Characterisation of secretome-based immune responses of human leukocytes infected with various Mycobacterium tuberculosis lineages
- Published
- Accepted
- Received
- Academic Editor
- Vasco Azevedo
- Subject Areas
- Biodiversity, Cell Biology, Microbiology, Immunology, Infectious Diseases
- Keywords
- East-Asian lineage, Euro-American lineage, Indo-Oceanic lineage, Mycobacterium tuberculosis, Proteomics
- Copyright
- © 2021 Kaewseekhao et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2021. Characterisation of secretome-based immune responses of human leukocytes infected with various Mycobacterium tuberculosis lineages. PeerJ 9:e11565 https://doi.org/10.7717/peerj.11565
Abstract
Background
Differences in immune responses against different lineages of Mycobacterium tuberculosis (Mtb), and by different types of immune cell, are still poorly understood. We aimed to compare the secretome-based immune responses among three Mtb lineages and among immune-cell types. The immune responses were also investigated during infection and when the bacilli had been eliminated from the immune cells.
Methods
Human primary leukocytes were infected with strains representing three lineages of Mtb (East-Asian, Indo-Oceanic and Euro-American). Label-free GeLC MS/MS proteomic analysis of secretomes was performed. The response of each immune-cell type was compared with the appropriate interactome database for each.
Results
The expression pattern of proteins secreted by Mtb-infected leukocytes differed among Mtb lineages. The ancestral lineage (IO lineage) had a greater ability to activate MMP14 (associated with leukocyte migration) than did the more recent lineages (EA and EuA). During infection, proteins secreted by macrophages, dendritic cells, neutrophils and B-cells were associated with cell proliferation. Following clearance of Mtb, proteins associated with interferon signaling were found in macrophages, dendritic cells and neutrophils: proteins associated with antigen processing were found in B-cells and regulatory T-cells. Expression of immune response-related proteins from many immune-cell types might be suppressed by Mtb infection. Our study has provided a better insight into the host-pathogen interaction and immune response against different Mtb lineages.
Introduction
Tuberculosis (TB) is the most important infectious disease globally, causing 10 million new cases and causing 1.5 million deaths annually (WHO, 2019). The only available vaccine is the Bacillus Calmette–Guérin (BCG) vaccine that can protect only against severe forms of TB in young children (Monteiro-Maia & Pinho, 2014). With the advance of research technology, knowledge regarding immune responses against Mycobacterium tuberculosis (Mtb) is progressing (Chai et al., 2020; De Martino et al., 2019). However, several gaps still remain. For example, it remains uncertain whether the immune response varies according to the lineage of Mtb involved, or according to the type of immune cell involved in the host response.
Major members of the Mtb complex have been classified, based on large sequence polymorphisms, into seven lineages that are associated with geographical regions: Indo-Oceanic (IO), East Asian (EA, including Beijing), East African Indian (EAI), Euro-American (EuA), West African1, West African2 and Ethiopian (Coscolla & Gagneux, 2014; Gagneux et al., 2006). EA, EuA and IO lineages of Mtb are commonly found in Southeast Asia (Gagneux et al., 2006). The Mtb lineages differ in virulence and transmissibility (Brites & Gagneux, 2012; Coscolla & Gagneux, 2014; Tientcheu et al., 2017). There have been few studies comparing immune responses against different Mtb strains using high-throughput analysis approaches (Li et al., 2017; Subbian et al., 2013). A transcriptomic study of rabbits infected with HN878 (a hyper-virulent strain of the EA lineage) and with CDC1551 (a hyper-immunogenic strain of the EuA lineage) found that the former induced higher gene expression of genes associated with macrophage activation as well as greater recruitment and activation of polymorphonuclear leukocytes (PMN) (Subbian et al., 2013). A proteomic study of a human monocytic cell line (THP-1) infected with H37Rv or H37Ra strains showed that H37Rv induced higher levels of expression of proteins associated with coagulations, inflammatory response and apoptosis, as well as oxidative phosphorylation (Li et al., 2017). However, no previous study has compared the immune responses mounted against the three main Mtb lineages (IO, EA, EuA).
Various host immune cells cooperate during immune responses against Mtb infection. Given that the pathogen is intracellular, macrophages and T-cells have been suggested as the major immune cells operating against Mtb infection (De Martino et al., 2019). Using classical cell-sorting techniques, the responses of particular immune cells, such as macrophages (Yuan et al., 2019) and T-cells (Yang et al., 2015), to Mtb infection have been demonstrated. However, the selective culture of particular types of immune cells diminishes our ability to observe interactions among these types. Using high-throughput techniques, the function of neutrophils during the Mtb infection process has been clarified (Gideon et al., 2019). However, no previous study has used a proteomic approach and human primary leukocytes (a mixture of several types of immune cells) to demonstrate the interactions among immune-cell types during active infection and following clearance of Mtb from the cells.
