Screening of candidate substrates and coupling ions of transporters by thermostability shift assays
Figures
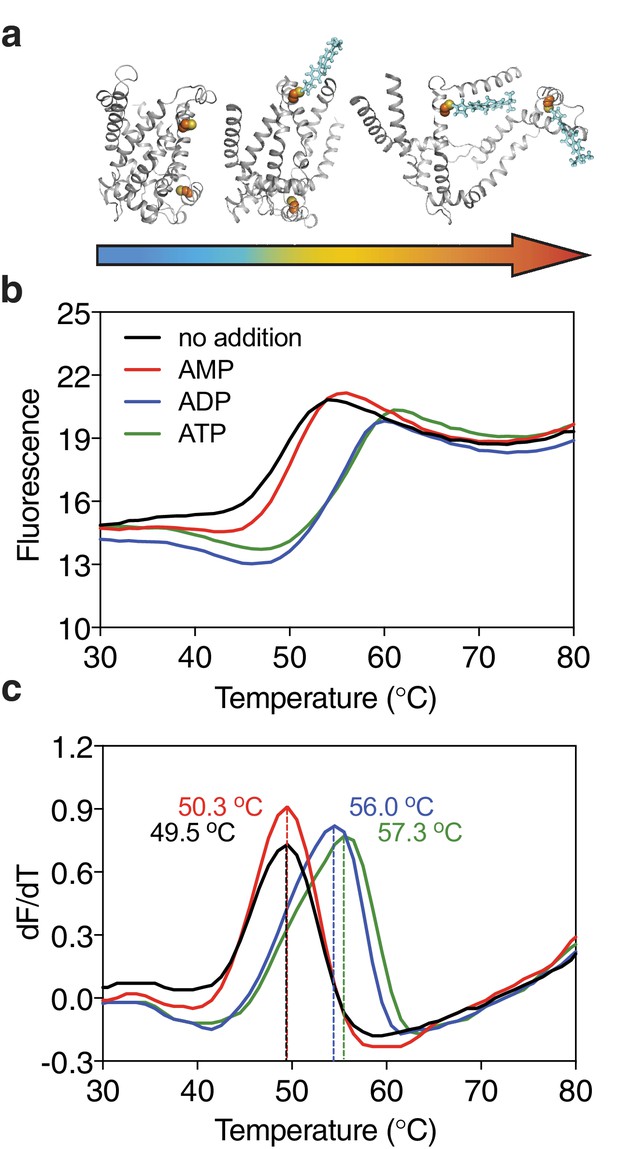
Substrate-induced stabilization of a mitochondrial ADP/ATP carrier.
(a) As protein molecules in a population unfold due to a gradual rise in temperature (25–90°C), buried cysteine residues become solvent-exposed and accessible to the thiol-specific probe CPM (blue stick representation) that becomes fluorescent upon reaction. (b) Typical unfolding curves of the mitochondrial ADP/ATP carrier of Thermothelomyces thermophila (2 μg) in the absence and presence of 2.5 mM AMP, ADP and ATP, shown in red, blue and green, respectively. (c) The apparent melting temperature (Tm) is the peak in the derivative of the unfolding curve (dF/dT), which is used as an indicator of thermal stability. The apparent melting temperatures reported in the text are from three independent protein purifications.
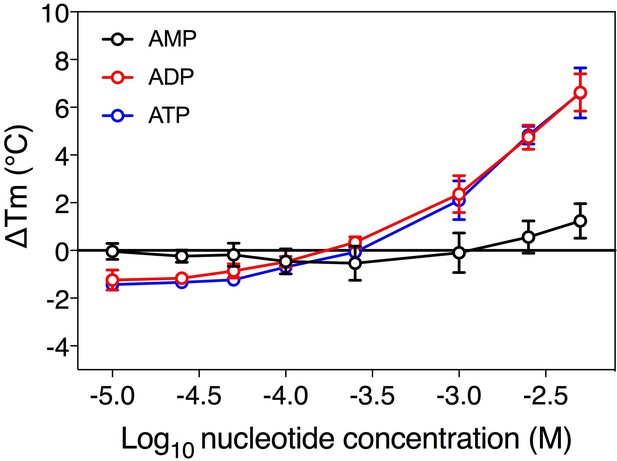
Substrate-induced stabilization of a mitochondrial ADP/ATP carrier.
