (Silva et al., 2021). Silva et al. (2021) employed an Endotrap HD resin in conjunction with the detergent Triton X-114 followed by a polishing step with SEC to remove endotoxins from the heavy chain of ferritin nanocages. Recovery of nanocages was 57% and the final concentration of endotoxins was 0.83 EU mL−1, which is lower than the maximum acceptable limit for in vitro and in vivo biomedical purposes. Additionally, Molino and his research group used a method that relied on successive washes of E2 nanocages with Triton X-114 to successfully decrease the concentration of endotoxins to acceptable values (Molino et al., 2013).
Vault nanocages whose production was performed both in P. pastoris SMD1168 and Sf9 insect cells were purified using a discontinuous density gradient ultracentrifugation after cell lysis (Stephen et al., 2001; Poderycki et al., 2006; Wang et al., 2018).
4.3.4 Representative downstream processes
To illustrate the diversity of options available in terms of producer hosts and unit operations, four different block diagrams of NVPNs manufacturing processes are schematized in Figure 5. These block diagrams were adapted from the literature and are representative of the current panorama in the downstream processing of protein nanocages.
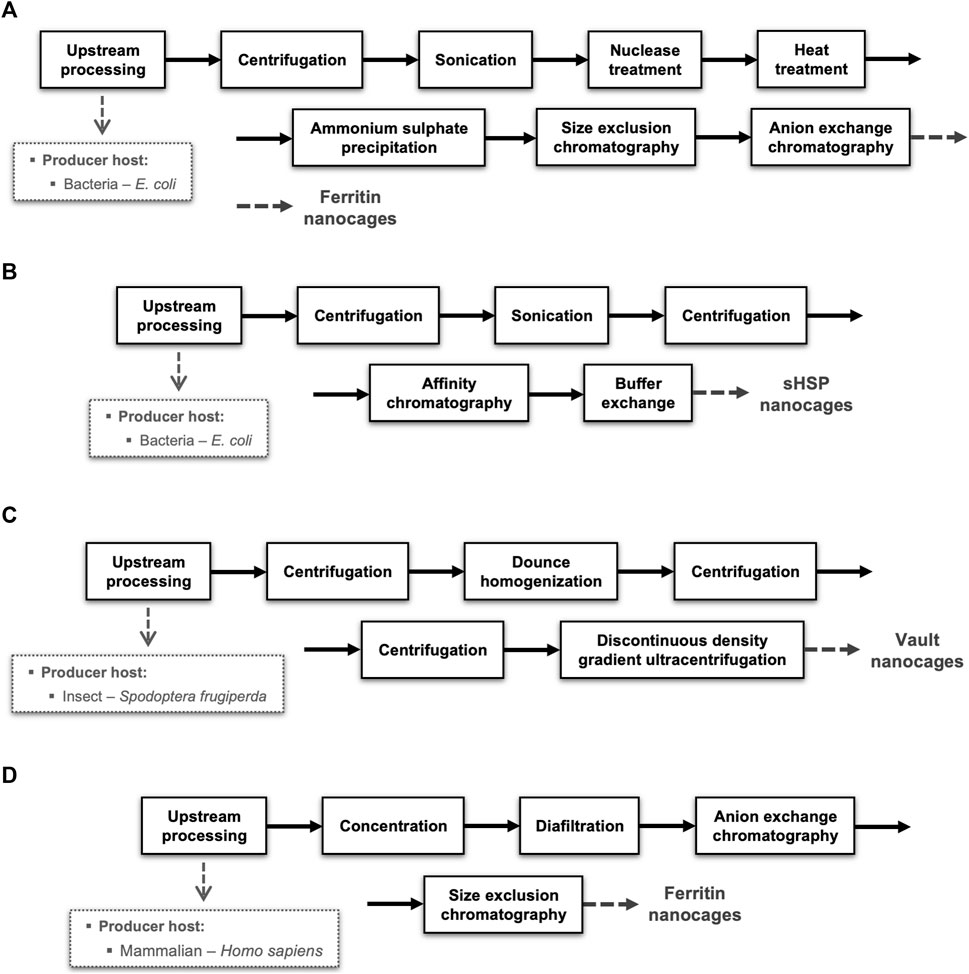
FIGURE 5. Representative examples of downstream processes implemented for distinct NVPNs produced in different organisms. Blocks diagrams (A,B) illustrate the downstream step respectively of ferritin and sHSP nanocages produced in E. coli and purified by chromatography. Block diagram (C) shows the production of vaults nanocages in insect (Spodoptera frugiperda) cells. Block diagram (D) exemplifies a downstream process of ferritin nanocages produced in mammalian (Homo sapiens) cells.
The majority of the downstream processing studies available refer to NVPNs produced in E. coli, which is clearly the most common producer microbial host. Blocks diagrams A (Figure 5A) and B (Figure 5B) exemplify the downstream processing respectively of ferritin (Santambrogio et al., 2000) and sHSP (Guan et al., 2018) nanocages produced in E. coli and purified by chromatography. On the other hand, block diagram C (Figure 5C) illustrates the production of vault nanocages in Spodoptera frugiperda insect cells (Stephen et al., 2001). Finally, block diagram D (Figure 5D) shows a downstream process of ferritin nanocages produced in mammalian (Homo sapiens) cells (Kanekiyo et al., 2013).
4.4 Analytical and characterization technologies
The establishment and validation of analytical and characterization technologies for assessing the structural and functional characteristics is crucial to confirm if the purified nanocage is within specifications. Furthermore, analytical techniques are also relevant to monitor the performance, robustness and consistency of the different manufacturing steps (João et al., 2021). Some well-established techniques routinely used for the determination and assessment of the biophysical characteristics of different types of NVPNs are summarized in Table 4. Dynamic light scattering (DLS) and transmission electron microscopy (TEM) of negatively stained preparations allow the estimation of the average hydrodynamic diameter of nanocages. Microscopy provides a means to observe the morphology of nanostructures and compare it with the corresponding theoretical 3D structure available in databases. Furthermore, TEM is also useful to visualize the biomineralization of nanocages. The use of analytical SEC (typical resins/columns are Superdex 200, Superose 6 and TSKgel G4000SW) enables the estimation of the molecular weight of the assembled nanostructure by reference to a calibration curve of high and low molecular weight proteins. Further, it can be used to evaluate the oligomeric state of the nanocages at the end of the bioprocess.
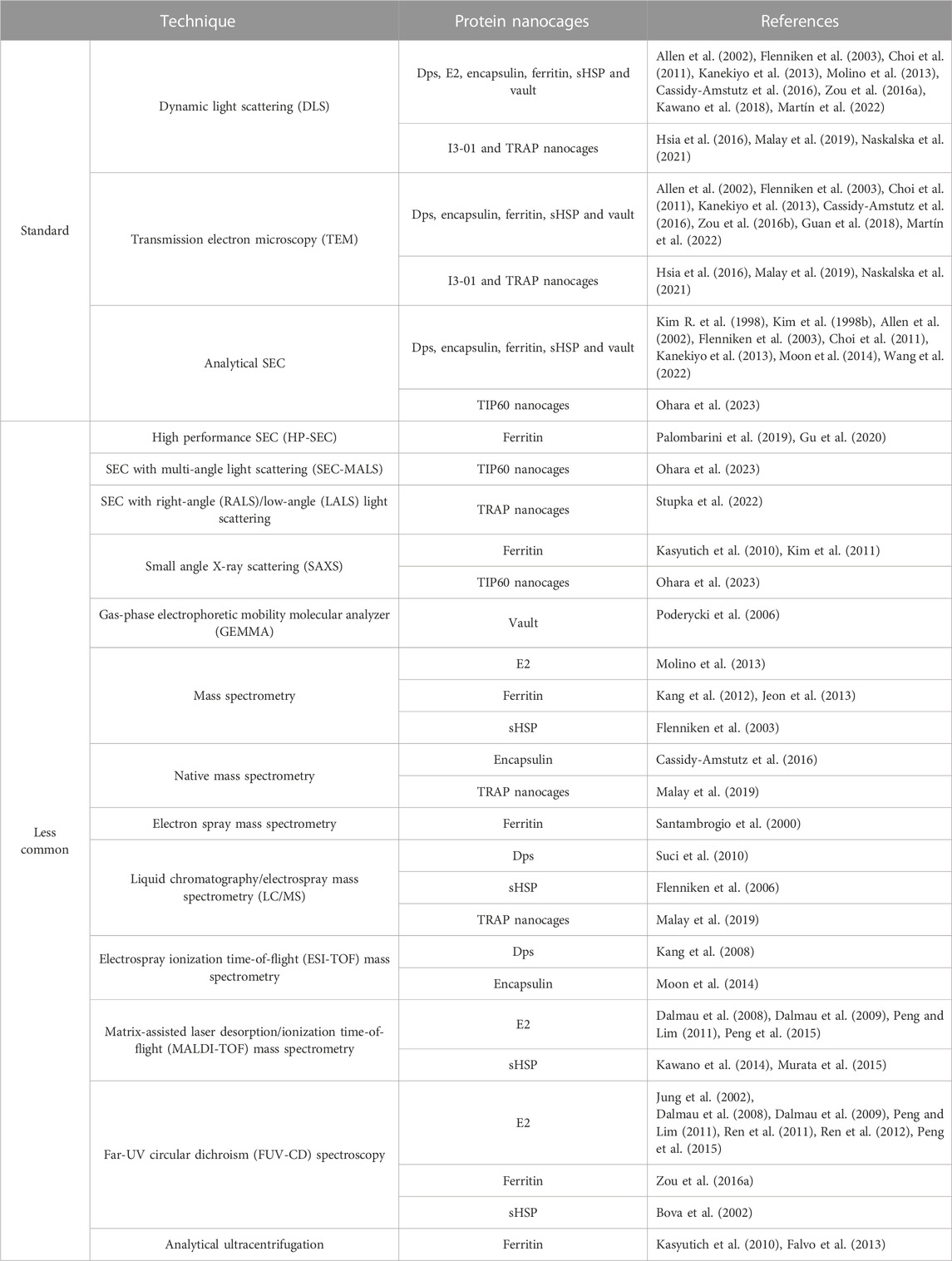
TABLE 4. A summary of well-established and less common analytical techniques used to determine and evaluate the biophysical characteristics of different types of NVPNs.