In this study, we aimed to characterise and compare the immune responses of human primary leukocytes infected with Mtb lineages including IO, EA and EuA using LC-MS/MS analysis. The immune responses were also compared among immune-cell types using protein databases specific for each cell type. Comparisons were also made between the responses during the infected state and after Mtb has been cleared from the cells.
Materials & Methods
Bacterial culture and inoculum preparation
Three clinical isolates of Mtb were selected from our biobank (Research and Diagnostic Center for Emerging Infectious Diseases (RCEID), Faculty of Medicine, Khon Kaen University). These represented three lineages commonly found in Southeast Asia: EA (strain number 15177), IO (strain number 15561) and EuA (strain number 19477). The genotypic characteristics of these isolates are described in Table S1. IO represents the ancestral lineage and the other strains represent modern lineages of Mtb. Bacterial cell suspensions were prepared as described previously (Kaewseekhao et al., 2020). Briefly, all bacterial strains were cultured in Middlebrook 7H9 with OADC for 14 days. Mtb H37Rv was used as the reference control strain. The concentrations of Mtb cells were measured and adjusted to 0.5 McFarland standards. Any clumping of Mtb cell suspensions was broken up by passing cultures through a 26-gauge needle set.
Infection experiment
Leukocytes, isolated from blood of three participants using HetaSep, were cultured in RPMI medium. The leukocyte culture from each host was used separately in three independent replicates of all experiments. All participants were healthy with an average age of 30.7 ± 3.1 years, Bacillus Calmette–Guérin (BCG) vaccinated and negative for TB according to the interferon gamma release assay (IGRA). The protocol used to infect leukocytes has been described previously (Kaewseekhao et al., 2020). Briefly, the leukocytes were activated by 50 nM phorbol myristate acetate (PMA) for 24 h then exposed to infection with Mtb (multiplicity of infection = 1) for 4 h, after which they were deemed to be infected. The Mtb-infected cells were then treated with a combination of isoniazid (INH: 3 µg/ml) and rifampicin (RIF: 9 µg/ml) starting 4 h after the start of exposure to Mtb. These drug concentrations were optimized for bacillary killing within 3 days (Kaewseekhao et al., 2015a). Medium containing the drugs was changed every 24 h. Uninfected cells treated with the combination of isoniazid and rifampicin were used as negative controls. The infection state at the beginning and the clearance state at the end of the experiments was confirmed using CFU counts on M7H11 plates, which were incubated for at least 4 weeks (Kaewseekhao et al., 2015b). Written informed consents were received from all participants. This study protocol (number HE581377) was approved by Ethics Committee in Human Research Ethics, Office of The Khon Kaen University.
Extracellular protein collection and preparation
The extracellular proteins from each replicate (from each host) were collected by pipetting the extracellular fluids and mixing these with sodium dodecyl sulfate (SDS: final concentration 0.5% w/v). The intact infected leukocytes were discarded. The protocol of protein preparation has been described previously (Kaewseekhao et al., 2020). Briefly, protein concentrations were measured using the Lowry method (Waterborg, 2009). Bovine serum albumin (BSA) was used as a standard (0, 2, 4, 6, 8, 10 µg) for estimation of sample protein concentration. The BSA standard and sample were transferred into 96-well plates (performed in triplicate). Each sample was incubated with 200 µl of solution A (2.5% SDS, 2.5% Na2CO3, 0.2 N NaOH, 0.025% CuSO4 and 0.05% tartaric acid) at room temperature for 30 min. Then, 50 µl of solution B (20% Folin-Ciocalteu phenol reagent) was added and the mixture incubated at room temperature for 30 min. The protein concentrations were measured at OD750 nm and compared with the standard curve for BSA.