The substrate concentration-dependency of the apparent melting temperature of purified mitochondrial ADP/ATP carrier. The apparent melting temperature (Tm) was determined from the peak in the derivative of the unfolding curve for different concentrations of AMP, ADP, and ATP in 20 mM HEPES pH 8.0, 100 mM NaCl, 0.1% dodecyl-maltoside, 0.1 mg ml−1 tetraoleoyl cardiolipin. The data are represented by the average and standard deviation of three biological repeats for ADP and ATP, and five biological repeats for AMP.
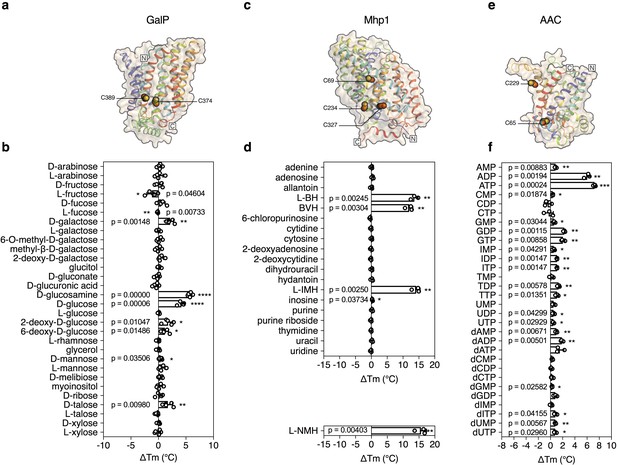
Validation of the method for determining substrate specificity using three unrelated proteins.
(a), (c) and (e) Structural models of GalP (based on GLUT5, PDB 4YBQ), Mhp1 (PDB 2JLN) and AAC (based on Aac2p, PDB 4C9G), respectively. The models are shown in rainbow cartoon and wheat surface representations. Cysteine residues are shown as spheres, except for Cys-19 of GalP, which could not be modeled. (b), (d) and (f) Thermostability screen of GalP (3 μg), Mhp1 (3 μg) and AAC (2 μg) against sugar (50 mM final concentration), nucleobase (2 mM) and nucleotide libraries (2.5 mM), respectively. The temperature shift (ΔTm) is the apparent melting temperature in the presence of compound minus the apparent melting temperature in the absence of compound. The data are represented by the standard deviation of five, three and three independent repeats, respectively. Two-tailed Student’s t-tests assuming unequal variances were performed for the significance analysis (0.05 < p-value: not significant; 0.01 < p-value<0.05: *; 0.001 < p-value<0.01: **; 0.0001 < p-value<0.001: ***; p-value<0.0001: ****). L-BH, 5-benzyl-L-hydantoin; BVH, 5-bromovinylhydantoin; L-IMH, 5-indolylmethyl-L-hydantoin, L-NMH 5- (2-naphthylmethyl)-L-hydantoin.
