Light-at-night exposure affects brain development through pineal allopregnanolone-dependent mechanisms
Figures
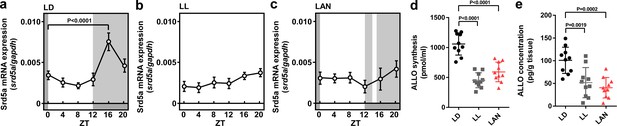
Light-at-night-induced disruption of diurnal variation of pineal ALLO synthesis during early life.
Diurnal changes in srd5a mRNA expression in the pineal gland under LD (a), LL (b), or light-at-night (c) conditions at P7 chicks (n = 10). ALLO synthesis (d) and concentration (e) in the pineal gland of ZT16 at P7 chicks (n = 10). LAN, light-at-night.
-
Figure 1—source data 1
Source data for diurnal changes in srd5a mRNA expression, and ALLO synthesis and concentration in the pineal gland.
- https://doi.org/10.7554/eLife.45306.003
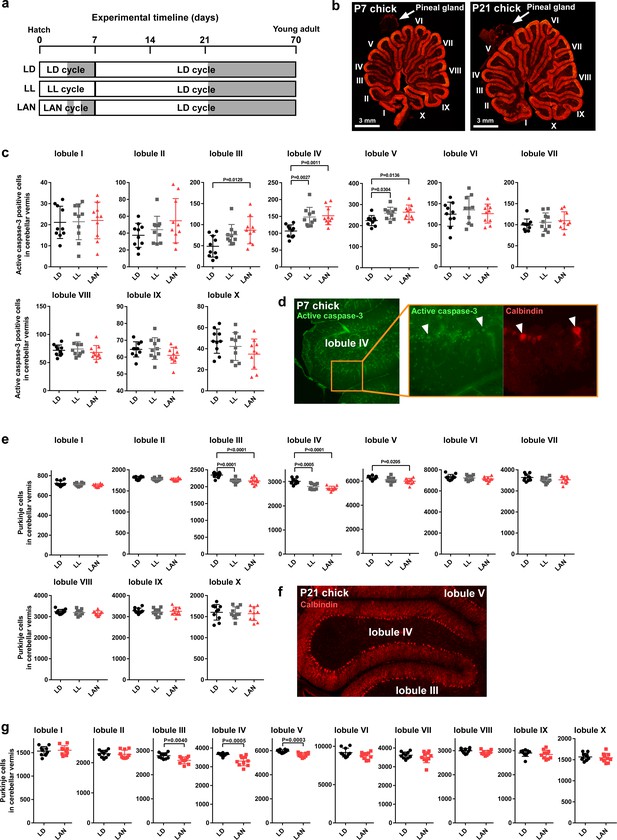
Light-at-night-induced Purkinje cell death during early life.
(a–g) Male chicks were incubated under LD, LL, or light-at-night cycle for 1 week, and then, all groups were housed under LD cycle for 2 or 9 weeks (a). (b) The lobular structure of the whole cerebellum of chick. Scale bar, 3 mm. (c) Number of Purkinje cells expressing active caspase-3 in each lobule at P7 (n = 10). (d) Purkinje cells expressing active caspase-3 in lobule IV at P7. (e) Number of Purkinje cells in each lobule at P21 (n = 10). (f) Purkinje cells in lobule IV at P21. (g) Number of Purkinje cells in each lobule at P70 (young adult; n = 10). LAN, light-at-night.
-
Figure 2—source data 1
Numbers of active caspase-3 positive cells, and Purkinje cells in male.
- https://doi.org/10.7554/eLife.45306.010
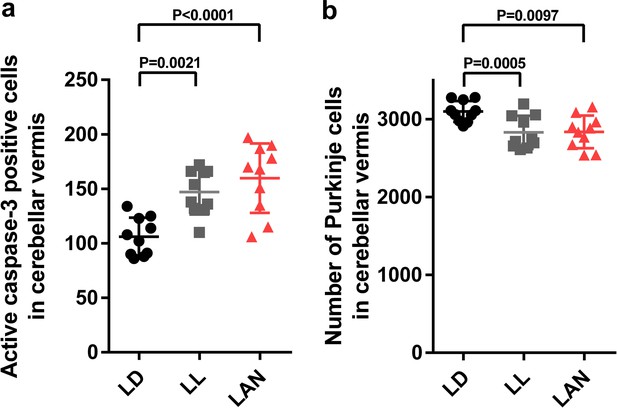
Light-at-night-induced Purkinje cell death during early life in female.
Female chicks were incubated under LD, LL, or light-at-night cycle for 1 week, and then, all groups were housed under LD cycle for 2 weeks. (a) Number of Purkinje cells expressing active caspase-3 in lobule IV at P7 (n = 10). (b) Number of Purkinje cells in lobule IV at P21 (n = 10). LAN, light-at-night.
-
Figure 2—figure supplement 1—source data 1
Numbers of active caspase-3 positive cells, and Purkinje cells in female.
- https://doi.org/10.7554/eLife.45306.006
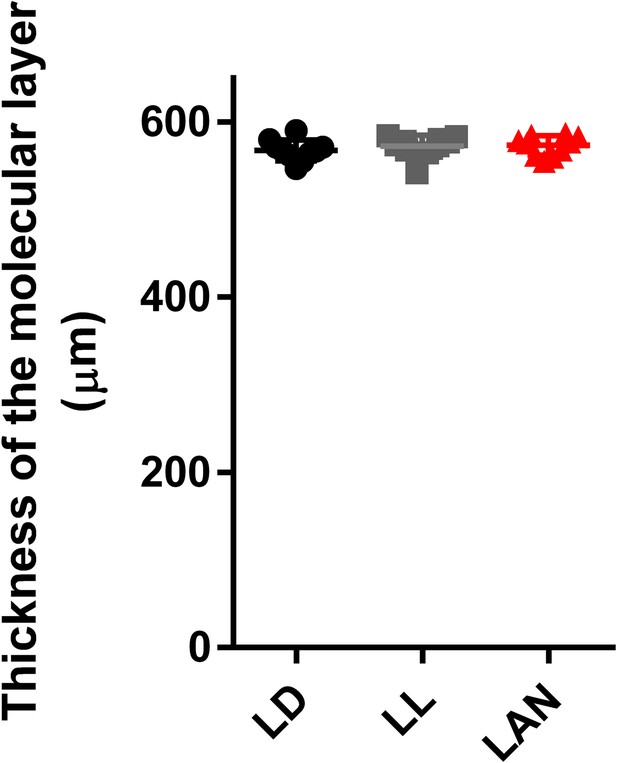
Effect of pineal ALLO on the thickness of the molecular layer.
Male chicks were incubated under LD, LL, or light-at-night cycle for 1 week, and then, all groups were housed under LD cycle for 2 weeks. The thickness of the molecular layer in lobule IV at P21 (n = 10).
-
Figure 2—figure supplement 2—source data 1
Source data for the thickness of the molecular layer of Purkinje cells.
- https://doi.org/10.7554/eLife.45306.008
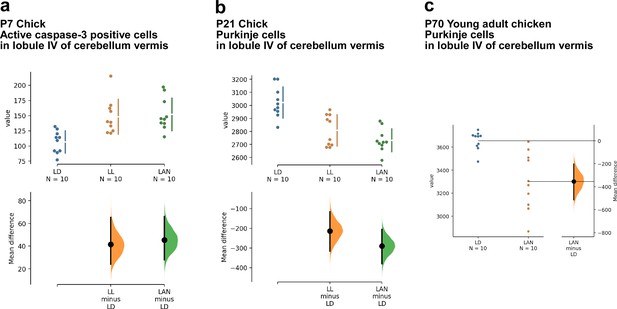
Estimation plots of the effects of light-at-night on cerebellar Purkinje cells.