Additionally, the literature reports the use of less common analytical techniques (summarized in Table 4). These techniques are used to estimate or determine structural properties of the target protein nanocages including the molecular weight, the particle size distribution, and the oligomeric state of each nanostructure. Furthermore, far-UV circular dichroism (FUV-CD) spectroscopy is used to characterize the secondary structure, the folding, and the thermostability of the protein nanocages.
Atomic force microscopy (AFM) is another promising tool for nanocage characterization. Apart from providing a means to visualize the nanostructures, AFM can be used to determine mechanical properties and study protein-protein interactions. Heddle and his group used AFM and high speed-AFM to visualize and characterize TRAP nanocages (Majsterkiewicz et al., 2022; Stupka et al., 2022). AFM was also used to analyse encapsulin nanocages (Boyton et al., 2022), ferritin nanocages (Stühn et al., 2019), lumazine synthase nanocages (Heinze et al., 2016), vault nanocages (Llauró et al., 2016) and O3-33 artificial nanocages (Heinze et al., 2016).
Although not extensively explored, methods based on fluorescence are promising due to the ease with which it is possible to label the exterior or interior of nanocages with fluorophores (e.g., Alexa Fluor 488-maleimide, Alexa Fluor 750-maleimide, fluorescein, Cys 5.5 dye) (Bova et al., 2002; Flenniken et al., 2003; 2006; Choi et al., 2011; Falvo et al., 2013; Kawano et al., 2014; Murata et al., 2015). For example, the detection and study of time-dependent fluctuations in fluorescence intensity afforded by fluorescence correlation spectroscopy (FCS) can be used to determine several physical and chemical parameters including translational and rotational diffusion coefficients (from which hydrodynamic diameters can be inferred), chemical kinetic rate constants and molecular aggregation (Schmitt et al., 2022).
Monitoring the concentration, homogeneity, and purity of the target protein nanocages throughout manufacturing is critical. Total protein is estimated by assays such as the bicinchoninic acid (BCA) (Wang et al., 2003; Wang et al., 2015; Kickhoefer et al., 2005; Heddle et al., 2006; Kar et al., 2011; Peng and Lim, 2011; Bruun et al., 2018; Malay et al., 2019; Naskalska et al., 2021; Martín et al., 2022), the Bradford (Choi et al., 2011; Zhen et al., 2013) and the biuret (Allen et al., 2002). Methods for the specific quantitation of nanocages in a mixture would be especially useful to monitor the performance of the difference steps of manufacturing. High performance liquid chromatography (HPLC) could be a potential approach, in particular based on a size exclusion chromatographic support, as it has been investigated and implemented for other large biological molecules, namely, VLPs (Effio et al., 2016; Steppert et al., 2017). SDS-PAGE is commonly used, but due to its denaturing characteristics it can only detect the presence of the nanocage monomers and other protein impurities (Flenniken et al., 2003; Kanekiyo et al., 2013; Martín et al., 2022). Native PAGE, on the other hand, can be used to check the native quaternary structure of nanocages (Santambrogio et al., 2000; Cassidy-Amstutz et al., 2016; Ohara et al., 2023).
4.5 Drawbacks and challenges
Developing a manufacturing process for NVPNs is a complex task, in part due to the structural and functional nature of protein nanocages. The initial challenge for process developers is to collect available data and information related to the features of the target nanocages (Moleirinho et al., 2020). Then, improving the efficiency of current processes and incorporating innovative unit operations into an integrated process is vital for improving both upstream and downstream processing of NVPNs (Moleirinho et al., 2020). The existing literature lacks data regarding the upstream processing steps, particularly concerning the optimization of the protein expression conditions and operating parameters that impact quality and concentration of protein nanocages. This optimization process is quite challenging due to the large number of involved factors and their potential interaction. To improve the purity of the final product, it is essential to establish a consistent and effective sequence of unit operations for purification. Furthermore, there should be a greater emphasis on investigating and exploring the conditions and parameters that impact purification, particularly in the chromatography stages. One example is conducting dynamic binding capacity studies. Efforts to improve the manufacturing process of NVPNs should also encompass an investment in the enhancement of current analytics and the exploration of newer techniques (Moleirinho et al., 2020). This is crucial since the analytical component of the manufacturing process is often challenging due to the complexity, time-consuming nature, or poor reliability of certain quantification methodologies.
Once a NVPNs manufacturing process is established in batch, there is potential for a shift towards continuous processing, which could bring several advantages, including high purity, increased productivity, and reduced overall process costs (Moleirinho et al., 2020; Papathanasiou and Kontoravdi, 2020). Improved analytical and monitoring methodologies that are capable of on-line and in-line analysis and can be integrated into the process will be required for continuous monitoring, process control, and product quantification (Moleirinho et al., 2020). For the upstream processing, this could entail the monitoring of standard physical and physicochemical cell culture parameters (e.g., temperature, pH, dissolved oxygen, optical density, and off-gas composition), as well as the implementation of spectroscopic (e.g., UV-visible, fluorescence, near-infrared, infrared, and Raman) and light scattering techniques [e.g., DLS and multi-angle light scattering (MALS)]. In the downstream processing, and apart from the more standard monitoring of UV, conductivity, and refractive index, light scattering sensors such as MALS should be considered (Patel et al., 2018; Aguilar et al., 2019; Tripathi and Shrivastava, 2019; Papathanasiou and Kontoravdi, 2020; João et al., 2021).
Overall, developing a robust, efficient, and scalable NVPNs manufacturing process with a reduced number of unit operations is crucial to maximize recovery yield. However, achieving this while maintaining the final quality of nanocages in a cost-effective manner remains a major challenge. As with other biological products, adjusting the manufacturing process for each type of NVPNs will be required (Moleirinho et al., 2020; João et al., 2021).
5 Conclusion
Protein-based nanoparticles, including natural NVPNs, have gained significant attention in bioengineering, biotechnology, and biomedicine due to their intrinsic characteristics. The design of protein-based nanocages based on natural nanostructures has also shown promise for practical applications. However, there is still a need for further research to fully understand the underlying characteristics and assembly mechanisms of nanocages. While implementing biomanufacturing processes suitable for a large-scale production is critical to bring nanocages closer to the market, few scientific studies have addressed the upstream and downstream processing of nanocages. E. coli is commonly used as the producer host but in the future, alternative host bacteria and other non-bacterial organisms could be explored. Further the standard production processes based on laboratory protocols should be modified and adapted for scale-up. The conceptual design of downstream processing of NVPNs is hampered by limited data and information on alternative steps and process yields. While the combination of AEX and SEC is the most used approach, complementary strategies such as aqueous two-phase extraction and crossflow ultrafiltration could be explored in the future. Finally, it is crucial to invest in more effective and simpler analytical and characterization techniques to determine and assess the structural and functional characteristics of nanocages. Clearly, further research is required to develop cost-effective NVPNs manufacturing processes.
Author contributions
JJ was responsible for conceptualization, writing–original draft, and writing–review and editing. DMFP was responsible for supervision, funding acquisition, conceptualization, validation, and writing–review and editing. All authors contributed to the article and approved the submitted version.
Funding
FCT–Fundação para a Ciência e a Tecnologia (Projects UIDB/04565/2020 and UIDP/04565/2020) and Programa Operacional Regional de Lisboa 2020 (Project Nr. 007317) for the iBB–Institute for Bioengineering and Biosciences. FCT for the i4HB–Associate Laboratory Institute for Health and Bioeconomy (Project LA/P/0140/2020). FCT through the PhD fellowship to JJ (PD/BD/150335/2019, BIOTECnico PhD Program).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2023.1200729/full#supplementary-material
References
Abedin, M. J., Liepold, L., Suci, P., Young, M., and Douglas, T. (2009). Synthesis of a cross-linked branched polymer network in the interior of a protein cage. J. Am. Chem. Soc. 131, 4346–4354. doi:10.1021/ja8079862
Aguilar, P. P., González-Domínguez, I., Schneider, T. A., Gòdia, F., Cervera, L., and Jungbauer, A. (2019). At-line multi-angle light scattering detector for faster process development in enveloped virus-like particle purification. J. Sep. Sci., jssc.201900441–2649. doi:10.1002/jssc.201900441
Allen, M., Willits, D., Mosolf, J., Young, M., and Douglas, T. (2002). Protein cage constrained synthesis of ferrimagnetic iron oxide nanoparticles. Adv. Mater. 14, 1562–1565. doi:10.1002/1521-4095(20021104)14:21<1562:aid-adma1562>3.0.co;2-d
Andar, A. U., Karan, R., Pecher, W. T., DasSarma, P., Hedrich, W. D., Stinchcomb, A. L., et al. (2017). Microneedle-assisted skin permeation by nontoxic bioengineerable gas vesicle nanoparticles. Mol. Pharm. 14, 953–958. doi:10.1021/acs.molpharmaceut.6b00859
Anderson, D. H., Kickhoefer, V. A., Sievers, S. A., Rome, L. H., and Eisenberg, D. (2007). Draft crystal structure of the vault shell at 9-Å resolution. PLoS Biol. 5, e318. doi:10.1371/journal.pbio.0050318
Bellini, M., Mazzucchelli, S., Galbiati, E., Sommaruga, S., Fiandra, L., Truffi, M., et al. (2014). Protein nanocages for self-triggered nuclear delivery of DNA-targeted chemotherapeutics in cancer cells. J. Control Release 196, 184–196. doi:10.1016/j.jconrel.2014.10.002
Belval, L., Hemmer, C., Sauter, C., Reinbold, C., Fauny, J., Berthold, F., et al. (2016). Display of whole proteins on inner and outer surfaces of grapevine fanleaf virus-like particles. Plant Biotechnol. J. 14, 2288–2299. doi:10.1111/pbi.12582
Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., et al. (2000). The protein data bank. Nucleic Acids Res. 28, 235–242. doi:10.1093/nar/28.1.235
Bhaskar, S., and Lim, S. (2017). Engineering protein nanocages as carriers for biomedical applications. NPG Asia Mater 9, e371. doi:10.1038/am.2016.128
Bova, M. P., Huang, Q., Ding, L., and Horwitz, J. (2002). Subunit exchange, conformational stability, and chaperone-like function of the small heat shock protein 16.5 from Methanococcus jannaschii. J. Biol. Chem. 277, 38468–38475. doi:10.1074/jbc.M205594200
Boyton, I., Goodchild, S. C., Diaz, D., Elbourne, A., Collins-Praino, L. E., and Care, A. (2022). Characterizing the dynamic disassembly/reassembly mechanisms of encapsulin protein nanocages. ACS Omega 7, 823–836. doi:10.1021/acsomega.1c05472
Bruun, T. U. J., Andersson, A.-M. C., Draper, S. J., and Howarth, M. (2018). Engineering a rugged nanoscaffold to enhance plug-and-display vaccination. ACS Nano 12, 8855–8866. doi:10.1021/acsnano.8b02805
Cassidy-Amstutz, C., Oltrogge, L., Going, C. C., Lee, A., Teng, P., Quintanilla, D., et al. (2016). Identification of a minimal peptide tag for in vivo and in vitro loading of encapsulin. Biochemistry 55, 3461–3468. doi:10.1021/acs.biochem.6b00294
Champion, C. I., Kickhoefer, V. A., Liu, G., Moniz, R. J., Freed, A. S., Bergmann, L. L., et al. (2009). A vault nanoparticle vaccine induces protective mucosal immunity. PLoS ONE 4, e5409. doi:10.1371/journal.pone.0005409
Chen, H., Zhang, S., Xu, C., and Zhao, G. (2016). Engineering protein interfaces yields ferritin disassembly and reassembly under benign experimental conditions. Chem. Commun. 52, 7402–7405. doi:10.1039/C6CC03108K
Chen, H., Tan, X., Fu, Y., Dai, H., Wang, H., Zhao, G., et al. (2021). The development of natural and designed protein nanocages for encapsulation and delivery of active compounds. Food Hydrocoll. 121, 107004. doi:10.1016/j.foodhyd.2021.107004
Choi, S.-H., Kwon, I. C., Hwang, K. Y., Kim, I.-S., and Ahn, H. J. (2011). Small heat shock protein as a multifunctional scaffold: Integrated tumor targeting and caspase imaging within a single cage. Biomacromolecules 12, 3099–3106. doi:10.1021/bm200743g
Colant, N., Melinek, B., Teneb, J., Goldrick, S., Rosenberg, W., Frank, S., et al. (2021). A rational approach to improving titer in Escherichia coli-based cell-free protein synthesis reactions. Biotechnol. Prog. 37, e3062. doi:10.1002/btpr.3062
Corsi, F., and Mazzucchelli, S. (2016). The potential of protein-based nanocages for imaging and drug delivery. Ther. Deliv. 7, 149–151. doi:10.4155/tde.15.95
Cristie-David, A. S., and Marsh, E. N. G. (2019). Metal-dependent assembly of a protein nano-cage. Protein Sci. 28, 1620–1629. doi:10.1002/pro.3676
Dalmau, M., Lim, S., Chen, H. C., Ruiz, C., and Wang, S.-W. (2008). Thermostability and molecular encapsulation within an engineered caged protein scaffold. Biotechnol. Bioeng. 101, 654–664. doi:10.1002/bit.21988
Dalmau, M., Lim, S., and Wang, S.-W. (2009). Design of a pH-dependent molecular switch in a caged protein platform. Nano Lett. 9, 160–166. doi:10.1021/nl8027069
DasSarma, S., and DasSarma, P. (2015). Gas vesicle nanoparticles for antigen display. Vaccines 3, 686–702. doi:10.3390/vaccines3030686
de Turris, V., Cardoso Trabuco, M., Peruzzi, G., Boffi, A., Testi, C., Vallone, B., et al. (2017). Humanized archaeal ferritin as a tool for cell targeted delivery. Nanoscale 9, 647–655. doi:10.1039/C6NR07129E
Des Soye, B. J., Davidson, S. R., Weinstock, M. T., Gibson, D. G., and Jewett, M. C. (2018). Establishing a high-yielding cell-free protein synthesis platform derived from Vibrio natriegens. ACS Synth. Biol. 7, 2245–2255. doi:10.1021/acssynbio.8b00252
Diaz, D., Care, A., and Sunna, A. (2018). Bioengineering strategies for protein-based nanoparticles. Genes 9, 370. doi:10.3390/genes9070370
Diekmann, Y., and Pereira-Leal, J. B. (2013). Evolution of intracellular compartmentalization. Biochem. J. 449, 319–331. doi:10.1042/BJ20120957
Domingo, G. J., Orru’, S., and Perham, R. N. (2001). Multiple display of peptides and proteins on a macromolecular scaffold derived from a multienzyme complex. J. Mol. Biol. 305, 259–267. doi:10.1006/jmbi.2000.4311
Doyle, C. M., Rumfeldt, J. A., Broom, H. R., Broom, A., Stathopulos, P. B., Vassall, K. A., et al. (2013). Energetics of oligomeric protein folding and association. Arch. Biochem. Biophys. 531, 44–64. doi:10.1016/j.abb.2012.12.005
Effio, C. L., Oelmeier, S. A., and Hubbuch, J. (2016). High-throughput characterization of virus-like particles by interlaced size-exclusion chromatography. Vaccine 34, 1259–1267. doi:10.1016/j.vaccine.2016.01.035
Ensign, D., Young, M., and Douglas, T. (2004). Photocatalytic synthesis of copper colloids from Cu(II) by the ferrihydrite core of ferritin. Inorg. Chem. 43, 3441–3446. doi:10.1021/ic035415a
Failmezger, J., Scholz, S., Blombach, B., and Siemann-Herzberg, M. (2018). Cell-free protein synthesis from fast-growing Vibrio natriegens. Front. Microbiol. 9, 1146–1210. doi:10.3389/fmicb.2018.01146
