Abstract
Eye-tracking is widely used throughout the scientific community, from vision science and psycholinguistics to marketing and human-computer interaction. Surprisingly, there is little consistency and transparency in preprocessing steps, making replicability and reproducibility difficult. To increase replicability, reproducibility, and transparency, a package in R (a free and widely used statistical programming environment) called gazeR was created to read and preprocess two types of data: gaze position and pupil size. For gaze position data, gazeR has functions for reading in raw eye-tracking data, formatting it for analysis, converting from gaze coordinates to areas of interest, and binning and aggregating data. For data from pupillometry studies, the gazeR package has functions for reading in and merging multiple raw pupil data files, removing observations with too much missing data, eliminating artifacts, blink identification and interpolation, subtractive baseline correction, and binning and aggregating data. The package is open-source and freely available for download and installation: https://github.com/dmirman/gazer. We provide step-by-step analyses of data from two tasks exemplifying the package’s capabilities.
Similar content being viewed by others
Introduction
Recent advances in eye-tracking technology make it a highly powerful and relatively inexpensive tool to gather fine-grained measures of the temporal dynamics of cognitive processing. Because of this, a growing number of fields, from vision science and psycholinguistics to marketing and human-computer interaction, have adopted this methodology. Despite its growing presence, there is considerable variability in how eye-tracking data are processed. With increased attention on replicability, reproducibility, and transparency, there is a need for a cross-platform, fully free implementation of standard practices in eye-tracking data processing. R (R Core Team, 2019) is a widely-used, free, cross-platform, and open-source statistical programming language that provides the tools needed to meet those needs. In R, there are few established pipelines for handling pupil and fixation data from the visual world paradigm and pupillometry, especially contained in one package (see Tables 1 and 2). To meet this need, we created the gazeR package. The gazeR package is meant to facilitate the end-to-end handling of eye-tracking data within a single programming environment (R) – from reading in raw data files to statistical analysis and generating figures. The gazeR package is also designed to be as familiar as possible for the regular R user, thus handling data in formats and functions that will be accessible for most users.
In this paper, we provide a step-by-step walk-through of how to use the gazeR package to analyze data from experiments in which the primary outcome measure is gaze position or pupil size. There are several conceptual or theoretical discussions on best practices when analyzing pupil and gaze data available elsewhere (see Mathôt et al., 2018; Winn, Wendt, Koelewijn, and Kuchinsky, 2018; Salverda & Tanenhaus, 2018). The main aim of the present paper is to illustrate and explain how to analyze gaze and pupil data in a more standardized way using gazeR, such that it may be used by researchers to analyze their own data. While there exist various packages and online resources to get started with eye-tracking, such materials are typically limited to the analysis of a single participant and do not represent what researchers typically want to do with their data. A secondary aim is to facilitate reproducible and transparent preprocessing of these types of data, using conventional practices in eye-tracking data processing, and smoothing the transition from data preprocessing to data analysis and visualization. In the remainder of this report, we provide a step-by-step walk through of the installation and core functionality of the gazeR package.
Package Installation and Setup
Reading in Data
GazeR is meant to work on data in a relatively raw format, where each row is a sample corresponding to the sampling rate of the eye tracker. This allows gazeR to maximize compatibility: data from any eye-tracker can be used as long as the file contains information such as X and Y coordinates, pupil size, and/or relevant event messages. For the examples contained herein, we will discuss how to preprocess data collected with one of the most popular commercial eye-trackers on the market, the SR EyeLink. Keeping with the spirit of open-access and transparency, however, we will highlight how to read raw EDF files for use with gazeR, so the proprietary SR software DataViewer is not necessaryFootnote 1.
Package Installation
The gazeR package can be installed along with helper packages using the remotes package:

Once this has been completed, gazeR can be loaded along with additional useful packages:

Once gazeR and other helper packages have been installed and loaded the user is ready to start preprocessing data.
Preprocessing Gaze Position Data from the Visual World Paradigm
In a typical instantiation of the Visual World Paradigm (VWP), participants hear spoken instructions to manipulate or select one of several images on a computer screen or objects in the real world (Cooper, 1974; Tanenhaus, Spivey-Knowlton, Eberhard, & Sedivy, 1995). Decades of research have shown that the time course of fixation proportions – that is, the probability of fixating a particular object at a particular time – reflects the activation of that object’s mental representation. Fig 1 illustrates a typical VWP task. In this example (from Mirman & Graziano, 2012), the study examined semantic competition: the display contained a critical distractor that was related to the target either thematically (associates; e.g., dog-leash; shown in the left panel of Fig. 1) or taxonomically (e.g., apple-pear). On each trial, the display contained a target object image, a semantic competitor (taxonomically or thematically related), and two unrelated distractors. The outcome measure was the probability of looks (fixation proportion) to a particular object at each point in time (example data shown in the right panel of Fig. 1).
Left: Example display from a VWP experiment. The target is dog, the critical semantic competitor is leash (thematically related to the target), and snowman and carriage are unrelated distractors. