Mechanotransduction: Two views of the same stimulus
The ability of sensory neurons to detect and respond to mechanical force using a process known as mechanotransduction allows us to interpret and navigate the physical world around us. Although a number of different proteins have been linked to mechanotransduction, how they interact with each other or with other signaling pathways in a single sensory neuron is still not well understood.
The study of mechanotransduction has focused primarily upon ion channels that span cell membranes and open in response to mechanical perturbation. In most cases, the channel protein itself is the mechanosensor, responding either to changes in the physical properties of the cell membrane or the tension in a molecular anchor within the cell (Ranade et al., 2015). This promotes the false impression that each type of channel operates independently. However, individual sensory neurons normally express a variety of ion channels and other proteins that allow them to respond to many different signals, such as touch, chemicals, and both hot and cold temperatures (Geffeney and Goodman, 2012; Abraira and Ginty, 2013).
Recent studies have begun to unravel the complex relationship between mechanosensory channels and their environment inside a single neuron. For example, some channels have been shown to interact with members of a superfamily of membrane proteins called the G-protein coupled receptors (GPCRs). The binding of an external signal molecule (ligand) to a GPCR stimulates signaling pathways inside the cell that are involved in a large variety of processes. Furthermore, some of the components in these pathways can interact with mechanosensory channels in response to pain or inflammation (Geppetti et al., 2015; Veldhuis et al., 2015).
All GPCRs contain a seven transmembrane domain embedded within the cell membrane, along with one or more domains inside the cell that trigger the downstream signaling pathways. Members of a subgroup known as the adhesion GPCRs also contain an unusual extracellular domain (ECD) that is thought to interact with components of the extracellular matrix, a scaffold-like structure that surrounds cells to provide structural support (Langenhan et al., 2016)
The ECD is linked to the transmembrane domains by another domain that allows some adhesion GPCRs to cut themselves into two pieces. It has been assumed that this “autoproteolysis” step, which splits the ECD away from the rest of the protein, is essential to activate adhesion GPCRs. Recent studies suggest that some adhesion GPCRs may detect mechanosensory information through the ECD when it is tethered to the extracellular matrix (Petersen et al., 2015; Scholz et al., 2015).
In 2015, a team of researchers led by Robert Kittel and Tobias Langenhan at the University of Würzburg reported that an adhesion GPCR called dCIRL may influence the activity of the NOMPC mechanosensory channel in the chordotonal organ of fruit fly larvae (Scholz et al., 2015). However, it was not clear whether the two proteins directly interact with each other. Now, in eLife, Kittel, Langenhan and co-workers – including Nicole Scholz as first author – report that dCIRL may modulate the activity of NOMPC by stimulating signaling pathways inside the neuron (Scholz et al., 2017).
The chordotonal organ plays crucial roles in a range of mechanosensory processes in fruit fly larvae, and Scholz et al. found that dCIRL (also known as latrophillin) and NOMPC co-localize to the same structures within the neurons in this organ. Mechanical stimuli trigger weaker responses in mutant larvae that are unable to produce dCIRL than they do in normal larvae. In addition, changing the length of the ECD modified the response of dCIRL to mechanical stimuli consistent with an essential role for the ECD in transducing the signal. Together, these results suggest that dCIRL interacts with unidentified ligands outside of the cell to modulate the activity of NOMPC and adjust how strongly a neuron responds to a mechanical stimulus.
Unlike the adhesion GPCRs studied in other animals, dCIRL does not require autoproteolysis to be correctly localized or activated in neurons. Moreover, dCIRL also appears to use different signaling pathways because it decreases the level of a signal molecule called cAMP in cells, whereas the adhesion GPCRs in other animals have the opposite effect (Müller et al., 2015). By using different signaling pathways, the various members of this GPCR subgroup may play different roles in different cell types or species.