Falvo, E., Tremante, E., Fraioli, R., Leonetti, C., Zamparelli, C., Boffi, A., et al. (2013). Antibody–drug conjugates: Targeting melanoma with cisplatin encapsulated in protein-cage nanoparticles based on human ferritin. Nanoscale 5, 12278. doi:10.1039/c3nr04268e
Fan, K., Cao, C., Pan, Y., Lu, D., Yang, D., Feng, J., et al. (2012). Magnetoferritin nanoparticles for targeting and visualizing tumour tissues. Nat. Nanotech 7, 459–464. doi:10.1038/nnano.2012.90
Ferrer-Miralles, N., Rodríguez-Carmona, E., Corchero, J. L., García-Fruitós, E., Vázquez, E., and Villaverde, A. (2015). Engineering protein self-assembling in protein-based nanomedicines for drug delivery and gene therapy. Crit. Rev. Biotechnol. 35, 209–221. doi:10.3109/07388551.2013.833163
Flenniken, M. L., Willits, D. A., Brumfield, S., Young, M. J., and Douglas, T. (2003). The small heat shock protein cage from Methanococcus jannaschii is a versatile nanoscale platform for genetic and chemical modification. Nano Lett. 3, 1573–1576. doi:10.1021/nl034786l
Flenniken, M. L., Willits, D. A., Harmsen, A. L., Liepold, L. O., Harmsen, A. G., Young, M. J., et al. (2006). Melanoma and lymphocyte cell-specific targeting incorporated into a heat shock protein cage architecture. Chem. Biol. 13, 161–170. doi:10.1016/j.chembiol.2005.11.007
Franco, E., Garcia-Recio, V., Jiménez, P., Garrosa, M., Girbés, T., Cordoba-Diaz, M., et al. (2018). Endotoxins from a pharmacopoeial point of view. Toxins 10, 331. doi:10.3390/toxins10080331
Fu, Y., and Li, J. (2016). A novel delivery platform based on bacteriophage MS2 virus-like particles. Virus Res. 211, 9–16. doi:10.1016/j.virusres.2015.08.022
Gao, R., Tan, H., Li, S., Ma, S., Tang, Y., Zhang, K., et al. (2022). A prototype protein nanocage minimized from carboxysomes with gated oxygen permeability. Proc. Natl. Acad. Sci. 119, e2104964119. doi:10.1073/pnas.2104964119
Giessen, T. W. (2016). Encapsulins: Microbial nanocompartments with applications in biomedicine, nanobiotechnology and materials science. Curr. Opin. Chem. Biol. 34, 1–10. doi:10.1016/j.cbpa.2016.05.013
Goldsmith, L. E., Pupols, M., Kickhoefer, V. A., Rome, L. H., and Monbouquette, H. G. (2009). Utilization of a protein “shuttle” to load vault nanocapsules with gold probes and proteins. ACS Nano 3, 3175–3183. doi:10.1021/nn900555d
Gregorio, N. E., Levine, M. Z., and Oza, J. P. (2019). A user’s guide to cell-free protein synthesis. MPs 2, 24. doi:10.3390/mps2010024
Gu, C., Zhang, T., Lv, C., Liu, Y., Wang, Y., and Zhao, G. (2020). His-mediated reversible self-assembly of ferritin nanocages through two different switches for encapsulation of cargo molecules. ACS Nano 14, 17080–17090. doi:10.1021/acsnano.0c06670
Guan, X., Chang, Y., Sun, J., Song, J., and Xie, Y. (2018). Engineered Hsp protein nanocages for siRNA delivery. Macromol. Biosci. 18, 1800013. doi:10.1002/mabi.201800013
Guimaraes, C. P., Witte, M. D., Theile, C. S., Bozkurt, G., Kundrat, L., Blom, A. E. M., et al. (2013). Site-specific C-terminal and internal loop labeling of proteins using sortase-mediated reactions. Nat. Protoc. 8, 1787–1799. doi:10.1038/nprot.2013.101
Haikarainen, T., and Papageorgiou, A. C. (2010). Dps-like proteins: Structural and functional insights into a versatile protein family. Cell. Mol. Life Sci. 67, 341–351. doi:10.1007/s00018-009-0168-2
Han, J.-A., Kang, Y. J., Shin, C., Ra, J.-S., Shin, H.-H., Hong, S. Y., et al. (2014). Ferritin protein cage nanoparticles as versatile antigen delivery nanoplatforms for dendritic cell (DC)-based vaccine development. Nanomedicine Nanotechnol. Biol. Med. 10, 561–569. doi:10.1016/j.nano.2013.11.003
Hassani-Mehraban, A., Creutzburg, S., van Heereveld, L., and Kormelink, R. (2015). Feasibility of Cowpea chlorotic mottle virus-like particles as scaffold for epitope presentations. BMC Biotechnol. 15, 80. doi:10.1186/s12896-015-0180-6
He, D., and Marles-Wright, J. (2015). Ferritin family proteins and their use in bionanotechnology. N. Biotechnol. 32, 651–657. doi:10.1016/j.nbt.2014.12.006
Heddle, J. G., Yokoyama, T., Yamashita, I., Park, S.-Y., and Tame, J. R. H. (2006). Rounding up: Engineering 12-membered rings from the cyclic 11-mer TRAP. Structure 14, 925–933. doi:10.1016/j.str.2006.03.013
Heddle, J. G., Fujiwara, I., Yamadaki, H., Yoshii, S., Nishio, K., Addy, C., et al. (2007). Using the ring-shaped protein TRAP to capture and confine gold nanodots on a surface. Small 3, 1950–1956. doi:10.1002/smll.200700400
Heinze, K., Sasaki, E., King, N. P., Baker, D., Hilvert, D., Wuite, G. J. L., et al. (2016). Protein nanocontainers from nonviral origin: Testing the mechanics of artificial and natural protein cages by AFM. J. Phys. Chem. B 120, 5945–5952. doi:10.1021/acs.jpcb.6b01464
Hempstead, P. D., Yewdall, S. J., Fernie, A. R., Lawson, D. M., Artymiuk, P. J., Rice, D. W., et al. (1997). Comparison of the three-dimensional structures of recombinant human H and horse L ferritins at high resolution 1 1Edited by R. Huber. J. Mol. Biol. 268, 424–448. doi:10.1006/jmbi.1997.0970
Hill, A. M., and Salmond, G. P. C. (2020). Microbial gas vesicles as nanotechnology tools: Exploiting intracellular organelles for translational utility in biotechnology, medicine and the environment. Microbiology 166, 501–509. doi:10.1099/mic.0.000912
Hsia, Y., Bale, J. B., Gonen, S., Shi, D., Sheffler, W., Fong, K. K., et al. (2016). Design of a hyperstable 60-subunit protein icosahedron. Nature 535, 136–139. doi:10.1038/nature18010
Ilari, A., Stefanini, S., Chiancone, E., and Tsernoglou, D. (2000). The dodecameric ferritin from Listeria innocua contains a novel intersubunit iron-binding site. Nat. Struct. Mol. Biol. 7, 38–43. doi:10.1038/71236
Izard, T., Ævarsson, A., Allen, M. D., Westphal, A. H., Perham, R. N., de Kok, A., et al. (1999). Principles of quasi-equivalence and Euclidean geometry govern the assembly of cubic and dodecahedral cores of pyruvate dehydrogenase complexes. Proc. Natl. Acad. Sci. U. S. A. 96, 1240–1245. doi:10.1073/pnas.96.4.1240
Jeon, J. O., Kim, S., Choi, E., Shin, K., Cha, K., So, I.-S., et al. (2013). Designed nanocage displaying ligand-specific peptide bunches for high affinity and biological activity. ACS Nano 7, 7462–7471. doi:10.1021/nn403184u
Ji, P., Wang, X., Yin, J., Mou, Y., Huang, H., and Ren, Z. (2022). Selective delivery of curcumin to breast cancer cells by self-targeting apoferritin nanocages with pH-responsive and low toxicity. Drug Deliv. 29, 986–996. doi:10.1080/10717544.2022.2056662
João, J., Lampreia, J., Prazeres, D. M. F., and Azevedo, A. M. (2021). Manufacturing of bacteriophages for therapeutic applications. Biotechnol. Adv. 49, 107758. doi:10.1016/j.biotechadv.2021.107758
Johnson, E., Cascio, D., Sawaya, M. R., Gingery, M., and Schröder, I. (2005). Crystal structures of a tetrahedral open pore ferritin from the hyperthermophilic archaeon Archaeoglobus fulgidus. Structure 13, 637–648. doi:10.1016/j.str.2005.01.019
Jung, H.-I., Cooper, A., and Perham, R. N. (2002). Identification of key amino acid residues in the assembly of enzymes into the pyruvate dehydrogenase complex of Bacillus stearothermophilus: A kinetic and thermodynamic analysis. Biochemistry 41, 10446–10453. doi:10.1021/bi020147y
