. Introduction
Orthotrichum Hedw. is a large genus mainly with temperate distribution in both the northern and southern hemispheres. However, as is the case with most of such moss genera, Orthotrichum has been recognized as a heterogeneous taxon. Extensive taxonomic and molecular investigations have confirmed its polyphyly, resulting in the separation of three distinct segregates from the genus: Nyholmiella Holmen & E. Warncke, Pulvigera Plášek, Sawicki & Ochyra and Lewinskya F. Lara, Garilleti & Goffinet (Lara et al., 2016; Sawicki et al., 2009, 2010, 2017).
Species belonging to the Orthotrichum genus are primarily found in temperate regions, with fewer occurrences in tropical areas where they are replaced by Macromitrium Brid., Schlotheimia Brid. and related genera. In Australia, the genus displays a widespread presence, primarily within the forests of the southeastern temperate regions. These mosses predominantly thrive as epiphytes on trees and shrubs, rarely as epilithic species on boulders and rocks (Lewinsky-Haapasaari & Ramsay, 2006). Lewinsky (1984a) provided detailed distribution maps and an overview of substrate preferences for all taxa, revealing that introduced woody plants, such as those within the genera Fraxinus L., Populus L., and Salix L., support the large diversity of Orthotrichum species.
Taxonomic revision of Orthotrichum genus (in a broadly taxonomic sense) in Australia was comprehensively treated by Lewinsky (1984a), and subsequently, the recent distribution was also documented by Lewinsky-Haapasaari and Ramsay (2006). According to these studies, the Orthotrichum s.l. genus in Australia is represented solely by two non-endemic species, O. cupulatum Hoffm. ex Brid. and O. assimile Müll. Hal. In neighboring New Zealand its diversity is one more species greater (moreover, endemic Orthotrichum calvum Hook. f. & Wilson grows here). It should be noted that in the past, O. calvum was also reported from Australia (Scott & Stone, 1976; Watts & Whitelegge, 1906); however, these reports were solely based on collection from Victoria, and no such specimen has been found during the revision process (cf. Lewinsky, 1984a). Consequently, O. calvum has been excluded from the Australian flora.
While identifying material collected during a bryofloristic survey in the Vale of Belvoir Conservation Area in northern Tasmania, a noteworthy epilithic moss from the genus Orthotrichum was discovered. Upon closer examination, it was determined that two specimens represented a new variety previously unknown to science. To confirm the distinctiveness and taxonomic status of the newly described variety, the complete chloroplast genomes were assembled using the latest 3rd generation sequencing technology.
. Material and methods
Plant material
Specimens were collected on two separate occasions: as part of a Bush Blitz survey of the Vale of Belvoir and on a separate visit to the area. Specimens were collected into packets, air dried and returned to the Tasmanian Herbarium for identification and incorporation into the herbarium collections. Plants were subsequently drawn dry and wet, dissected and sectioned for illustration of morphological features.
Morphological evaluation
Leaves, sections and sporophytes were mounted on glass microscope slides, cleared using lactic acid, and all components drawn using an Olympus SMZ10 dissecting microscope and Zeiss Axioskop compound microscope, both fitted with drawing attachments.
An Olympus SZ61 trinocular microscope was used to take macrophotographs of plants. Detailed SEM photo-documentation of leaves and peristome structures was prepared using a Jeol SEM microscope.
Molecular analyses
Total genomic DNA from a single individual was extracted using Qiagen Mini Spin Plant Kit (Qiagen, Germany). Details concerning ptDNA enrichment, library construction and nanopore sequencing are identical to those in the previous study (Sawicki et al., 2023). Obtained raw reads were trimmed using porechop 0.2.4 and assembled using Flye 2.91 assembler (Kolmogorov et al., 2019) which produced complete, circularized plastome contigs. Complete chloroplast genomes were annotated using previously published Orthotrichum Hedw. sequences (Frangedakis et al., 2021; Mizia et al., 2019) as references in Geneious Prime 2023.2.1 software (Biomatters, Auckland, New Zealand).
The newly sequenced plastomes were aligned with those available in GenBank Orthotrichaceae (Supplementary material, Table S1) plastomes using MAFFT 7.52 (Katoh & Standley, 2013). The second copy of IR was removed from subsequent analyses and ambiguously aligned regions were trimmed by Gblocks 0.91 (Talavera & Castresana, 2007).
Chloroplast sequences of 21 specimens of Orthotrichaceae, including seven from Orthotrichum were used for phylogenetic analysis. Macrocoma tenuis (Hook. & Grev.) Vitt (MT591413) was selected as an outgroup based on earlier studies (Draper et al., 2022).