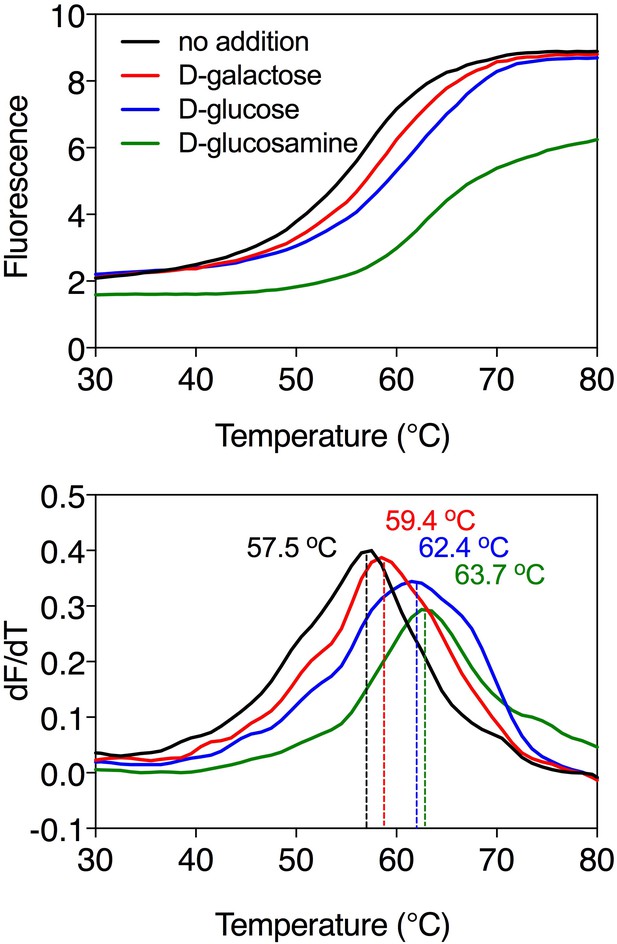
Substrate-induced stabilization of the galactose transporter GalP.
Typical thermostability profiles of purified GalP (top) and their derivatives (dF/dT; bottom) in the absence and presence of stabilizing substrates D-galactose (red trace), D-glucose (blue trace) and D-glucosamine (green trace), which was also causing a significant shift. The apparent melting temperature (Tm) is the peak in the derivative of the unfolding curve (dF/dT) and is indicated for each curve. For each reaction, 2.5 μg purified protein was assayed in the presence of 50 mM compound. The apparent melting temperature reported in the text is from five independent protein purifications.
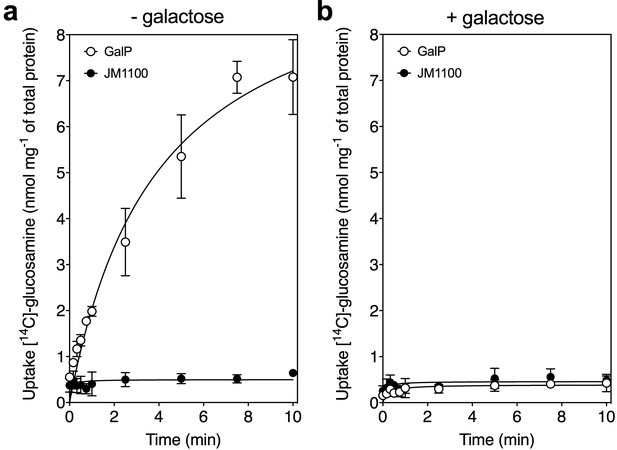
Glucosamine is a transported substrate of the galactose transporter GalP.
Transport assays with [14C]-labeled D-glucosamine in whole cells of the GalP-expressing plasmid pGP1 in the host E. coli strain JM1100 (open circles) and control JM1100 without plasmid (closed circles). Transport in (a) the absence or (b) presence of 4000-fold excess of unlabeled D-galactose. The data are represented by the average and standard deviation of four technical repeats.
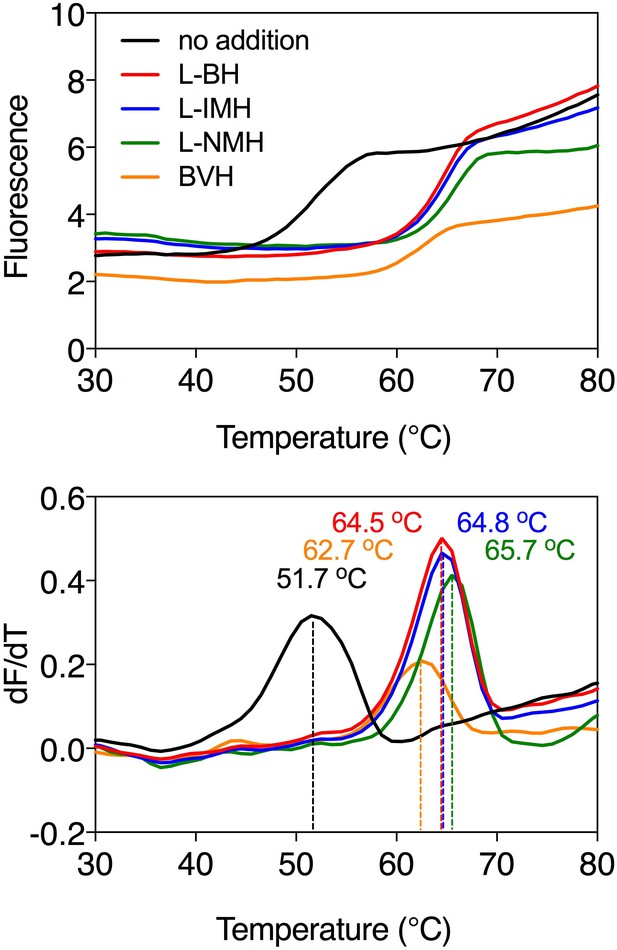
Ligand-induced stabilization of Mhp1 in the presence of sodium ions.
Typical thermostability profiles of purified Mhp1 (top) and their derivatives (dF/dT; bottom) in the absence or presence of stabilizing compounds 5-benzyl-L-hydantoin (L-BH, red trace), 5-indolylmethyl-L-hydantoin (L-IMH, blue trace), 5-(2-naphthylmethyl)-L-hydantoin (L-NMH, green trace) and 5-bromovinylhydantoin (BVH, orange trace). The apparent melting temperature (Tm) is the peak in the derivative of the unfolding curve (dF/dT) and is indicated for each curve. For each reaction, 2.5 μg purified protein was assayed in the presence of 140 mM NaCl and 2 mM compound in buffer containing 10 mM Tris-HCl pH 8.0, 2.5% glycerol and 0.05% dodecyl-maltoside. The apparent melting temperature reported in the text is from three independent protein purifications.
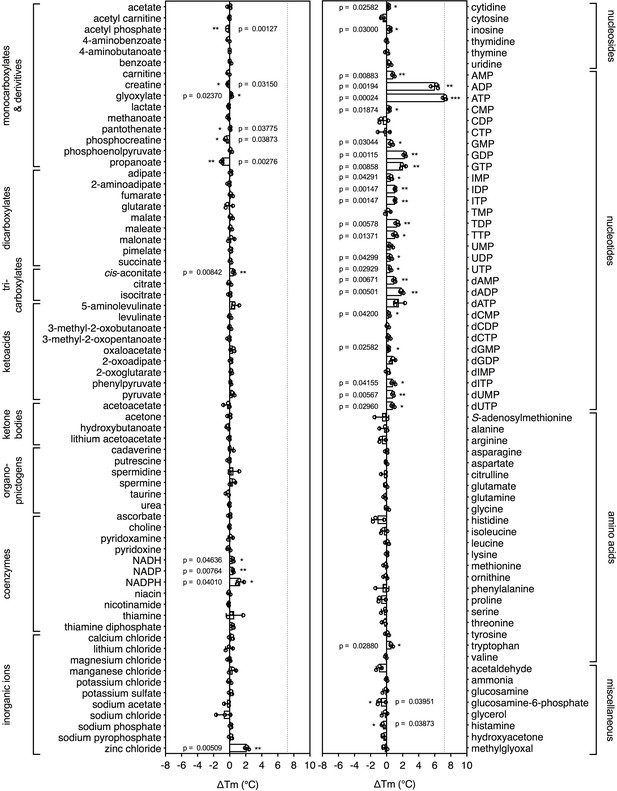
Substrate screening of the mitochondrial ADP/ATP carrier.