(a, b) The mean difference for two comparisons against the shared control LD are shown in the above Cumming estimation plot. The raw data are plotted on the upper axes. On the lower axes, mean differences are plotted as bootstrap sampling distributions. Each mean difference is depicted as a dot. Each 95% confidence interval is indicated by the ends of the vertical error bars. (c) The mean difference between LD and LAN is shown in the above Gardner-Altman estimation plot. Both groups are plotted on the left axes; the mean difference is plotted on a floating axes on the right as a bootstrap sampling distribution. The mean difference is depicted as a dot; the 95% confidence interval is indicated by the ends of the vertical error bar. LAN, light-at-night.
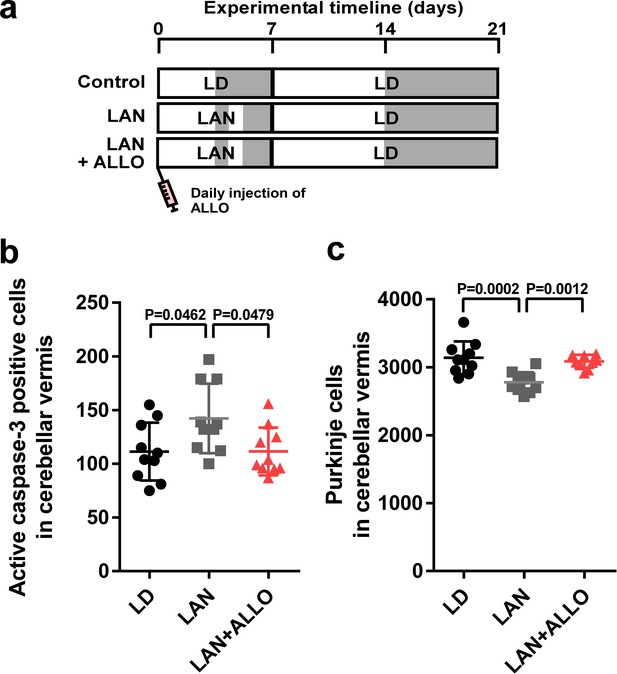
Light-at-night-induced Purkinje cell death was rescued by ALLO injection during early life.
(a) Male chicks were incubated under LD or light-at-night cycle for 1 week, and then, all groups were housed under LD cycle for 2 weeks. Half of the light-at-night chicks were treated with a daily injection of ALLO from P1 to P7. (b) Number of Purkinje cells expressing active caspase-3 in lobule IV at P7 (n = 10). (c) Number of Purkinje cells in lobule IV at P21 (n = 10). LAN, light-at-night.
-
Figure 3—source data 1
Numbers of active caspase-3 positive cells, and Purkinje cells in male.
- https://doi.org/10.7554/eLife.45306.012
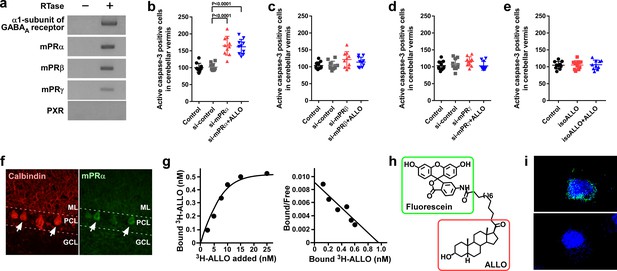
Pineal ALLO prevented Purkinje cell death through mPRα in cerebellar Purkinje cells.
(a) Shown are the expressions of the α1-subunit of the GABAA receptor, mPRα, mPRβ, mPRγ, and PXR in the cerebellum of P1 chicks (similar results were obtained in repeated experiments using three different samples). (b–e) Number of Purkinje cells expressing active caspase-3 in lobule IV at P7 by receptor gene silencing as a candidate of the ALLO receptor in the cerebellar cortex at P1 (n = 10). (f) Immunohistochemistry of mPRα in the cerebellum of P1 chicks (similar results were obtained in repeated experiments using three different samples). (g) Representative saturation curve and Scatchard plot of specific [3H]-ALLO binding to plasma membranes of mPRα transfected cells (n = 4). (h) Structure of fluorescein-labeled ALLO. (i) Confocal images of mPRα-expressing cells stained with fluorescein-labeled ALLO under permeabilized conditions in the absence (upper panel) or presence (lower panel) of excess competing ALLO (similar results were obtained in repeated experiments using three different samples). ML, molecular layer; PCL, Purkinje cell layer; GCL, granule cell layer.