Kanekiyo, M., Wei, C.-J., Yassine, H. M., McTamney, P. M., Boyington, J. C., Whittle, J. R. R., et al. (2013). Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature 499, 102–106. doi:10.1038/nature12202
Kang, S., Lucon, J., Varpness, Z. B., Liepold, L., Uchida, M., Willits, D., et al. (2008). Monitoring biomimetic platinum nanocluster formation using mass spectrometry and cluster-dependent H2 production. Angew. Chem. Int. Ed. 47, 7845–7848. doi:10.1002/anie.200802481
Kang, H. J., Kang, Y. J., Lee, Y.-M., Shin, H.-H., Chung, S. J., and Kang, S. (2012). Developing an antibody-binding protein cage as a molecular recognition drug modular nanoplatform. Biomaterials 33, 5423–5430. doi:10.1016/j.biomaterials.2012.03.055
Kar, U. K., Srivastava, M. K., Andersson, Å., Baratelli, F., Huang, M., Kickhoefer, V. A., et al. (2011). Novel CCL21-vault nanocapsule intratumoral delivery inhibits lung cancer growth. PLoS ONE 6, e18758. doi:10.1371/journal.pone.0018758
Kar, U. K., Jiang, J., Champion, C. I., Salehi, S., Srivastava, M., Sharma, S., et al. (2012). Vault nanocapsules as adjuvants favor cell-mediated over antibody-mediated immune responses following immunization of mice. PLoS ONE 7, e38553. doi:10.1371/journal.pone.0038553
Karan, R., Renn, D., Nozue, S., Zhao, L., Habuchi, S., Allers, T., et al. (2023). Bioengineering of air-filled protein nanoparticles by genetic and chemical functionalization. J. Nanobiotechnol 21, 108. doi:10.1186/s12951-023-01866-7
Kasyutich, O., Ilari, A., Fiorillo, A., Tatchev, D., Hoell, A., and Ceci, P. (2010). Silver ion incorporation and nanoparticle formation inside the cavity of pyrococcus furiosus ferritin: Structural and size-distribution analyses. J. Am. Chem. Soc. 132, 3621–3627. doi:10.1021/ja910918b
Kawakami, N., Kondo, H., Matsuzawa, Y., Hayasaka, K., Nasu, E., Sasahara, K., et al. (2018). Design of hollow protein nanoparticles with modifiable interior and exterior surfaces. Angew. Chem. Int. Ed. 57, 12400–12404. doi:10.1002/anie.201805565
Kawano, T., Murata, M., Piao, J., Narahara, S., Hamano, N., Kang, J.-H., et al. (2014). Systemic delivery of protein nanocages bearing CTT peptides for enhanced imaging of MMP-2 expression in metastatic tumor models. Int. J. Mol. Sci. 16, 148–158. doi:10.3390/ijms16010148
Kawano, T., Murata, M., Kang, J.-H., Piao, J. S., Narahara, S., Hyodo, F., et al. (2018). Ultrasensitive MRI detection of spontaneous pancreatic tumors with nanocage-based targeted contrast agent. Biomaterials 152, 37–46. doi:10.1016/j.biomaterials.2017.10.029
Kickhoefer, V. A., Vasu, S. K., and Rome, L. H. (1996). Vaults are the answer, what is the question? Trends Cell Biol. 6, 174–178. doi:10.1016/0962-8924(96)10014-3
Kickhoefer, V. A., Garcia, Y., Mikyas, Y., Johansson, E., Zhou, J. C., Raval-Fernandes, S., et al. (2005). Engineering of vault nanocapsules with enzymatic and fluorescent properties. Proc. Natl. Acad. Sci. U.S.A. 102, 4348–4352. doi:10.1073/pnas.0500929102
Kickhoefer, V. A., Han, M., Raval-Fernandes, S., Poderycki, M. J., Moniz, R. J., Vaccari, D., et al. (2009). Targeting vault nanoparticles to specific cell surface receptors. ACS Nano 3, 27–36. doi:10.1021/nn800638x
Kim, K. K., Kim, R., and Kim, S.-H. (1998a). Crystal structure of a small heat-shock protein. Nature 394, 595–599. doi:10.1038/29106
Kim, K. K., Yokota, H., Santoso, S., Lerner, D., Kim, R., and Kim, S.-H. (1998b). Purification, crystallization, and preliminary X-ray crystallographic data analysis of small heat shock protein homolog from Methanococcus jannaschii, a hyperthermophile. J. Struct. Biol. 121, 76–80. doi:10.1006/jsbi.1998.3969
Kim, R., Lai, L., Lee, H.-H., Cheong, G.-W., Kim, K. K., Wu, Z., et al. (2003). On the mechanism of chaperone activity of the small heat-shock protein of Methanococcus jannaschii. Proc. Natl. Acad. Sci. U.S.A. 100, 8151–8155. doi:10.1073/pnas.1032940100
Kim, D., San, B. H., Moh, S. H., Park, H., Kim, D. Y., Lee, S., et al. (2010). Structural basis for the substrate specificity of PepA from Streptococcus pneumoniae, a dodecameric tetrahedral protease. Biochem. Biophys. Res. Commun. 391, 431–436. doi:10.1016/j.bbrc.2009.11.075
Kim, M., Rho, Y., Jin, K. S., Ahn, B., Jung, S., Kim, H., et al. (2011). pH-dependent structures of ferritin and apoferritin in solution: disassembly and reassembly. Biomacromolecules 12, 1629–1640. doi:10.1021/bm200026v
Kim, J., Heu, W., Jeong, S., and Kim, H.-S. (2017). Genetically functionalized ferritin nanoparticles with a high-affinity protein binder for immunoassay and imaging. Anal. Chim. Acta 988, 81–88. doi:10.1016/j.aca.2017.07.060
Kim, J.-M., Kim, Y.-S., Kim, Y.-R., Choi, M.-J., DasSarma, P., and DasSarma, S. (2022). Bioengineering of Halobacterium sp. NRC-1 gas vesicle nanoparticles with GvpC fusion protein produced in E. coli. Appl. Microbiol. Biotechnol. 106, 2043–2052. doi:10.1007/s00253-022-11841-1
Kim, R., Kim, K. K., Yokota, H., and Kim, S.-H. (1998). Small heat shock protein of Methanococcus jannaschii, a hyperthermophile. Proc. Natl. Acad. Sci. U.S.A. 95, 9129–9133. doi:10.1073/pnas.95.16.9129
Kong, L. B., Siva, A. C., Rome, L. H., and Stewart, P. L. (1999). Structure of the vault, a ubiquitous celular component. Structure 7, 371–379. doi:10.1016/S0969-2126(99)80050-1
Kuruppu, A. I., Zhang, L., Collins, H., Turyanska, L., Thomas, N. R., and Bradshaw, T. D. (2015). An apoferritin-based drug delivery system for the tyrosine kinase inhibitor gefitinib. Adv. Healthc. Mater. 4, 2816–2821. doi:10.1002/adhm.201500389
Kwon, Y.-C., and Jewett, M. C. (2015). High-throughput preparation methods of crude extract for robust cell-free protein synthesis. Sci. Rep. 5, 8663. doi:10.1038/srep08663
Ladenstein, R., Fischer, M., and Bacher, A. (2013). The lumazine synthase/riboflavin synthase complex: Shapes and functions of a highly variable enzyme system. FEBS J. 280, 2537–2563. doi:10.1111/febs.12255
Lai, C.-Y., Wiethoff, C. M., Kickhoefer, V. A., Rome, L. H., and Nemerow, G. R. (2009). Vault nanoparticles containing an adenovirus-derived membrane lytic protein facilitate toxin and gene transfer. ACS Nano 3, 691–699. doi:10.1021/nn8008504
Lai, Y.-T., Cascio, D., and Yeates, T. O. (2012a). Structure of a 16-nm cage designed by using protein oligomers. Science 336, 1129. doi:10.1126/science.1219351
Lai, Y.-T., King, N. P., and Yeates, T. O. (2012b). Principles for designing ordered protein assemblies. Trends Cell Biol. 22, 653–661. doi:10.1016/j.tcb.2012.08.004
Laplagne, D. A., Zylberman, V., Ainciart, N., Steward, M. W., Sciutto, E., Fossati, C. A., et al. (2004). Engineering of a polymeric bacterial protein as a scaffold for the multiple display of peptides. Proteins 57, 820–828. doi:10.1002/prot.20248
Lawson, D. M., Artymiuk, P. J., Yewdall, S. J., Smith, J. M. A., Livingstone, J. C., Treffry, A., et al. (1991). Solving the structure of human H ferritin by genetically engineering intermolecular crystal contacts. Nature 349, 541–544. doi:10.1038/349541a0
Lee, E. J., Lee, S. J., Kang, Y.-S., Ryu, J. H., Kwon, K. C., Jo, E., et al. (2015a). Engineered proteinticles for targeted delivery of siRNA to cancer cells. Adv. Funct. Mater. 25, 1279–1286. doi:10.1002/adfm.201403680
Lee, W., Seo, J., Kwak, S., Park, E. J., Na, D. H., Kim, S., et al. (2015b). A double-chambered protein nanocage loaded with thrombin receptor agonist peptide (TRAP) and γ-carboxyglutamic acid of protein C (PC-Gla) for sepsis treatment. Adv. Mater. 27, 6637–6643. doi:10.1002/adma.201503093