Phylogenetic analysis was carried out using the Bayesian inference (BI). The optimal model for the plastome dataset was identified as GTR + F + I + G4 by ModelFinder based on the Bayesian information criterion (BIC). Bayesian analysis (BA) was conducted using MrBayes 3.2.7 (Huelsenbeck & Ronquist, 2001), and the MCMC algorithm was run for 20,000,000 generations (sampling every 1,000) with four incrementally heated chains (starting from random trees). The visual inspection of Tracer 1.7 (Rambaut et al., 2018) plots was used to examine the parameters and to determine the number of generations needed to reach stationarity, which occurred at approximately 400,000 generations. Therefore, the first 600 trees were discarded as burn-in, and the remaining trees were used to develop a Bayesian consensus tree.
The discrete molecular diagnostic characters (MDCs) for each species of Orthotrichum, Lewinskya and Ulota D. Mohr (genera with more than two known plastome sequences) were calculated according to the Jörger and Schrödl (2013) approach using FASTACHAR 0.2.4 (Merckelbach & Borges, 2020).
. Results
Taxonomic treatment
Orthotrichum cupulatum Hoffm. ex Brid. var. lithophilum Plášek & Sawicki, var. nov.
Diagnosis:Plantae olivacea, obscure viridis vel ferrugineae, usque ad 20 mm altae. Foliis erecto-adpressis, lanceolatis vel ovato-lanceolatis, carinatis, apicibus acuminatis. Lamina foliorum saepius bipartita. Cellulae superiores cum (1–) 3 papillis ramosis; basales glabris. Capsulae exserta, ovoideae-cylindricae. Stomata cryptopora. Vaginula dense pilosa cum capilli longi. Peristomium simplicibus, dentes exostomi 16. Calyptra conico-oblonga, dense pilosa. Sporae 15–19 µm, leniter papillosae.
Type: Australia, Tasmania: Middlesex, Vale of Belvoir Conservation Area, on exposed limestone outcrop in grazed pasture, 800 m a.s.l., GSP = 41° 32′ 51.27″ S, 145° 52′ 59.98″ E, leg. L.H. Cave #1124, 14 Mar 2010, holotype (HO #556429); isotype (OSTR #7129).
Additional specimen examined (Paratype): Australia, Tasmania: Middlesex, Vale of Belvoir Conservation Area, on limestone rocks, 800 m a.s.l., GSP = 41° 33′ 12″ S, 145° 52′ 48″ E, leg. J. Jarman, 25 May 2000 (HO #505285).
Description: Plants olive green, dark green to rusty brown, up to 20 mm tall. Stem moderately branched, branches up to 5 mm long. Rhizoids well-developed, reddish brown, densely distributed mainly at the base of the stem. Stem leaves erect-appressed when dry, erect-spreading when moist, lanceolate to ovate-lanceolate in shape, 1.5–2.0 × 0.3–0.6 mm, keeled, acuminate, sharply acute; lamina rather bistratose (sometimes unistratose), margins entire, recurved from base to near apex, at least on one side (Figure 1). Upper laminal cells isodiametric to short elongate, (7-)10–12 × 8–11 µm, fairly thick-walled, with 1-2(-3) branched papillae; basal laminal cells elongate rectangular to rhomboidal, thick-walled, (18-)20–45 × 10–13 µm, smooth. Alar cells slightly differentiated, forming small auricles with a row of almost rounded cells along the margin. Costa ending below the apex. Sexual condition goniautoicous. Seta 1.2–1.5 mm long, ochrea up to 1/5 of the seta, vaginula densely hairy with 0.8–1.4 mm long, papillose hairs, which usually reach the base of the urn. Capsule exserted; cylindric ovoid to oblong-ovoid, about 1.2–1.4 mm long, yellowish brown to red brown, not constricted below the mouth when dry. Exothecial cells differentiated mainly in the upper part of capsule, urn grooved alternately with 8 long and 8 shorter furrows. Stomata cryptopore, scattered throughout the length of the capsule, half covered by subsidiary cells. Peristome single. Exostome teeth 16, orange-yellow to light brown, erect-spreading when dry, OPL and PPL covered with vermicular or net-like lines, slightly ornamented also with low papillae. Endostome segments completely absent. Preperistome present, covering the outer base of the teeth. Calyptra conic-oblong, light brown with red-brown apex, densely hairy with long, yellowish, strongly papillose hairs. Lid with a short beak. Spores 15–19 µm, finely papillose. Asexual reproduction not observed.