Purified ADP/ATP carrier in dodecyl-maltoside was separately mixed with 2.5 mM of library compounds in 20 mM HEPES pH 8.0, 100 mM NaCl, 0.1% dodecyl-maltoside, 0.1 mg ml−1 tetraoleoyl cardiolipin and subjected to a melting regime. The shifts in the apparent Tm were recorded and are shown as bars. The data are represented by the average and standard deviation of three independent assays. The significance tests were carried out as in Figure 2.
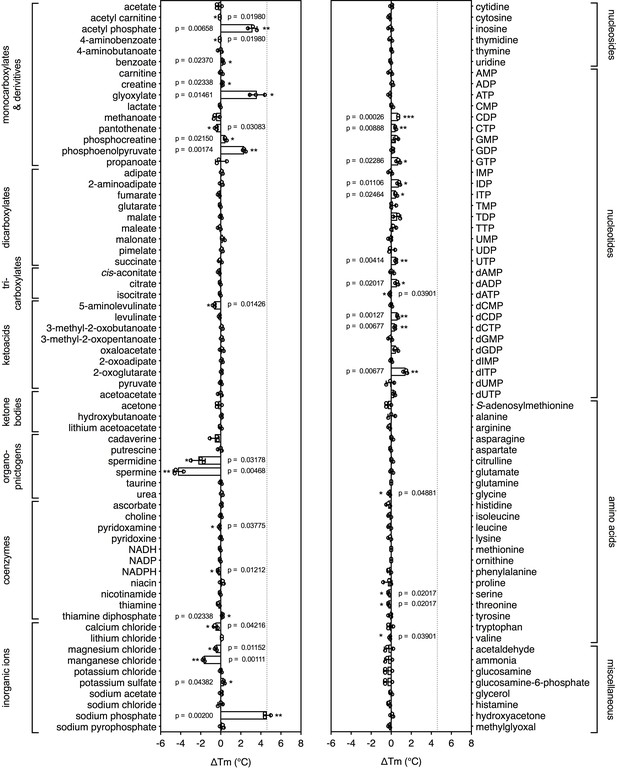
Identification of substrates for a mitochondrial phosphate carrier from the thermophilic ciliate Tetrahymena thermophila.
Purified carrier (1 μg) in lauryl maltose neopentyl glycol was incubated at pH 6.0 with 2.5 mM of each library compound separately and the ΔTm was determined, which is the difference between the apparent melting temperatures in the presence and absence of the tested compound. The data are represented by the average and standard deviation of three independent assays. The significance tests were performed as described in the legend to Figure 2.
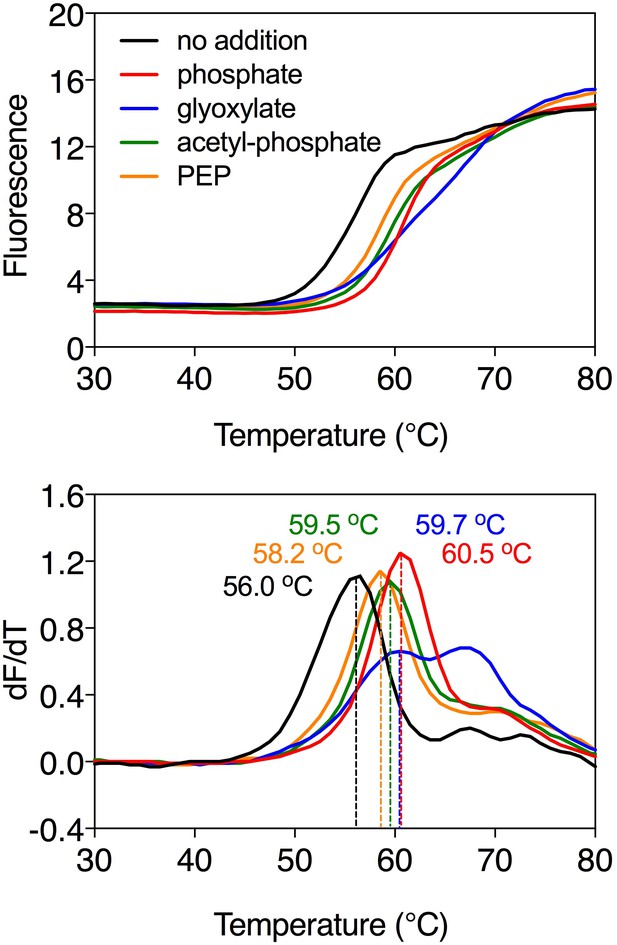
Substrate-induced stabilization of the mitochondrial phosphate carrier.
Typical thermostability curves of purified phosphate carrier (top) and their derivatives (dF/dT; bottom) in the absence and presence of stabilizing compounds phosphate (red trace), glyoxylate (blue trace), acetyl-phosphate (green trace) and phosphoenolpyruvate (PEP, orange trace). The apparent melting temperature (Tm) is the peak in the derivative of the unfolding curve (dF/dT) and is indicated for each curve. For each reaction, 2 μg purified protein was assayed in the presence of 2.5 mM compound in 20 mM MES pH 6.0, 100 mM NaCl, 0.1% lauryl maltose neopentyl glycol, 0.1 mg ml−1 tetraoleoyl cardiolipin. The apparent melting temperatures reported in the text are from three independent protein purifications.
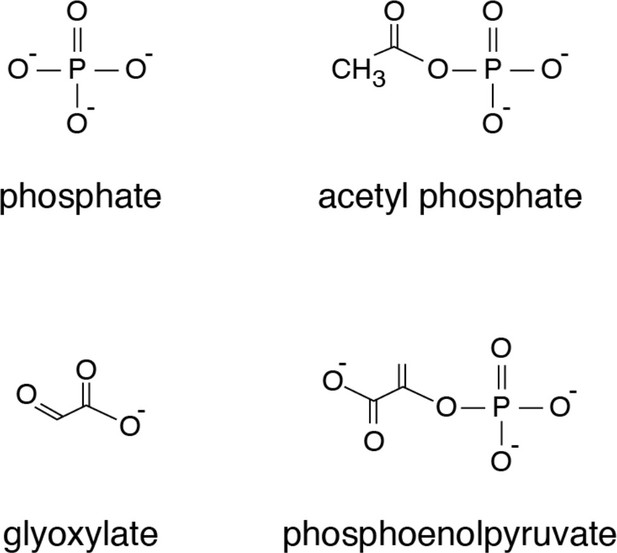
Chemical structures of compounds that showed significant thermostability shifts with the mitochondrial phosphate carrier.
https://doi.org/10.7554/eLife.38821.011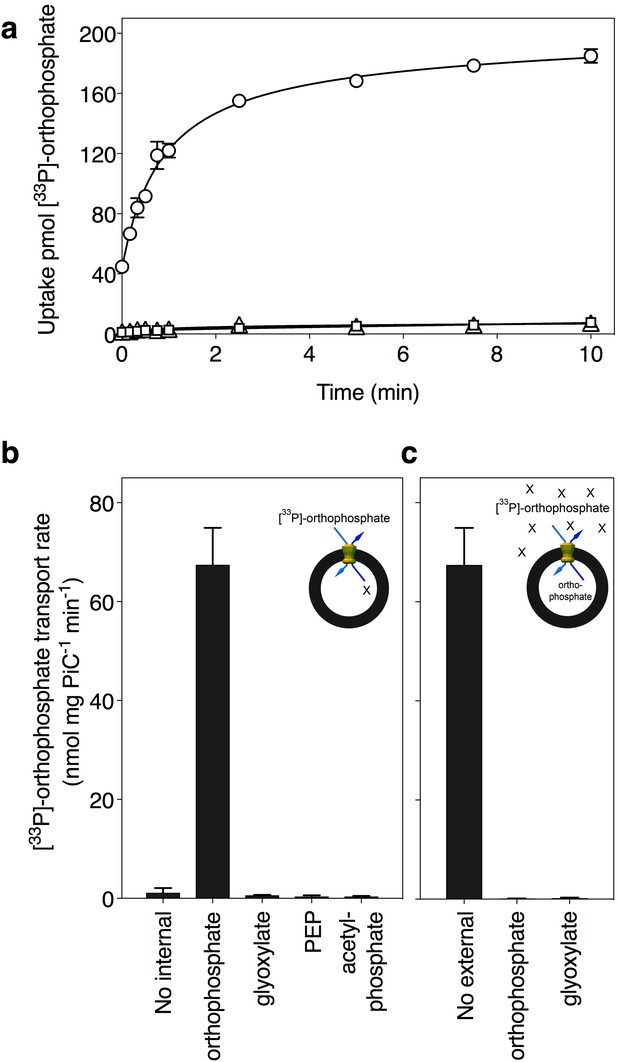
Phosphate is transported by the putative phosphate carrier of Tetrahymena thermophila.
(A) Transport assays were initiated by the addition of 20 μM. [33P]-labeled orthophosphate in proteoliposomes with reconstituted carrier in the absence of internal orthophosphate (triangles) or the presence of 20 mM internal orthophosphate (circles), and a control with liposomes without reconstituted protein (squares). (B) Proteoliposomes were loaded with 20 mM compound (as indicated), and transport was initiated by the addition of 20 μM [33P]-labeled orthophosphate. (C) Competition experiments in which uptake was assayed in the presence of 20 mM external unlabeled compound (1,000-fold excess). All data are represented by the average and the standard deviation of four technical repeats, except for [33P]-orthophosphate/orthophosphate homoexchange, which is represented by the average and the standard deviation of two biological repeats, each with four technical repeats (n = 8). PEP; phosphoenolpyruvate.
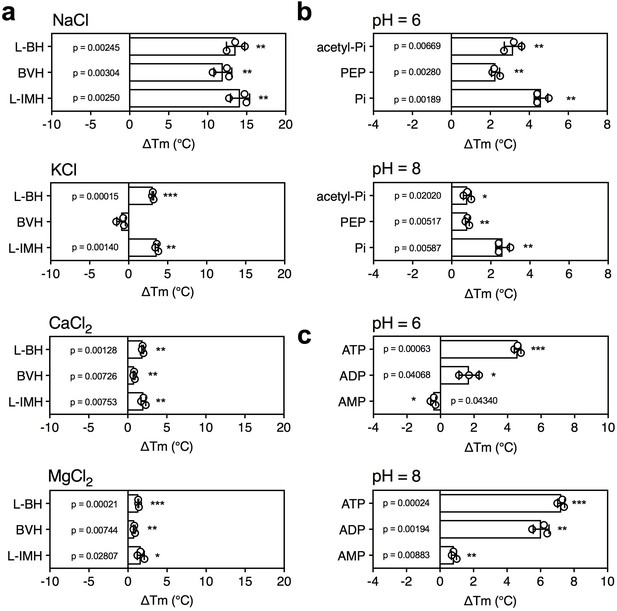
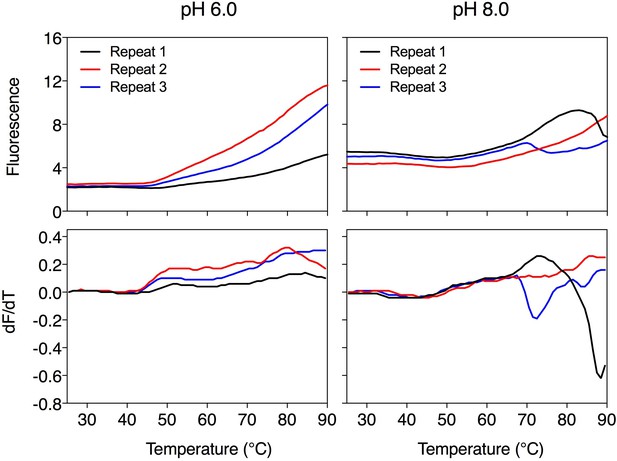
The effect of coupling ions on the stabilization of transporters by substrate binding.