-
Figure 4—source data 1
Numbers of active caspase-3 positive cells in male.
- https://doi.org/10.7554/eLife.45306.016
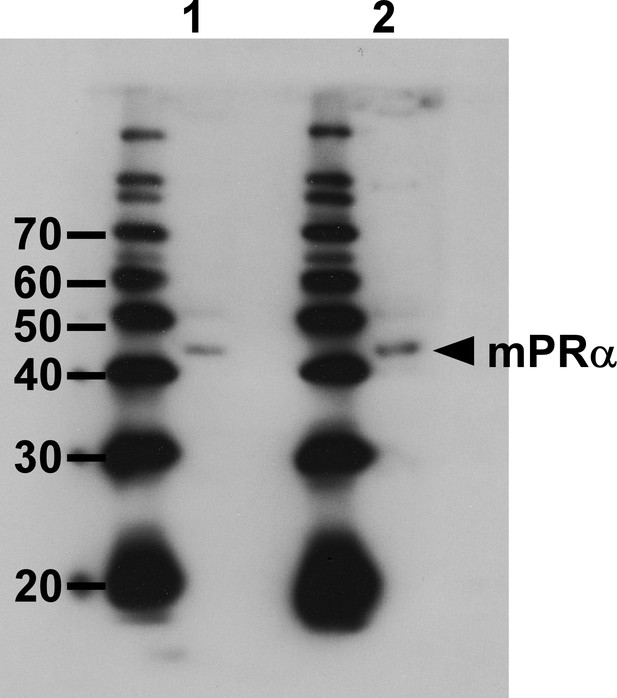
Western blot analysis with the anti-goldfish mPRα antibody.
lane 1, brain protein. Lane 2, Extract of COS-7 cells that were transfected with the chicken mPRα cDNA.
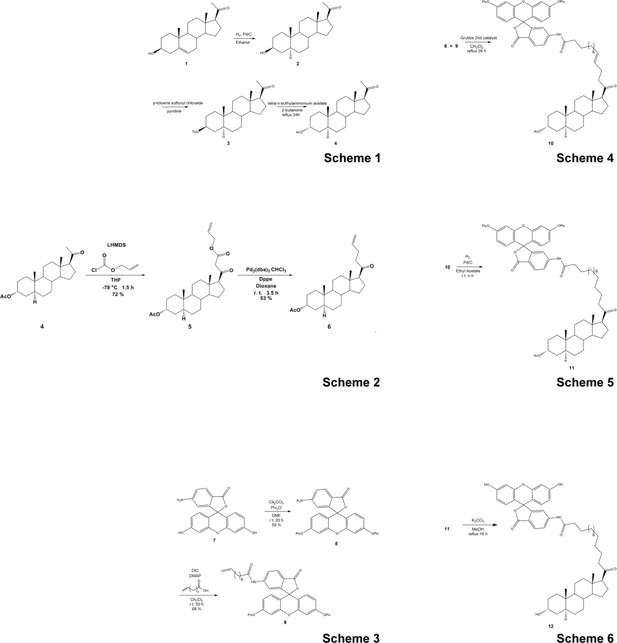
Synthetic scheme for the synthesis of the Fluorescein-labeled ALLO.