SDS PAGE and in-gel digestion
The protein samples (100 µg from each of the three donors) were pooled and mixed by pipetting for 1 min. Then, 50 µg of the pooled protein was separated by SDS-PAGE and the gels were stained with Coomassie blue. In-gel digestion was done as described previously (Kaewseekhao et al., 2020). Briefly, the stained gel in each lane was cut into 11 pieces and then further cut into one mm3 cubes. The region of the gel with high molecular-weight proteins including BSA was excluded. All gel cubes from each sample were transferred into a 96-well plate and tryptic digestion was performed. The gel pieces were incubated with 25 mM NH4HCO3 for 10 min. Then, 200 µl acetonitrile (ACN) was added and the plate incubated for 10 min at room temperature. The supernatants were discarded. Then, 10 mM DTT in 10 mM NH4HCO3 was added and the plate incubated at 56 °C for 1 hr. Next, 100 mM iodoacetamide in 10 mM NH4HCO3 was added and incubated at room temperature for 1 hr in the dark. Then, 200 µl of ACN was added twice and all liquid was removed from each well. Tryptic digestion was performed by incubation of the gel pieces with 10 ng/µl trypsin in 10 mM NH4HCO3 at 37 °C for 3 h. The protein extraction was done by adding 50% ACN and shaking the plate at room temperature for 10 min for 3 cycles. Peptide solutions were dried at 40 °C and kept at −20 °C until analysis.
LC-MS/MS analysis
LC-MS/MS analysis was done as previously described (Kaewseekhao et al., 2020). Briefly, the peptide samples were resuspended in 10 µl of 0.1% formic acid and transferred into low-protein-binding tubes. Large proteins were precipitated by centrifugation at 8,000×g for 10 min and the solution was transferred into vial tubes. Then, 4.5 µl of peptide sample was injected into a LC MS/MS analyser (hybrid quadrupole Q-TOF Impact II™, Bruker Daltonics). Separation of tryptic peptides was done using an Ultimate3000 Nano/Capillary LC System (Thermo Scientific, UK) coupled with a nano-CaptiveSpray™ ion source (Bruker Daltonics). Mobile phase A (0.1% formic acid) was used to transfer the samples at a flow rate of 15 µl/min for 1 min. Mobile phase B (5–50% solution of 0.1% formic acid in 80% acetonitrile) was used for solutions containing separated peptides with a flow rate of 600 nl/min for 15 min. Electrospray ionisation was carried out at 1.6 kV using the CaptiveSpray ion source. The column was rinsed with 80% mobile phase B for 3 min and column temperature was maintained at 35 °C. The MS/MS masses were screened over the range 150 to 2,200 Da and 0.5 s scan time. The LC MS/MS raw data have been deposited in the MassIVE database (https://massive.ucsd.edu) with accession No. MSV000084863.
Bioinformatics and data analyses
LC-MS/MS data analysis was done as previously described (Kaewseekhao et al., 2020). Briefly, LC MS/MS raw data files (.mzXML) were analysed using DeCyderMS 2.0 differential analysis software (DeCyderMS, GE Healthcare Life Science, UK) and Mascot software (Matrix Science, London, UK) for protein identification based on the NCBI database. In brief, the taxonomy search specified human or eukaryote, enzyme (trypsin), variable modifications (carbamidomethyl, oxidation of methionine residues), mass values (monoisotopic), protein mass (unrestricted), peptide mass tolerance (± 1.2 Da), fragment mass tolerance (± 0.6 Da), peptide charge state (1+, 2+, and 3+), and max missed cleavages. Log2 expression levels of the proteins were calculated. Proteins induced by infection (any lineage of Mtb) were identified by disregarding those also expressed in uninfected controls.
Protein network analysis
The detected proteins were classified into five groups according to their pattern of presence/ absence among conditions, i.e., whether they were upregulated or suppressed during initial infection (Day 1), sustained infection (Day 1 and Day 5), or following clearance from infected cells (Day 5 only). The criteria for this classification are described (Table S2). The candidate proteins in each group were used for protein network analysis. The union set of the leukocyte proteins was used for network analyses using NetworkAnalyst program (Xia, Benner & Hancock, 2014) based on entrez ID converted from GI number by db2db database. Network construction was done using IMEx Interactome and analysed by reference to the Reactome database (Orchard et al., 2012). The secretomes of leukocytes infected with each Mtb lineage were used for multi immune-cell type analyses based on the Immuno-Navigator database and the protein pathway analysis based on the Reactome database (Vandenbon et al., 2016). The expression levels of proteins in each group were plotted as heat maps using the ComplexHeatmap package in R.