Lee, E. J., Lee, N. K., and Kim, I.-S. (2016). Bioengineered protein-based nanocage for drug delivery. Adv. Drug Deliv. Rev. 106, 157–171. doi:10.1016/j.addr.2016.03.002
Lee, E. J. (2018). Recent advances in protein-based nanoparticles. Korean J. Chem. Eng. 35, 1765–1778. doi:10.1007/s11814-018-0102-0
Li, H., Onbe, K., Liu, Q., Iijima, M., Tatematsu, K., Seno, M., et al. (2017). Synthesis and assembly of Hepatitis B virus envelope protein-derived particles in Escherichia coli. Biochem. Biophys. Res. Commun. 490, 155–160. doi:10.1016/j.bbrc.2017.06.015
Lim, S., Peshenko, I. V., Dizhoor, A. M., and Ames, J. B. (2013). Protein nanocages: The versatile molecular shell. Asia Pac. Biotech. News 17, 39–42. doi:10.1007/s12104-012-9373-2
Llauró, A., Guerra, P., Kant, R., Bothner, B., Verdaguer, N., and de Pablo, P. J. (2016). Decrease in pH destabilizes individual vault nanocages by weakening the inter-protein lateral interaction. Sci. Rep. 6, 34143. doi:10.1038/srep34143
Lv, C., Zhang, X., Liu, Y., Zhang, T., Chen, H., Zang, J., et al. (2021). Redesign of protein nanocages: The way from 0D, 1D, 2D to 3D assembly. Chem. Soc. Rev. 50, 3957–3989. doi:10.1039/D0CS01349H
Majsterkiewicz, K., Biela, A. P., Maity, S., Sharma, M., Piette, B. M. A. G., Kowalczyk, A., et al. (2022). Artificial protein cage with unusual geometry and regularly embedded gold nanoparticles. Nano Lett. 22, 3187–3195. doi:10.1021/acs.nanolett.1c04222
Malay, A. D., Miyazaki, N., Biela, A., Chakraborti, S., Majsterkiewicz, K., Stupka, I., et al. (2019). An ultra-stable gold-coordinated protein cage displaying reversible assembly. Nature 569, 438–442. doi:10.1038/s41586-019-1185-4
Martín, F., Carreño, A., Mendoza, R., Caruana, P., Rodriguez, F., Bravo, M., et al. (2022). All-in-one biofabrication and loading of recombinant vaults in human cells. Biofabrication 14, 025018. doi:10.1088/1758-5090/ac584d
McCarthy, S., and Gonen, S. (2022). Improved interface packing and design opportunities revealed by CryoEM analysis of a designed protein nanocage. Heliyon 8, e12280. doi:10.1016/j.heliyon.2022.e12280
Milne, J. L. S., Wu, X., Borgnia, M. J., Lengyel, J. S., Brooks, B. R., Shi, D., et al. (2006). Molecular structure of a 9-MDa icosahedral pyruvate dehydrogenase subcomplex containing the E2 and E3 enzymes using cryoelectron microscopy. J. Biol. Chem. 281, 4364–4370. doi:10.1074/jbc.M504363200
Moleirinho, M. G., Silva, R. J. S., Alves, P. M., Carrondo, M. J. T., and Peixoto, C. (2020). Current challenges in biotherapeutic particles manufacturing. Expert Opin. Biol. Ther. 20, 451–465. doi:10.1080/14712598.2020.1693541
Molino, N. M., and Wang, S.-W. (2014). Caged protein nanoparticles for drug delivery. Curr. Opin. Biotechnol. 28, 75–82. doi:10.1016/j.copbio.2013.12.007
Molino, N. M., Anderson, A. K. L., Nelson, E. L., and Wang, S.-W. (2013). Biomimetic protein nanoparticles facilitate enhanced dendritic cell activation and cross-presentation. ACS Nano 7, 9743–9752. doi:10.1021/nn403085w
Moon, H., Lee, J., Min, J., and Kang, S. (2014). Developing genetically engineered encapsulin protein cage nanoparticles as a targeted delivery nanoplatform. Biomacromolecules 15, 3794–3801. doi:10.1021/bm501066m
Mrazek, J. (2016). Cell-free methods of producing vault particles and vault particles resulting therefrom. Available at: https://patents.google.com/patent/WO2016049122A1/en (Accessed January 5, 2023).
Murata, M., Narahara, S., Kawano, T., Hamano, N., Piao, J. S., Kang, J.-H., et al. (2015). Design and function of engineered protein nanocages as a drug delivery system for targeting pancreatic cancer cells via neuropilin-1. Mol. Pharm. 12, 1422–1430. doi:10.1021/mp5007129
Naskalska, A., Borzęcka-Solarz, K., Różycki, J., Stupka, I., Bochenek, M., Pyza, E., et al. (2021). Artificial protein cage delivers active protein cargos to the cell interior. Biomacromolecules 22, 4146–4154. doi:10.1021/acs.biomac.1c00630
Obata, J., Kawakami, N., Tsutsumi, A., Nasu, E., Miyamoto, K., Kikkawa, M., et al. (2021). Icosahedral 60-meric porous structure of designed supramolecular protein nanoparticle TIP60. Chem. Commun. 57, 10226–10229. doi:10.1039/D1CC03114G
Ohara, N., Kawakami, N., Arai, R., Adachi, N., Moriya, T., Kawasaki, M., et al. (2023). Reversible assembly of an artificial protein nanocage using alkaline Earth metal ions. J. Am. Chem. Soc. 145, 216–223. doi:10.1021/jacs.2c09537
Owczarek, B., Gerszberg, A., and Hnatuszko-Konka, K. (2019). A brief reminder of systems of production and chromatography-based recovery of recombinant protein biopharmaceuticals. BioMed Res. Int. 2019, 1–13. doi:10.1155/2019/4216060
Palombarini, F., Ghirga, F., Boffi, A., Macone, A., and Bonamore, A. (2019). Application of crossflow ultrafiltration for scaling up the purification of a recombinant ferritin. Protein Expr. Purif. 163, 105451. doi:10.1016/j.pep.2019.105451
Palombarini, F., Di Fabio, E., Boffi, A., Macone, A., and Bonamore, A. (2020). Ferritin nanocages for protein delivery to tumor cells. Molecules 25, 825. doi:10.3390/molecules25040825
Papathanasiou, M. M., and Kontoravdi, C. (2020). Engineering challenges in therapeutic protein product and process design. Curr. Opin. Chem. Eng. 27, 81–88. doi:10.1016/j.coche.2019.11.010
Patel, B. A., Gospodarek, A., Larkin, M., Kenrick, S. A., Haverick, M. A., Tugcu, N., et al. (2018). Multi-angle light scattering as a process analytical technology measuring real-time molecular weight for downstream process control. mAbs 10, 945–950. doi:10.1080/19420862.2018.1505178
Patterson, D. P., Prevelige, P. E., and Douglas, T. (2012). Nanoreactors by programmed enzyme encapsulation inside the capsid of the bacteriophage P22. ACS Nano 6, 5000–5009. doi:10.1021/nn300545z
Peng, T., and Lim, S. (2011). Trimer-based design of pH-responsive protein cage results in soluble disassembled structures. Biomacromolecules 12, 3131–3138. doi:10.1021/bm2005438
Peng, T., Lee, H., and Lim, S. (2015). Design of a reversible inversed pH-responsive caged protein. Biomater. Sci. 3, 627–635. doi:10.1039/C4BM00313F
Pfeifer, F. (2022). Recent advances in the study of gas vesicle proteins and application of gas vesicles in biomedical research. Life 12, 1455. doi:10.3390/life12091455
Phippen, S. W., Stevens, C. A., Vance, T. D. R., King, N. P., Baker, D., and Davies, P. L. (2016). Multivalent display of antifreeze proteins by fusion to self-assembling protein cages enhances ice-binding activities. Biochemistry 55, 6811–6820. doi:10.1021/acs.biochem.6b00864
Pieters, B. J. G. E., van Eldijk, M. B., Nolte, R. J. M., and Mecinović, J. (2016). Natural supramolecular protein assemblies. Chem. Soc. Rev. 45, 24–39. doi:10.1039/c5cs00157a
Poderycki, M. J., Kickhoefer, V. A., Kaddis, C. S., Raval-Fernandes, S., Johansson, E., Zink, J. I., et al. (2006). The vault exterior shell is a dynamic structure that allows incorporation of vault-associated proteins into its interior. Biochemistry 45, 12184–12193. doi:10.1021/bi0610552
Prazeres, D. M. de F. (2009). “Chromatographic separation of plasmid DNA using macroporous beads,” in Macroporous polymers: Production, properties and biotechnological/biomedical applications (New York: CRC Press), 335–361.