https://doi.org/10.7554/eLife.45306.015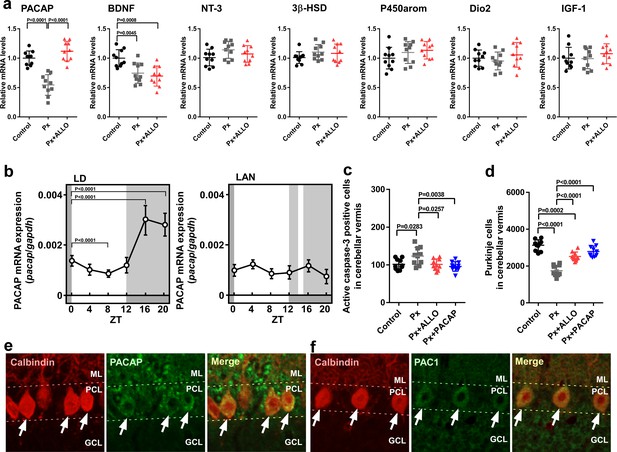
Pineal ALLO-induced expression of PACAP, an endogenous neuroprotective factor, in cerebellar Purkinje cells.
(a) Shown are the effects of Px or Px plus ALLO on the expression of neuroprotective and neurotoxic factors (n = 10). (b) Diurnal changes in PACAP mRNA expression in the cerebellum under LD or light-at-night conditions in P7 chicks (n = 10). (c) Number of Purkinje cells expressing active caspase-3 in lobule IV at P7 (n = 10). (d) Number of Purkinje cells in lobule IV at P21 (n = 10). Immunohistochemistry of PACAP (e) and PAC1 (f) in the cerebellum of P1 chicks (similar results were obtained in repeated experiments using three different samples). Calbindin is used as a marker for Purkinje cells. ML, molecular layer; PCL, Purkinje cell layer; GCL, granule cell layer; LAN, light-at-night.
-
Figure 5—source data 1
source data for mRNA expressions (a), diurnal changes in PACAP mRNA expression (b), and numbers of Purkinje cells (c and d) in male.
- https://doi.org/10.7554/eLife.45306.018
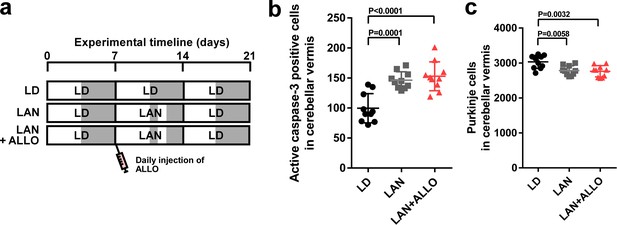
ALLO did not rescue Purkinje cells from death during the second week posthatch.
Male chicks were housed under LD cycle for 1 week during the first week posthatch. They were then incubated under light-at-night cycle and injected with ALLO or a solvent daily for 1 week during the second week posthatch. Finally, the chicks were housed under LD cycle for a week during the third week posthatch (a). (b) Number of Purkinje cells expressing active caspase-3 in lobule IV at P14 (n = 10). (c) Number of Purkinje cells in lobule IV at P21 (n = 10). LAN, light-at-night.
-
Figure 6—source data 1
Numbers of active caspase-3 positive cells, and Purkinje cells in male.
- https://doi.org/10.7554/eLife.45306.020
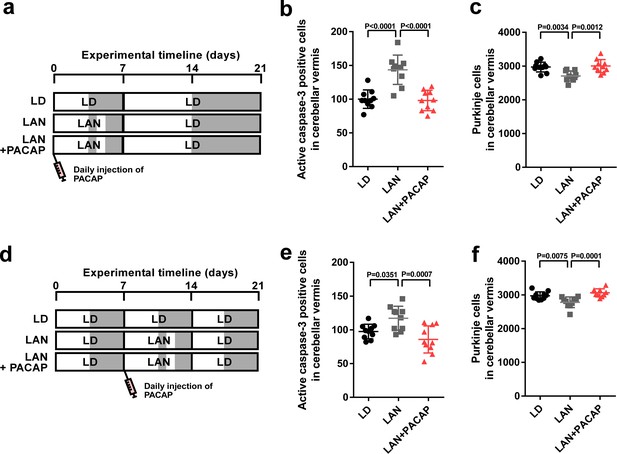
Neuroprotective effects of PACAP were not restricted during early life.