Figure 1
Drawing of Orthotrichum cupulatum var. lithophilum. A–D – shoots with sporophytes (B, D dry), E–L – stem leaves, M, N – perichaetial leaves, O–S – leaf cells: apex (O, P), margin, mid lamina (Q, R), basal angle (S), T – axillary hairs. U–CC – leaf sections, DD – part TS of stem, EE – sporophyte with hairs on the vaginula and calyptra, FF – cells of a calyptra hair, GG – Exothecial cells below capsule mouth, HH – stoma (immersed) on capsule urn. Figures A, B, E–J, P, Q, S, U–Z were drawn based on the holotype (HO556429), figures C, D, K–O, R, T, AA–HH were drawn based on the paratype (HO505285).

Name of the new variety (lithophilum) indicates its ecological characteristic – it was found growing on rocks.
Molecular survey
Chloroplast genome characteristics
The circular plastomes of the four newly assembled specimens have a typical quadripartite structure with one small single-copy (SSC), one large single-copy (LSC), and two inverted repeats (IR). Application of the 3rd generation sequencing did not reveal structural heteroplasmy connected with SSC region inversion. The total length of the plastomes ranged from 122,895 bp to 123,536 bp. The plastome contains 82 protein-coding genes (including the hypothetical chloroplast reading frames: ycf1, 2, 3, 4, 12, and 66), 32 tRNA, and four rRNA. The rps12 gene is divided into two independent transcription units (5′-rps12 and 3′-rps12) whose transcripts are trans-spliced. The gene structure and order are identical to those of other Orthotrichum sensu stricto plastomes (Frangedakis et al., 2021; Mizia et al., 2019).
Phylogeny and molecular diagnostics characters
Phylogenetic analysis based on complete plastome sequences resolved all clades with maximum values of posterior probabilities (Figure 2). The species of Orthotrichaceae formed two distinct clades, one grouping genera belonging to Lewinskyinae (Lewinskya, Pulvigera and Ulota) and the second formed by members of Orthotrichinae (Nyholmiella, Stoneobryum D.H. Norris & H. Rob. and Orthotrichum s.str.). Analyzed genera were revealed as being monophyletic, with Ulota sister to the Pulvigera/Lewinskya clade and Nyholmiella as sister to Orthotrichum.
Figure 2
The Bayesian inference tree based on complete chloroplast genomes. All clades have maximum PP values (1.0). The numbers in square brackets indicate the number of MDCs.
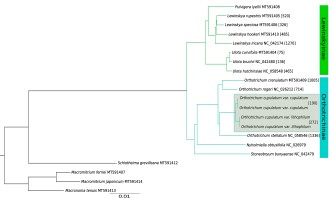
The number of detected MDCs ranged from 75 for Ulota curvifolia (Wahlenb.) Sw. to 1,336 for Orthotrichum stellatum Brid. (Figure 2). The remaining Ulota species revealed 135 (U. bruchii Hornsch. ex Brid.) and 465 (U. hutchinsiae (Sm.) Hammar) MDCs. Among species of Orthotrichum s.str. the lowest number of MDCs at the species level was found for O. rogeri Brid.
(714). The similar number of MDCs was detected for the O. cupulatum complex (1,002) and O. crenulatum Mitt. (1,005). The numbers were lower for the variety range of O. cupulatum: 190 MDCs were found for var. cupulatum and 272 for the newly described var. lithophilum. The lower number of MDCs were found for species of Lewinskya. With the exception of L. incana (Müll. Hal.) F. Lara, Garilleti & Goffinet (1,270), the analyzed species have at least a twofold lower number including 326 for L. speciosa (Nees) F. Lara, Garilleti & Goffinet, 485 for L. hookeri (Wilson ex Mitt.) F. Lara, Garilleti & Goffinet and 520 for L. rupestris (Schleich. ex Schwägr.) F. Lara, Garilleti & Goffinet.
. Discussion
Three species of the genus Lewinskya (L. hortensis (Bosw.) F. Lara, Garilleti & Goffinet, L. rupestris, and L. tasmanica (Hook.f. & Wilson) F. Lara, Garilleti & Goffinet) and two from Orthotrichum (O. cupulatum and O. longitheca R. Br. bis., subsequently synonymized with O. assimile Müll. Hal.) have previously been recorded from Tasmania (Lewinsky, 1984a,b). The differences in characteristics among the cryptoporus species are significant. Orthotrichum cupulatum typically forms cushions on rock surfaces, while O. assimile (syn. O. longitheca) thrives on tree trunks. In the former species, the endostome is mostly rudimentary or entirely absent (with the exception of var. austro-cupulatum), whereas the latter species possesses eight well-developed segments in its endostome. Additionally, the newly described variety differs from these aforementioned taxa due to the presence of a bistratose lamina and a notably hairy vaginula.