Results
Overall proteomic response of primary leukocytes infected with Mtb and protein classification
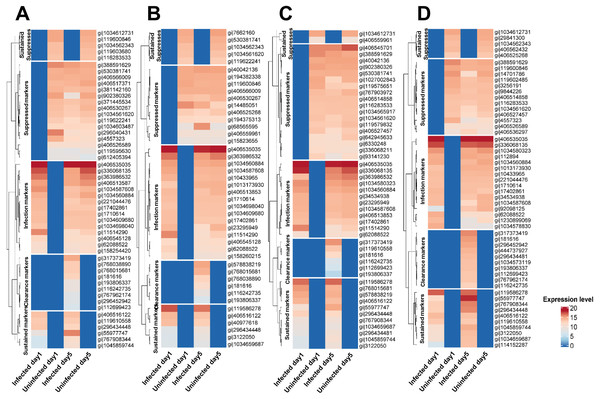
Secreted proteins were detected from leukocytes cells infected with the various Mtb lineages (union set; detected from at least one Mtb lineage) (n = 1,052) and following clearance of infection (n = 1,050). Based on the pattern of their expression among experimental conditions, these proteins were separated into five groups: markers of infection and of sustained infection, markers suppressed in infection and sustained infection, and markers of infection clearance (Fig. 1 and Table S3). These proteins were also classified according to the lineage of Mtb involved. The CFU plate count showed that Mtb cells were present in the leukocytes during the infection stage of the experiment but that no Mtb cells remained at the clearance stage (Day 5) (Table S4).
Figure 1: Classification of proteins secreted by primary leukocytes infected with different M. tuberculosis lineages.
Five categories of protein markers were recognized among Mtb lineages: markers of initial infection and of sustained infection, markers suppressed during initial infection and during sustained infection, and markers differentially expressed after clearance of infection. Classification criteria are shown in Table S2. Proteins secreted by primary leukocytes infected with EA lineage (A). Proteins secreted by primary leukocytes infected with IO lineage (B). Proteins secreted by primary leukocytes infected with EUA lineage (C). Proteins secreted by primary leukocytes infected with H37Rv lineage (D). The elevated expression of some infection markers in control cells at Day 5 (compared to Day 1) could be due to cell aging and/or the background response to the long-term anti-TB drug treatment.Proteomics during infection and clearance states of leukocytes infected with Mtb
Using stringent criteria (Table S2), subsets of proteins expressed by leukocytes during infection or after clearance of infection with Mtb were identified (Table S3). There were 35 proteins (based on GI number) exhibiting enhanced expression during infection with Mtb (Table S5). There were 67 proteins (based on GI number) suppressed at Day 1 or at Days 1 and 5 of Mtb infection (Table S5). Similarly, there were 42 (based on GI number) over-expressed following clearance of infection with Mtb (Table S5).
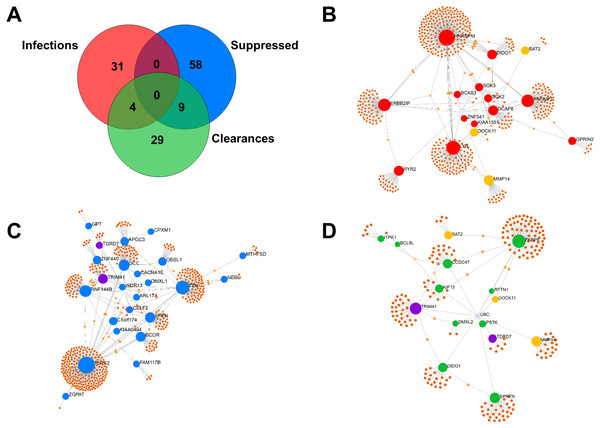
Of the 35 Mtb-infection proteins, 16 had matches in the interactome database and were used for network analysis (Fig. 2B and Table S6). Of the 67 proteins suppressed by Mtb during initial infection, 24 had matches in the interactome database and were used for network analysis (Fig. 2C and Table S6). Of the 42 Mtb-clearance proteins, 15 had matches in the interactome database and were used for network analysis (Fig. 2D and Table S6). SAT2, DOCK11 and MMP14 were found in both infection and clearance states of Mtb (Figs. 2C and 2D) but were not present in control cells. TRD7 and TRIM41 were suppressed during initial infection and also found in the clearance states of Mtb (Figs. 2B and 2D). TRIM41 and RNF144B were the major Mtb-suppressed proteins. Expression of TRIM41 was 12.44-fold lower in H37Rv infection compared to uninfected controls. Expression levels of RNF144B were 14.35-fold lower in leukocytes infected with H37Rv, IO and EuA strains compared to uninfected controls (Table S3).
Figure 2: Comparison of proteins expressed in infection and clearance state of leukocytes infected with M. tuberculosis (union set among EA, IO and EUA linages and H37Rv control).