Puetz, J., and Wurm, F. M. (2019). Recombinant proteins for industrial versus pharmaceutical purposes: A review of process and pricing. Processes 7, 476. doi:10.3390/pr7080476
Ra, J.-S., Shin, H.-H., Kang, S., and Do, Y. (2014). Lumazine synthase protein cage nanoparticles as antigen delivery nanoplatforms for dendritic cell-based vaccine development. Clin. Exp. Vaccine Res. 3, 227. doi:10.7774/cevr.2014.3.2.227
Rahmanpour, R., and Bugg, T. D. H. (2013). Assembly in vitro of Rhodococcus jostii RHA1 encapsulin and peroxidase DypB to form a nanocompartment. FEBS J. 280, 2097–2104. doi:10.1111/febs.12234
Ren, D., Kratz, F., and Wang, S.-W. (2011). Protein nanocapsules containing doxorubicin as a pH-responsive delivery system. Small 7, 1051–1060. doi:10.1002/smll.201002242
Ren, D., Dalmau, M., Randall, A., Shindel, M. M., Baldi, P., and Wang, S.-W. (2012). Biomimetic design of protein nanomaterials for hydrophobic molecular transport. Adv. Funct. Mater. 22, 3170–3180. doi:10.1002/adfm.201200052
Ritsert, K., Huber, R., Turk, D., Ladenstein, R., Schmidt-Bäse, K., and Bacher, A. (1995). Studies on the lumazine synthase/riboflavin synthase complex of Bacillus subtilis: Crystal structure analysis of reconstituted, icosahedral β-subunit capsids with bound substrate analogue inhibitor at 2.4 Å resolution. J. Mol. Biol. 253, 151–167. doi:10.1006/jmbi.1995.0542
Roldão, A., Mellado, M. C. M., Castilho, L. R., Carrondo, M. J., and Alves, P. M. (2010). Virus-like particles in vaccine development. Expert Rev. Vaccines 9, 1149–1176. doi:10.1586/erv.10.115
Ryu, S. J., An, H. J., Oh, Y. S., Choi, H. R., Ha, M. K., and Park, S. C. (2008). On the role of major vault protein in the resistance of senescent human diploid fibroblasts to apoptosis. Cell Death Differ. 15, 1673–1680. doi:10.1038/cdd.2008.96
San, B. H., Kim, S., Moh, S. H., Lee, H., Jung, D.-Y., and Kim, K. K. (2011). Platinum nanoparticles encapsulated by aminopeptidase: A multifunctional bioinorganic nanohybrid catalyst. Angew. Chem. Int. Ed. 50, 11924–11929. doi:10.1002/anie.201101833
Sana, B., Johnson, E., Sheah, K., Poh, C. L., and Lim, S. (2010). Iron-based ferritin nanocore as a contrast agent. Biointerphases 5, FA48–FA52. doi:10.1116/1.3483216
Sánchez-Sánchez, L., Cadena-Nava, R. D., Palomares, L. A., Ruiz-Garcia, J., Koay, M. S. T., Cornelissen, J. J. M. T., et al. (2014). Chemotherapy pro-drug activation by biocatalytic virus-like nanoparticles containing cytochrome P450. Enzyme Microb. Technol. 60, 24–31. doi:10.1016/j.enzmictec.2014.04.003
Santambrogio, P., Cozzi, A., Levi, S., Rovida, E., Magni, F., Albertini, A., et al. (2000). Functional and immunological analysis of recombinant mouse H- and L-ferritins from Escherichia coli. Protein Expr. Purif. 19, 212–218. doi:10.1006/prep.2000.1212
Schmitt, S., Nuhn, L., Barz, M., Butt, H., and Koynov, K. (2022). Shining light on polymeric drug nanocarriers with fluorescence correlation spectroscopy. Macromol. Rapid Commun. 43, 2100892. doi:10.1002/marc.202100892
Schott, K., Ladenstein, R., König, A., and Bacher, A. (1990). The lumazine synthase-riboflavin synthase complex of Bacillus subtilis. Crystallization of reconstituted icosahedral beta-subunit capsids. J. Biol. Chem. 265, 12686–12689. doi:10.1016/S0021-9258(19)38398-X
Schreiber, A., and Schiller, S. M. (2013). Nanobiotechnology of protein-based compartments: Steps toward nanofactories. Bioinspired, Biomim. Nanobiomaterials 2, 154–172. doi:10.1680/bbn.13.00008
Schwarz, H., Schmittner, M., Duschl, A., and Horejs-Hoeck, J. (2014). Residual endotoxin contaminations in recombinant proteins are sufficient to activate human CD1c+ dendritic cells. PLoS ONE 9, e113840. doi:10.1371/journal.pone.0113840
Seebeck, F. P., Woycechowsky, K. J., Zhuang, W., Rabe, J. P., and Hilvert, D. (2006). A simple tagging system for protein encapsulation. J. Am. Chem. Soc. 128, 4516–4517. doi:10.1021/ja058363s
Sehnal, D., Bittrich, S., Deshpande, M., Svobodová, R., Berka, K., Bazgier, V., et al. (2021). Mol* viewer: Modern web app for 3D visualization and analysis of large biomolecular structures. Nucleic Acids Res. 49, W431–W437. doi:10.1093/nar/gkab314
Shen, L., Zhou, J., Wang, Y., Kang, N., Ke, X., Bi, S., et al. (2015). Efficient encapsulation of Fe3O4 nanoparticles into genetically engineered Hepatitis B core virus-like particles through a specific interaction for potential bioapplications. Small 11, 1190–1196. doi:10.1002/smll.201401952
Shenton, W., Mann, S., Cölfen, H., Bacher, A., and Fischer, M. (2001). Synthesis of nanophase iron oxide in lumazine synthase capsids. Angew. Chem. Int. Ed. 40, 442–445. doi:10.1002/1521-3773(20010119)40:2<442:aid-anie442>3.0.co;2-2
SIB Swiss Institute of Bioinformatics (2023). ExPASy - ProtParam tool. Available at: https://web.expasy.org/protparam/(Accessed January 5, 2023).