The presence of intraspecific taxa that distinguish Lewinskya tasmanica and Orthotrichum cupulatum is also known in the region. For the former, two varieties have been described: L. tasmanica var. tasmanica (Hook.f. & Wilson) F. Lara, Garilleti & Goffinet and L. tasmanica var. parvitheca (R.Br.bis) F. Lara, Garilleti & Goffinet. For the latter, they are O. cupulatum var. cupulatum and O. cupulatum var. austro-cupulatum. Now, a third variety is newly introduced, O. cupulatum var. lithophilum.
All three above-mentioned varieties share the same ecological requirements, as they are epilithic species that grow on limestone rocks or stones in exposed habitats. Morphologically, however, they exhibit distinct differences and are, therefore, easily recognizable. For a quick assessment of their distinguishing characteristics, refer to Table 1, which compares individual critical traits. The unique differences that are typical of the newly described variety are highlighted in bold. Even at a macroscopic level, the prominently hairy vaginula is clearly visible, with papillose hairs extending to the base of the capsule (Figure 3A, Figure 3D–E). The vaginula, whether hairy or naked, serves as a commonly used diagnostic feature across the broadly understood genus Orthotrichum s.l. A comprehensive analysis discussing the presence or absence of the vaginula in selected species, as well as its genetic determination, was conducted by Plášek and Sawicki (2010).
Table 1
Comparison of different characters of individual varieties occur in Australasia. Relevant different characters of the new variety are highlighted in bold.
Figure 3
Macro photographs of Orthotrichum cupulatum var. lithophilum. A, B – habit, C – mature capsules, D, E – fertile plants with uncovered long vaginula hairs. Scale bars: A–C, E – 1 mm, D – 1 cm. Photographs were taken from the holotype (HO556429).

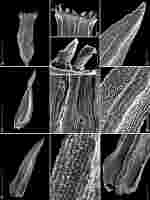
Under examination with a microscope, a simple peristome, occasionally accompanied by fragments of the preperistome (Figure 4C) can be seen. Furthermore, the new variety is characterized by the presence of a bistratose leaf lamina primarily occurring in the upper half of the leaves. The new variety also exhibits differences in spore size, particularly when compared to O. cupulatum var. cupulatum. However, the spore size for O. cupulatum var. austro-cupulatum was not indicated in the description (cf. Lewinsky, 1984a).
Figure 4
SEM photographs of Orthotrichum cupulatum var. lithophilum. A – capsule, B – single peristome, C – detailed OPL view on exostome teeth and one preperistome fragment covering the outer base of the tooth (left edge), D – cryptoporous stomata, E – adaxial view of stem leaf, F – abaxial view on stem leaf, G – I – detail views of laminar cells, abaxial views (G - upper part, H - middle part, and I - base of leaf), J – branched papillae on laminar cells. Scale bars: A – 0.5 mm, B – 100 µm, C, D – 50 µm, E, F – 0.5 mm, G–I – 50 µm, J – 10 µm. Photographs were taken from the holotype (HO556429).
For the sake of completeness, it should be noted that a fourth variety, O. cupulatum Brid. var. austro-americanum Lewinsky, grows in the southern hemisphere. This taxon was described from Peru (Lewinsky, 1984b) but is also present in mountainous regions of Colombia and Argentina. It is distinguished from all other varieties by the exostome teeth being smooth, never striolate or papillose (Lewinsky, 1984b).
The obtained phylogenetic tree is mostly congruent with previous studies on Orthotricheae phylogeny, which clearly separate Lewinskyinea and Orthotrichinae clades (Draper et al., 2021, 2022; Sawicki et al., 2009, 2017). The main difference between the plastid and mitogenomics datasets is in the relationships of Nyholmiella and Stoneobryum to Orthotrichum s.str. The phylogenomics analysis based on complete mitogenomes poorly supported the common clade of Stoneobryum and Orthotrichum in ML analysis and unresolved Orthotrichinae intergeneric relationships in the case of the BI method (Sawicki et al., 2017), while the plastomic dataset relates Nyholmiella as a sister to Orthotrichum and Stoneobryum as basal for Orthotrichinae with maximal node support (Figure 2).
The number of detected MDCs for Orthotricheae taxa is hard to compare, since none of the other genera have been analyzed to date. Studies on plastome-based delimitation of liverworts revealed comparable to Orthotrichinae the number of MDCS in the leafy genus Calypogeia (Ślipiko et al., 2020, 2022), which ranged from 159 (C. muelleriana) to 1,369 (C. neesiana), but lower than those detected in thalloid genera like Conocephalum (over 2,300) and Pellia in which the plastome revealed the presence of over 4,700 MDCs between P. epiphylla and P. neesiana (Paukszto et al., 2023; Sawicki et al., 2020).