(a) Thermostability shifts of Mhp1 (3 μg) induced by binding of hydantoins in the presence of 140 mM NaCl, KCl, CaCl2 and MgCl2. L-BH: 5-benzyl-L-hydantoin, BVH: 5-bromovinylhydantoin, L-IMH: 5-indolmethyl-L-hydantoin. (b) Thermostability shifts of the phosphate carrier (1 μg) induced by binding of 2.5 mM phosphate-containing compounds at pH 6.0 and 8.0. Acetyl-Pi: acetyl-phosphate, Pi: phosphate. (c) Thermostability shifts of the (2 μg) induced by 2.5 mM ADP and ATP binding at pH 6.0 and 8.0, using AMP as control.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Chemical compound, drug | Dodecyl-β-D-maltoside | Glycon Biochemicals GmbH | Glycon:D97002 | |
| Chemical compound, drug | Dodecyl maltose neopentyl glycol | Anatrace | Anatrace:NG310 | |
| Chemical compound, drug | N-[4-(7-diethylamino-4-methyl-3- coumarinyl)phenyl] maleimide (CPM) | Sigma | Sigma:C1484 | |
| Chemical compound, drug | Complete Mini EDTA-free protease inhibitor tablets | Roche | Roche:5056489001 | |
| Chemical compound, drug | Nickel Sepharose (High Performance) | GE Healthcare | GE Healthcare:17526802 | |
| Chemical compound, drug | Nickel-NTA Superflow | Qiagen | Qiagen:30430 | |
| Chemical compound, drug | Factor Xa protease | NEB | NEB:P8010L | |
| Chemical compound, drug | Tetraoleoyl cardiolipin (TOCL) | Avanti Polar Lipids | Avanti Polar Lipids:710335C | |
| Chemical compound, drug | E. coli polar lipid extract | Avanti Polar Lipids | Avanti Polar Lipids:100600C | |
| Chemical compound, drug | [14C]-galactose | American Radiolabeled Chemicals | American Radiolabeled Chemicals:ARC0117 | |
| Chemical compound, drug | [14C]-glucosamine | American Radiolabeled Chemicals | American Radiolabeled Chemicals:ARC0118A | |
| Chemical compound, drug | [33P]-orthophosphate | Hartmann Analytic | Hartmann Analytic:FF-01 | |
| Chemical compound, drug | BioBeads | BioRad | BioRad:152–3920 | |
| Chemical compound, drug | C10E5 | Sigma | Sigma:76436 | |
| Chemical compound, drug | Hydantoin compounds | Other | A gift from Marta Sans, Maria Kokkinidou and Arwen Pearson, University of Hamburg | |
| Software, algorithm | Prism | GraphPad | ||
| Strain, strain background (S. cerevisiae) | Saccharomyces cerevisiae W303-1B | ATCC | ATTC:201238 | |
| Strain, strain background (S. cerevisiae) | Saccharomyces cerevisiae WB12 | PMID:9878703 | A gift from Dr H. Terada, Tokyo University of Science | |
| Strain, strain background (E. coli) | Escherichia coli JM1100 | PMID:15558 | ||
| Recombinant DNA reagent | Modified pYES3 vector | PMID:26453935 |
Additional files
-
Supplementary file 1
Current status of identification of transport proteins in different archaeal, eubacterial and eukaryotic species (April 2018).
- https://doi.org/10.7554/eLife.38821.014
-
Supplementary file 2
Key resources table.
- https://doi.org/10.7554/eLife.38821.015