(a) Male chicks were incubated under LD or light-at-night cycle for 1 week during the first week posthatch and injected either with PACAP or solvent daily, and then, all groups were housed under LD cycle for 2 weeks during the second and third weeks posthatch. (b) Number of Purkinje cells expressing active caspase-3 in lobule IV at P7 (n = 10). (c) Number of Purkinje cells in lobule IV at P21 (n = 10). Male chicks were housed under LD cycle for 1 week during the first week posthatch and then incubated under light-at-night cycle and injected PACAP or solvent daily for 1 week during the second week posthatch. Finally, chicks were housed under LD cycle for 1 week during the third week posthatch (d). (e) Number of Purkinje cells expressing active caspase-3 in lobule IV at P14 (n = 10). (f) Number of Purkinje cells in lobule IV at P21 (n = 10). LAN, light-at-night.
-
Figure 7—source data 1
Numbers of active caspase-3 positive cells, and Purkinje cells in male.
- https://doi.org/10.7554/eLife.45306.022
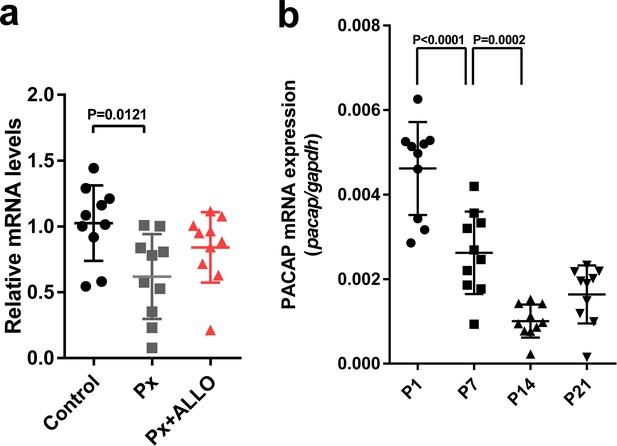
Pineal ALLO did not induce the expression of PACAP in the cerebellum during the second week posthatch.
(a) The effects of Px or Px plus ALLO on the expression of PACAP in the cerebellum during the second week posthatch (n = 10). (b) Changes in the expression of PACAP during early life.
-
Figure 8—source data 1
Source data for mRNA expressions.
- https://doi.org/10.7554/eLife.45306.024
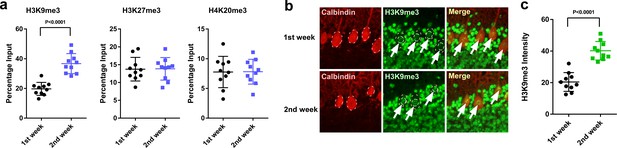
H3K9me3 levels at the Adcyap1 promoter increased in Purkinje cells during development.
(a) Changes in the levels of histone modification between the first and second weeks of posthatch. (b) Immunohistochemistry of H3K9me3 in the cerebellum of P1 chicks. Calbindin is used as a marker for Purkinje cells. (c) The relative intensity of H3K9me3 in the nucleus of Purkinje cells (n = 10).
-
Figure 9—source data 1
Source data for the levels of histone modification.
- https://doi.org/10.7554/eLife.45306.026
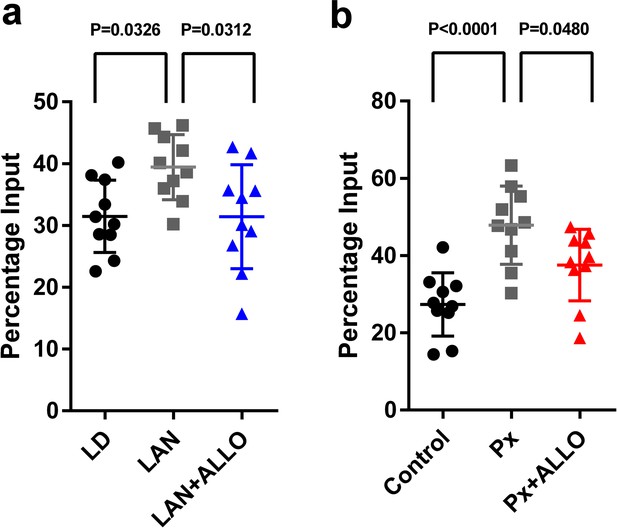
Light-at-night and Px increased in H3K9me3 levels at the Adcyap1 promoter.