The number of proteins (not found in the controls) detected during initial infection, clearance state of Mtb and proteins that were suppressed during initial infection (A). Network analysis of proteins enhanced in Mtb-infected cells (B). Network analysis of Mtb-suppressed proteins during initial infection (C). Network analysis of Mtb-clearance proteins (D). Red dots represent proteins expressed during the infection state with levels at least four-fold higher than in uninfected controls, yellow dots represent proteins expressed in both infection and clearance states, blue dots represent proteins suppressed during the infection state with levels at least four-fold lower than in uninfected controls, purple dots represent proteins expressed in both suppression and clearance states, green dots represent proteins expressed in the clearance state with levels at least four-fold higher than in uninfected controls and brown dots represent proteins linked to those proteins expressed in each condition.Proteomic patterns from infected leukocytes differ according to the lineage of Mtb involved
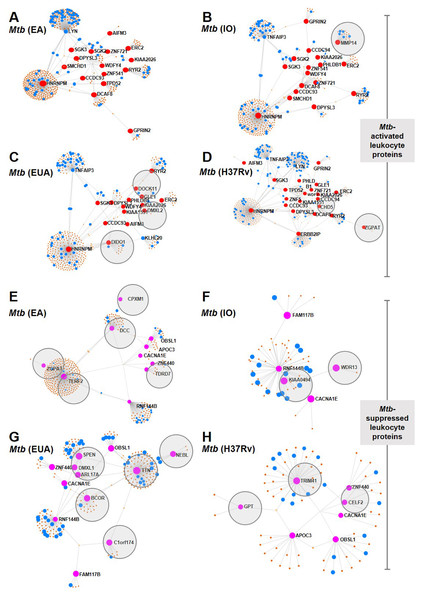
Leukocyte proteins enhanced or suppressed by Mtb infection were compared among Mtb lineages (Fig. 3). Among the upregulated proteins, MMP14 was only found in IO infections (Fig. 3B) and DOCK11, DIDO1 and DMXL2 were only found in leukocytes infected with the EuA lineage (Fig. 3C). No protein was found exclusively in cells infected with the EA lineage (Fig. 3A). ZGPAT and CHD5 were found only in leukocytes with H37Rv strain infection (Fig. 3D). A list of proteins from these networks expressed during Mtb infection is provided (Table S7).
Figure 3: Network analysis of leukocyte proteins activated or suppressed by different lineages of M. tuberculosis.
Network diagrams of Mtb-activated leukocyte proteins (present at Day 1 or (present at Day 1 and Day 5) (A–D) and of Mtb-suppressed leukocyte proteins (suppressed at Day 1 or (suppressed at Day 1 and Day 5) (E–H) compared among Mtb lineages. Protein networks of leukocytes infected with Mtb EA lineage are shown in Figs. 4A and 4E, IO lineage (B, F), EU lineage (C, G) and H37Rv strain (D, H). Red dots represent proteins expressed more than four-fold higher during infection compared to uninfected controls, blue dots represent proteins associated with the immune response, brown dots represent proteins linked to those expressed during infection and gray circles represent proteins uniquely expressed in response to a particular Mtb lineage.Proteins that were suppressed by infection differed among Mtb lineages. CPXM1, DCC, ZGPAT, TERF2 and TDRD7 were suppressed only in leukocytes infected with the EA lineage (Fig. 3E). WDR13 and KIAA0494 proteins were suppressed only in IO-lineage infections (Fig. 3F). SPEN, DMXL1, APL17A, BCOR TTN, NEBL and C1orf174 were suppressed only in EuA-lineage infections (Fig. 3G). GPT, TRIM41 and CELF2 were suppressed only in cells infected with the H37Rv strain (Fig. 3H). A list of suppressed proteins from these networks is provided (Table S8).
Proteomic patterns differ among immune-cell types in response to Mtb infection
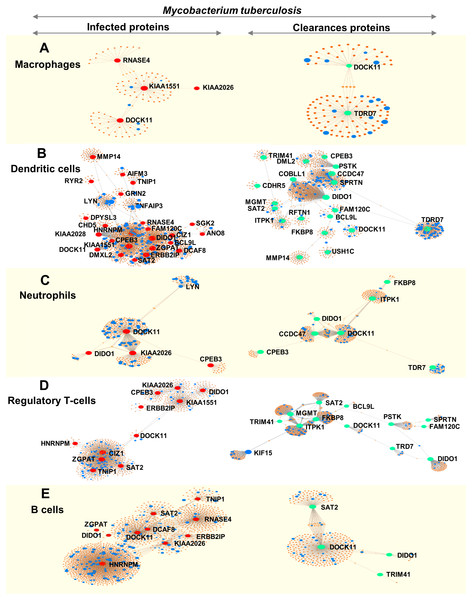
We analysed immune cell-specific molecules based on the Immuno-Navigator database of the secretome. Proteins specific to macrophages, dendritic cells, neutrophils, regulatory T-cells and B-cells were detected (Fig. 4).
Figure 4: Networks of proteins secreted by leukocytes while infected with, and following clearance of, M. tuberculosis (any lineage) classified by immune-cell type (macrophages, dendritic cells, neutrophils, regulatory T-cells and B-cells).
Networks are shown for macrophage-specific proteins (A), dendritic cell-specific proteins (B), neutrophil-specific proteins (C), regulatory T-cell-specific proteins (D) and B-cell-specific proteins (E). Red dots represent proteins expressed by infected leukocytes, green dots represent proteins expressed following clearance of infection, blue dots represent proteins associated with the immune response and orange-brown dots represent additional proteins complementing the pathways of those found in this study.During infection, macrophages secreted proteins involved in cell growth, mitosis and cell survival. Dendritic cells produced molecules involved in interleukin signaling, gene expression and cell-cycle progression. Neutrophils secreted proteins associated with signaling by interleukins and cell mitosis. Regulatory T-cells produced proteins involved in antigen processing and presentation to the immune system whereas proteins secreted by B-cells were associated with cell-cycle progression (Fig. 4 and Table S9).
Following clearance of infection, macrophages produced proteins involved in interferon-gamma signaling, phagocytosis and hemostasis. Proteins secreted by dendritic cells and neutrophils were involved in interferon signaling and cytokine signaling in the immune system. Regulatory T-cells secreted proteins involved in MHC class-II antigen presentation and cell-cycle progression, whereas B-cells produced proteins involved in cell-cycle activation and antigen presentation (peptide folding, assembly and loading of MHC class I) (Fig. 4 and Table S9).
Discussion
In this study, we characterised and compared the secreted proteome of human primary leukocytes (containing mixed populations of immune cells) and infected with strains representing three major Mtb lineages: uninfected cells acted as controls. Mtb H37Rv, a widely studied laboratory stain with relatively low virulence compared to clinical isolates (Sarkar et al., 2012; Van Soolingen et al., 2004), was additionally used for comparisons with the clinical strains. The specific interactions of Mtb with the various host immune cell types are still unclear. We feel that study of the range of immune responses among the immune cell types during infection and following pathogen clearance from the host cells will provide insights into the mechanisms of host-pathogen interaction. We compared the three main lineages (EA, IO and EuA) of Mtb as these are commonly found in Southeast Asia. The EA lineage of Mtb is reportedly more virulent than other lineages (Lopez et al., 2003), and especially more than the ancestral IO lineage (Tram et al., 2018).
The overall immune-response proteins secreted by leukocytes infected with any lineage of Mtb, after disregarding those also secreted in similar quantities by in uninfected controls, were compared. We found that leukocytes infected with Mtb mainly secreted proteins involved in gene expression, translation and metabolism, reflecting the role of these cells in inducing an immune response against the pathogen. After Mtb was eliminated from the leukocytes, proteins from three main pathways—cellular protein degradation, antigen presentation and B-cell activation—were expressed. This is consistent with previous work from our group showing that clearance proteins from a monocytic cell line were associated with the cell cycle, RNA post-transcriptional modification, antimicrobial responses, cell proliferation, migration and movement (Kaewseekhao et al., 2015a). The current study used primary mixed leukocytes and found that most proteins secreted after clearance of Mtb and associated with the immune response belonged to two groups. First, those associated with cellular protein degradation and homeostasis, which might allow the leukocytes to adjust and return to a normal state following the challenge posed by infection. Second, proteins of the immune-response pathways, especially antigen presentation and B-cell activation, were found. This suggested that B-cells and the function of antigen presentation were initially suppressed during an active infection and were then restored after pathogen clearance. Alternatively, experience of infection might lead to trained immunity of innate cells (Lerm & Netea, 2016). This restored or trained function, and especially the ability to effectively process and present antigens to T-cells, is crucial to mount an adaptive immune response, and hence provide a long-term immune response to Mtb (Cooper, 2009). Further studies should be done to investigate functions of B-cells after the clearance of Mtb from the host. To better demonstrate possible trained immunity in cells that have experienced infection, other pathogen-killing methods, such as accelerated killing by immune cells themselves, might be used instead of anti-TB drugs.
Mtb has many immune-suppression strategies (De Martino et al., 2019; Zhai et al., 2019). We found that several immune-response pathways were suppressed by Mtb including MHC class I antigen processing and presentation, cytokine signaling, signaling by NGF and FCGR-dependent phagocytosis. Our study has provided more insight into these suppression strategies as follows. First, we found that tripartite motif 41 (TRIM41) was suppressed during Mtb infection. Mtb might suppress MHC class I antigen presentation via TRIM41. The TRIM protein family is involved in innate immune inflammatory responses and has the potential to be used as a TB diagnostic marker (Chen et al., 2018; Li, 2019). TRIM14 is a key regulator of the type I interferon response against Mtb infection (Hoffpauir et al., 2020). Second, Mtb might suppress cytokine signaling via RNF144B, a protein which plays an important role in activation of NF-κB, cytokines and chemokines (Baek, Huang & Chang, 2016). RNF144B was suppressed in Mtb-infected macrophages, leading to increased intracellular survival of the pathogen (Banks et al., 2019). Therefore, the RNF144B-associated pathway might be a target for suppression during Mtb infection.
Different lineages of Mtb differ in virulence and in host-pathogen immune interactions. The lack of the TbD1 genome region in the more recent lineages (EA, EAI, EuA) is associated with a decrease of immune response level compared to the ancestral lineage (IO) (Portevin et al., 2011). Based on patient serum analysis and macrophage culture experiments, the IO lineage of Mtb induces higher levels of immune activation compared to the EA and EuA lineages (Amin et al., 2019; Portevin et al., 2011). Our study using primary cells in culture has also found that immune responses to the IO lineage differ from those to other lineages. MMP14 was highly expressed only in cells during active infection with the IO lineage. Following clearance, high expression levels of MMP14 were found in cells infected with the EA and EuA lineages as well as the H37Rv strain. MMP14 is a key protein required for leukocyte migration to the site of infection (Sathyamoorthy et al., 2015). The failure by the recent lineages of Mtb to promote production of MMP14 during active infection might benefit the pathogen by host immune suppression. DOCK11, DIDO1 and DMXL2 were only found in leukocytes during infection with the EuA lineage: these are associated with innate immunity and inflammation activation (Namekata et al., 2020). SPEN, DMXL1, APL17A, BCOR TTN, NEBL and C1orf174 were suppressed only in the EuA-lineage. Some of these proteins, such as BCOR, are related to apoptosis repression in Mtb-infected non-human primates (Mehra et al., 2010). The EuA lineage was found to induce high levels of cytokines and is known to have a high growth rate in human macrophages (Reiling et al., 2013). These responses might benefit the EuA lineage by upregulation of inflammation and suppression of apoptosis. Notably, the EA lineage (including the Beijing sub-lineage) is more strongly associated with increased transmissibility, relapse and drug resistance compared to other Mtb lineages (Burman et al., 2009; Feyisa et al., 2019; Glynn et al., 2002; Karmakar et al., 2019). In experiments using human monocyte-derived macrophages, strains of the EA lineage (including the Beijing sub-lineage) induced lower cytokine levels but exhibited higher replication rates than did other strains (Reiling et al., 2013). We found no protein uniquely expressed in the secretome of leukocytes infected with the EA lineage. However, some proteins (CPXM1, DCC, ZGPAT, TERF2 and TDRD7) were suppressed only in cells infected with EA lineages. Some of these proteins (e.g., CPXM1) are involved in protective immune responses against TB via interleukin-17, Th17 activation and cell homeostasis (Wareham et al., 2014). Therefore, the EA lineage might benefit from the suppression of host immune responses that promote pathogenesis. Similarly, no proteins were uniquely up-regulated or suppressed by the H37Rv strain. It is clear that immune responses differed when leukocytes were challenged with different lineages of Mtb. Such differences might explain the variability in vaccine efficacy among lineages of M. tuberculosis (Chae & Shin, 2018; Huang et al., 2020). Further comparisons among lineages, such as using intracellular proteomic analysis and animal-infection models, might clarify this situation.