Silva, F., Sitia, L., Allevi, R., Bonizzi, A., Sevieri, M., Morasso, C., et al. (2021). Combined method to remove endotoxins from protein nanocages for drug delivery applications: The case of human ferritin. Pharmaceutics 13, 229. doi:10.3390/pharmaceutics13020229
Smith, M. T., Varner, C. T., Bush, D. B., and Bundy, B. C. (2012). The incorporation of the A2 protein to produce novel Qβ virus-like particles using cell-free protein synthesis. Biotechnol. Prog. 28, 549–555. doi:10.1002/btpr.744
Stephen, A. G., Raval-Fernandes, S., Huynh, T., Torres, M., Kickhoefer, V. A., and Rome, L. H. (2001). Assembly of vault-like particles in insect cells expressing only the major vault protein. J. Biol. Chem. 276, 23217–23220. doi:10.1074/jbc.C100226200
Steppert, P., Burgstaller, D., Klausberger, M., Tover, A., Berger, E., and Jungbauer, A. (2017). Quantification and characterization of virus-like particles by size-exclusion chromatography and nanoparticle tracking analysis. J. Chromatogr. A 1487, 89–99. doi:10.1016/j.chroma.2016.12.085
Stühn, L., Auernhammer, J., and Dietz, C. (2019). pH-depended protein shell dis- and reassembly of ferritin nanoparticles revealed by atomic force microscopy. Sci. Rep. 9, 17755. doi:10.1038/s41598-019-53943-3
Stupka, I., Azuma, Y., Biela, A. P., Imamura, M., Scheuring, S., Pyza, E., et al. (2022). Chemically induced protein cage assembly with programmable opening and cargo release. Sci. Adv. 8, eabj9424. doi:10.1126/sciadv.abj9424
Suci, P., Kang, S., Gmür, R., Douglas, T., and Young, M. (2010). Targeted delivery of a photosensitizer to Aggregatibacter actinomycetemcomitans biofilm. Antimicrob. Agents Chemother. 54, 2489–2496. doi:10.1128/AAC.00059-10
Sutter, M., Boehringer, D., Gutmann, S., Günther, S., Prangishvili, D., Loessner, M. J., et al. (2008). Structural basis of enzyme encapsulation into a bacterial nanocompartment. Nat. Struct. Mol. Biol. 15, 939–947. doi:10.1038/nsmb.1473
Swift, J., Butts, C. A., Cheung-Lau, J., Yerubandi, V., and Dmochowski, I. J. (2009). Efficient self-assembly of Archaeoglobus fulgidus ferritin around metallic cores. Langmuir 25, 5219–5225. doi:10.1021/la8040743
Tamura, A., Fukutani, Y., Takami, T., Fujii, M., Nakaguchi, Y., Murakami, Y., et al. (2015). Packaging guest proteins into the encapsulin nanocompartment from rhodococcus erythropolis N771: Packaging guest proteins into the encapsulin nanocompartment. Biotechnol. Bioeng. 112, 13–20. doi:10.1002/bit.25322
Tanaka, H., Kato, K., Yamashita, E., Sumizawa, T., Zhou, Y., Yao, M., et al. (2009). The structure of rat liver vault at 3.5 angstrom resolution. Science 323, 384–388. doi:10.1126/science.1164975
Theil, E. C. (2012). Ferritin protein nanocages-the story. Nanotechnol. Percept. 8, 7–16. doi:10.4024/n03th12a.ntp.08.01
Tiainen, P., Gustavsson, P.-E., Ljunglöf, A., and Larsson, P.-O. (2007). Superporous agarose anion exchangers for plasmid isolation. J. Chromatogr. A 1138, 84–94. doi:10.1016/j.chroma.2006.10.029
Toita, R., Murata, M., Abe, K., Narahara, S., Piao, J. S., Kang, J.-H., et al. (2013). A nanocarrier based on a genetically engineered protein cage to deliver doxorubicin to human hepatocellular carcinoma cells. Chem. Commun. 49, 7442. doi:10.1039/c3cc44508a
Tran, K., Gurramkonda, C., Cooper, M. A., Pilli, M., Taris, J. E., Selock, N., et al. (2018). Cell-free production of a therapeutic protein: Expression, purification, and characterization of recombinant streptokinase using a CHO lysate. Biotechnol. Bioeng. 115, 92–102. doi:10.1002/bit.26439
Tripathi, N. K., and Shrivastava, A. (2019). Recent developments in bioprocessing of recombinant proteins: Expression hosts and process development. Front. Bioeng. Biotechnol. 7, 420. doi:10.3389/fbioe.2019.00420
Tripathi, N. K. (2016). Production and purification of recombinant proteins from Escherichia coli. ChemBioEng Rev. 3, 116–133. doi:10.1002/cben.201600002
Uchida, M., Klem, M. T., Allen, M., Suci, P., Flenniken, M., Gillitzer, E., et al. (2007). Biological containers: Protein cages as multifunctional nanoplatforms. Adv. Mater. 19, 1025–1042. doi:10.1002/adma.200601168
Varpness, Z., Peters, J. W., Young, M., and Douglas, T. (2005). Biomimetic synthesis of a H2 catalyst using a protein cage architecture. Nano Lett. 5, 2306–2309. doi:10.1021/nl0517619
Vázquez, E., and Villaverde, A. (2010). Engineering building blocks for self-assembling protein nanoparticles. Microb. Cell Fact. 9, 101. doi:10.1186/1475-2859-9-101
Votteler, J., Ogohara, C., Yi, S., Hsia, Y., Nattermann, U., Belnap, D. M., et al. (2016). Designed proteins induce the formation of nanocage-containing extracellular vesicles. Nature 540, 292–295. doi:10.1038/nature20607
Wang, X.-Y., Chen, X., Manjili, M. H., Repasky, E., Henderson, R., and Subjeck, J. R. (2003). Targeted immunotherapy using reconstituted chaperone complexes of heat shock protein 110 and melanoma-associated antigen gp100. Cancer Res. 63, 2553–2560.
Wang, M., Abad, D., Kickhoefer, V. A., Rome, L. H., and Mahendra, S. (2015). Vault nanoparticles packaged with enzymes as an efficient pollutant biodegradation technology. ACS Nano 9, 10931–10940. doi:10.1021/acsnano.5b04073
Wang, Z., Huang, P., Jacobson, O., Wang, Z., Liu, Y., Lin, L., et al. (2016). Biomineralization-inspired synthesis of copper sulfide–ferritin nanocages as cancer theranostics. ACS Nano 10, 3453–3460. doi:10.1021/acsnano.5b07521
Wang, Z., Gao, H., Zhang, Y., Liu, G., Niu, G., and Chen, X. (2017). Functional ferritin nanoparticles for biomedical applications. Front. Chem. Sci. Eng. 11, 633–646. doi:10.1007/s11705-017-1620-8
Wang, M., Kickhoefer, V. A., Rome, L. H., Foellmer, O. K., and Mahendra, S. (2018). Synthesis and assembly of human vault particles in yeast. Biotechnol. Bioeng. 115, 2941–2950. doi:10.1002/bit.26825
Wang, Z., Zhao, Y., Zhang, S., Chen, X., Sun, G., Zhang, B., et al. (2022). Re-engineering the inner surface of ferritin nanocage enables dual drug payloads for synergistic tumor therapy. Theranostics 12, 1800–1815. doi:10.7150/thno.68459
Wen, A. M., Shukla, S., Saxena, P., Aljabali, A. A. A., Yildiz, I., Dey, S., et al. (2012). Interior engineering of a viral nanoparticle and its tumor homing properties. Biomacromolecules 13, 3990–4001. doi:10.1021/bm301278f
Yu, K., Yau, Y. H., Sinha, A., Tan, T., Kickhoefer, V. A., Rome, L. H., et al. (2017). Modulation of the vault protein-protein interaction for tuning of molecular release. Sci. Rep. 7, 14816. doi:10.1038/s41598-017-12870-x
Zhang, Y., and Orner, B. P. (2011). Self-assembly in the ferritin nano-cage protein superfamily. IJMS 12, 5406–5421. doi:10.3390/ijms12085406
Zhang, X., Meining, W., Fischer, M., Bacher, A., and Ladenstein, R. (2001). X-Ray structure analysis and crystallographic refinement of lumazine synthase from the hyperthermophile aquifex aeolicus at 1.6 Å resolution: Determinants of thermostability revealed from structural comparisons. J. Mol. Biol. 306, 1099–1114. doi:10.1006/jmbi.2000.4435
Zhang, C., Zhang, X., and Zhao, G. (2020). Ferritin nanocage: A versatile nanocarrier utilized in the field of food, nutrition, and medicine. Nanomaterials 10, 1894. doi:10.3390/nano10091894
Zhen, Z., Tang, W., Chen, H., Lin, X., Todd, T., Wang, G., et al. (2013). RGD-modified apoferritin nanoparticles for efficient drug delivery to tumors. ACS Nano 7, 4830–4837. doi:10.1021/nn305791q
Zou, W., Liu, X., Chen, D., Wang, J., Zhao, X., Li, J., et al. (2016a). Expression, purification, and characterization of recombinant human H-chain ferritin. Prep. Biochem. Biotechnol. 46, 833–837. doi:10.1080/10826068.2016.1141300
Zou, W., Liu, X., Zhao, X., Wang, J., Chen, D., Li, J., et al. (2016b). Expression, purification, and characterization of recombinant human L-chain ferritin. Protein Expr. Purif. 119, 63–68. doi:10.1016/j.pep.2015.11.018
Glossary
Keywords: biomanufacturing, bottom-up synthesis, downstream processing, drug delivery, nanostructure engineering, protein nanocages, self-assembly, upstream processing
Citation: João J and Prazeres DMF (2023) Manufacturing of non-viral protein nanocages for biotechnological and biomedical applications. Front. Bioeng. Biotechnol. 11:1200729. doi: 10.3389/fbioe.2023.1200729
Received: 05 April 2023; Accepted: 05 July 2023;
Published: 13 July 2023.
Edited by:
Luan Luong Chu, Phenikaa University, VietnamReviewed by:
Ram Karan, King Abdullah University of Science and Technology, Saudi ArabiaZhi-Gang Jeff Qian, Shanghai Jiao Tong University, China
Copyright © 2023 João and Prazeres. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Duarte Miguel F. Prazeres, miguelprazeres@tecnico.ulisboa.pt
 Jorge João
Jorge João Duarte Miguel F. Prazeres
Duarte Miguel F. Prazeres