(a) The effects of light-at-night and light-at-night plus ALLO on the level of H3K9me3 at the Adcyap1 gene promoter (n = 10). (b) The effects of Px and Px plus ALLO on the level of H3K9me3 at the Adcyap1 gene promoter (n = 10). LAN, light-at-night.
-
Figure 10—source data 1
source data for H3K9me3 levels at the Adcyap1 promoter.
Link to datasets: https://doi.org/10.5061/dryad.k6g8b53.
- https://doi.org/10.7554/eLife.45306.028
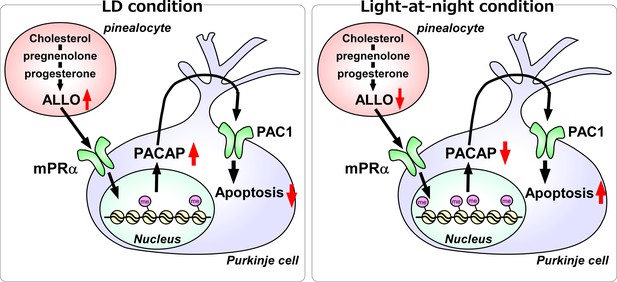
A schematic model of the effect of pineal ALLO on Purkinje cell survival during the first week posthatch under LD or light-at-night conditions.
(Left panel) A schematic model of normal development of the cerebellum under LD during the first week posthatch (that is early posthatch life-stage). Pineal ALLO-induced the expression of PACAP, a neuroprotective factor, through mPRα mechanism in Purkinje cells. Then, PACAP inhibited the expression of active caspase-3 that may facilitate the apoptosis of Purkinje cells in the cerebellum. (Right panel) A schematic model of the abnormal development of the cerebellum under light-at-night conditions during the first week posthatch (that is early posthatch life-stage). Light-at-night conditions disrupted the diurnal rhythm in pineal ALLO synthesis. Decreased pineal ALLO synthesis induced H3K9me3 histone tail modifications of the Adcyap1 gene promoter and then decreased the expression of PACAP in Purkinje cells. Following this, increased amounts of caspase-3 facilitated the apoptosis of Purkinje cells in the cerebellum.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Cercopithecus aethiops) | COS-7 | JCRB | Cat# JCRB9127 RRID:CVCL_0224 | |
| Recombinant DNA reagent | membrane progesterone receptor α (mPRα)-Histag | This study | Materials and methods subsection ‘Conformation of the specificity of mPRα antisera’ | |
| Antibody | Anti-Cleaved Caspase-3 (Asp175) , (Rabbit polyclonal) | Cell Signaling Technology | Cat# 9661, RRID:AB_2341188 | 1:300, IHC |
| Antibody | Anti-Calbindin D-28k, (Mouse monoclonal) | Swant Swiss antibodies | Cat# 300, RRID:AB_10000347 | 1:1000, IHC |
| Antibody | Anti-Pituitary adenylate cyclase-activating polypeptide (PACAP), (Chicken polyclonal) | Nakamachi et al., 2018 | doi: 10.1016/j.peptides.2018.03.006 | 1:100, IHC |
| Antibody | Anti-membrane progesterone receptor α (mPRα),(Rabbit polyclonal) | This study | Materials and methods subsection ‘Conformation of the specificity of mPRα antisera’; Against Goldfish mPRα aa 19–34 and53–66; 1:100, IHC | |
| Antibody | Anti-ADCYAP1R1 (PAC1), (Mouse monoclonal) | Santa Cruz Biotechnology | Cat# sc-100315, RRID:AB_1126992 | 1:100, IHC |