Several immune-cell types participate in host-Mtb interactions. In this study, we demonstrated the role of each type during Mtb infection and following clearance. We did this by comparing the secretome with the specific database for each cell type. During active infection, the cell type-specific proteins reflected cell proliferation including macrophage cell growth and survival, cell-cycle progression of dendritic cell and B-cell cycle and neutrophil cell mitosis. Only proteins from regulatory T-cells were associated with an immune response against Mtb infection via proteins involved with antigen processing and presentation. Following pathogen clearance, the proteins secreted by each cell type differed from those found during the infection stage. Interferon-signaling proteins were commonly found from macrophages, dendritic cells and neutrophils. Other immune-response proteins were found following clearance, including those associated with macrophage phagocytosis, B-cell antigen presentation and T-cell antigen processing. However, proteins involved in cell proliferation (the key activity found during active infection) were found only in regulatory T-cells following clearance. These results might reflect suppression of the immune response caused by Mtb. A previous study suggested that immune suppression by Mtb occurred mainly in macrophages as the primary host (Zhai et al., 2019). Our study suggests that immune suppression involves many cell types, as reflected by the immune-response proteins, especially those associated with the interferon-signaling pathway and antigen processing, found following clearance of Mtb from the host-cells. Trained immunity (which can involve many cell types) of these immune cells might be an alternative explanation (Lerm & Netea, 2016). It can be argued that the effect of BCG vaccination might mimic or confound the effect of trained immunity (Lalor et al., 2010). All our leukocyte donors were BCG-vaccinated and leukocytes from the same donors were compared in infection and control situations, thus controlling for the effect of BCG vaccination. It is possible that BCG vaccination could alter our secretome analysis result when compared to the unvaccinated condition. On the other hand, our participants, now around 30 years of age, received their BCG vaccine at birth. The protective effect of BCG is minimal in adults (Andersen & Doherty, 2005) and perhaps had no effect on our results.
A strength of using a mixed co-cultured cell population is that interaction among cell types can still occur, as would happen in intact immune-cell networks. We were then able to identify, using the IMEx interactome database, the proteins secreted by each type of immune cell. The specific response of individual cell types during infection and after clearance needs to be confirmed in the future. This could be done, for example, by excluding one type of immune cell at a time from the co-culture system and determining what proteins are missing. Uninfected cells activated with PMA and treated with INH and RIF were used as negative (background) controls. The secretome signals from the analysis were obtained after subtraction of the background control values and hence were not confounded by PMA or anti-TB drugs.
Limitations of this study should be noted. Heterogeneity in responses among individuals might be a confounding factor, hence we used leukocytes from three donors rather than a single donor. For logistical reasons, although we separately infected the leukocytes from each host in individual experiments, we pooled the extracellular proteins from all donors rather than analysing them individually. This could affect the results and interpretation. We did not determine the leukocyte population in each healthy donor: the proportions of WBC types were assumed to be in within the normal range. The leukocytes that we used were from human blood: leukocytes in lung tissues might exhibit different behaviours. Natural infection with Mtb is chronic, persisting for long periods: our in-vitro experiment was performed over only 5 days. Therefore, we used a low dose of PMA to activate the immune cells in an effort to mimic the natural activation of leukocytes (Sullivan et al., 2000). Thus primed, the cells should respond strongly and quickly to infection. However, PMA activation might skew this response. Previously, we have used a macrophage cell line (THP-1 cells) in an infection experiment and confirmed the results with western blotting (Kaewseekhao et al., 2015b). However, a single cell type cannot reflect the actual host immune environment. Here, we used primary leukocytes, a mixture of all immune cells, from human donors. Nonetheless, the host secretome was investigated in a cellular context which could distort the evaluation and interpretation of the actual host-pathogen interactions. We exposed our uninfected control cells to the same Mtb-killing drugs as the infected cells. Inclusion of an additional uninfected control group without drugs would have demonstrated whether use of these drugs influenced our results. Furthermore, the secretome of the clearance state could also reflect long-lasting or late responses to the infection. Due to budget constraints and the large number of proteins found, we did not confirm the results using western blotting as we have previously done (Kaewseekhao et al., 2015b). The high abundance of host protein interfered with detection of the secretome of the pathogen: we therefore did not analyse the pathogen secretome in our study.
Conclusion
We demonstrated that the proteins secreted by Mtb-infected leukocytes differed according to the Mtb lineage involved. The ancestral lineage (IO lineage) had a greater ability to activate MMP14 (related to leukocyte migration) than did the more recent lineages (EA and EuA). During active infection, proteins from each type of immune cell were associated with cell proliferation whereas interferon-signaling and antigen-processing proteins were found after the pathogen was cleared from host cells. Immune response-related proteins from many immune cell types might be suppressed by Mtb. Our study has provided an improved insight into the host-pathogen interaction showing that immune response differ according to Mtb lineage.