The findings of Scholz et al. provide a potential process by which multiple signaling events (such as the complementary mechanical inputs from NOMPC and dCIRL) can interact within a single sensory neuron to precisely modulate the neuron's response to a mechanical perturbation (Figure 1). The diversity of the adhesion GPCR proteins across different cell types and species supports the hypothesis that the way adhesion GPCRs modulate mechanotransduction may also depend upon the type of mechanical signal they receive.
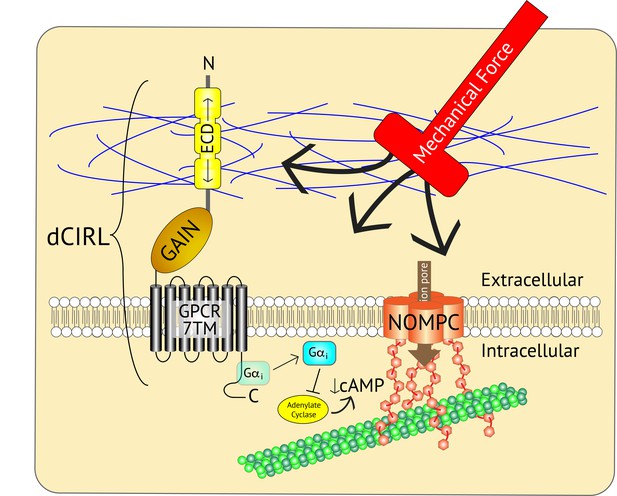
Adjusting the mechanosensory response of sensory neurons.
The ion channel NOMPC (orange) is a transmembrane protein that is anchored to the neuron’s cytoskeleton (green circles), allowing it to detect mechanical force. This stimulates the pore of the channel (brown arrow) to open, leading to the generation of an electrical signal within the neuron. At the same time, the extracellular domain (ECD) of another transmembrane protein, the adhesion GPCR dCIRL, detects the mechanical force differently as a result of being anchored to the extracellular matrix (blue strings). Activation of dCIRL decreases the level of cAMP in the cell, possibly due to the activation of a G protein (Gαi) that inhibits the enzyme that makes cAMP (known as adenylate cyclase). Unidentified signaling components downstream of cAMP alter NOMPC activity to modulate the strength of the overall response. In this manner, both extracellular and intracellular views of the same stimulus are combined to precisely adjust the neuronal response. GAIN, GPCR autoproteolytic domain. 7TM: seven transmembrane domain.
References
-
Adhesion G protein-coupled receptors in nervous system development and diseaseNature Reviews Neuroscience 17:550–561.https://doi.org/10.1038/nrn.2016.86
Article and author information
Author details
Publication history
- Version of Record published: August 14, 2017 (version 1)
- Version of Record updated: August 18, 2017 (version 2)
Copyright
© 2017, Johnson
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 2,091
- views
-
- 211
- downloads
-
- 5
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Cell Biology
- Neuroscience
Alternative RNA splicing is an essential and dynamic process in neuronal differentiation and synapse maturation, and dysregulation of this process has been associated with neurodegenerative diseases. Recent studies have revealed the importance of RNA-binding proteins in the regulation of neuronal splicing programs. However, the molecular mechanisms involved in the control of these splicing regulators are still unclear. Here, we show that KIS, a kinase upregulated in the developmental brain, imposes a genome-wide alteration in exon usage during neuronal differentiation in mice. KIS contains a protein-recognition domain common to spliceosomal components and phosphorylates PTBP2, counteracting the role of this splicing factor in exon exclusion. At the molecular level, phosphorylation of unstructured domains within PTBP2 causes its dissociation from two co-regulators, Matrin3 and hnRNPM, and hinders the RNA-binding capability of the complex. Furthermore, KIS and PTBP2 display strong and opposing functional interactions in synaptic spine emergence and maturation. Taken together, our data uncover a post-translational control of splicing regulators that link transcriptional and alternative exon usage programs in neuronal development.
-
- Genetics and Genomics
- Neuroscience
Single-cell RNA sequencing reveals the extent to which marmosets carry genetically distinct cells from their siblings.