| Antibody | Anti-trimethyl-Histone H3 (Lys9), (Rabbit polyclonal) | Merck Millipore | Cat# 07–442, RRID:AB_310620 | 1:50, ChIP |
| Antibody | Anti-trimethyl-Histone H3 (Lys27), (Rabbit polyclonal) | Merck Millipore | Cat# 07–449, RRID:AB_310624 | 1:50, ChIP |
| Antibody | Anti-trimethyl-Histone H4 (Lys20), (Rabbit polyclonal) | Merck Millipore | Cat# 07–463, RRID:AB_310636 | 1:50, ChIP |
| Antibody | Anti-Mouse IgG (H+L), F(ab’)2 Fragment, Alexa Fluor 555 Conjugate | Cell Signaling Technology | Cat# 4409, RRID:AB_1904022 | 1:1000, IHC |
| Antibody | Anti-rabbit IgG (H+L), F(ab')2 Fragment, Alexa Fluor 488 Conjugate | Cell Signaling Technology | Cat# 4412, RRID:AB_1904025 | 1:1000, IHC |
| Peptide, recombinant protein | Chicken PACAP | This study | ||
| Commercial assay kit | SimpleChIP plus sonication chromatin IP kit | Cell Signaling Technology | Cat# 56383 | |
| Chemical compound | Allopregnanolone | Cayman Chemical | 16930, CAS RN: 516-54-1 | |
| Chemical compound, drug | Fluorescein-labeled allopregnanolone | This study | CAS RN: 2294937-67-8 | Materials and methods subsection ‘Fluorescein-labeled ALLO synthesis’ |
| Software, algorithm | GraphPad Prism6 | GraphPad Software | RRID:SCR_002798 |
Oligonucleotide sequence of PCR.
https://doi.org/10.7554/eLife.45306.030| Target genes | Forward primer 5’- > 3’ | Reverse primer 5’- > 3’ |
|---|---|---|
| Srd5a | AGAAAACCCGGGGAAGTCAC | AGCGATGGCAAAACCAAACC |
| α1-subunit of GABAAR | TCGTGGCAGTCTCCTTTGTC | CTCATGCCCACAAGTGTCCT |
| mPRα | TCTGCCCTGTGTGTCTTCAC | TTTGTCCCTCACCTTCCGTG |
| mPRβ | AGGGCCTTGTGGGAAAGATG | TGCCAGATTCAAAGCCCCAT |
| mPRγ | CGTGCGCTCGATGAGAAATG | TTTCATAACCCACCCCCAGC |
| PXR | CCCATAACCAAAGCCAAGCG | ATCATGTCCTTCCGCATCCC |
| PACAP | CACGCCGATGGGATCTTCA | GTGCAGGTATTTCCTTGCGG |
| BDNF | ACATCACTGGCGGACACTTT | CAGCATGACTCGGGATGTGT |
| NT-3 | ACCACCACCACTGTACCTCA | TCGGTGGCTCTTGTGTTCTG |
| 3β-HSD | CACTCTGCTGAACACCCCTT | GCTGGTGTACCTCTTTGCCT |
| P450arom | CGGGGCTGTGTAGGAAAGTT | TGTCTGTACTCTGCACCGTC |
| Dio2 | TGACCACCATTCACAAGCCA | CAACAGAAAGTCAGCCACGC |
| IGF-1 | ATGGATCCAGCAGTAGACGC | GCCTCCTCAGGTCACAACTC |
| Adcyap1 gene promoter | CAGTTTCATGGTAAGGACCCG | ACGACCCACCGAGCG |
Oligonucleotide sequence of siRNA.
https://doi.org/10.7554/eLife.45306.031| Target genes | Sequence-sense | Sequence-anti-sense |
|---|---|---|
| mPRα | CGGAGCUGGGCUGGUUUCUUCCCA | UGGGAAGAAACCAGCCCAGCUCCG |
| mPRβ | GAGGAGGAUGCUGCUUGGUACCAU | AUGGUNCCAAGCAGCAUCCUCCUC |
| mPRγ | CCGACAGAGUUUGGCUGCUGCGAU | AUCGCAGCAGCCAAACUCUGUCGG |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.45306.032





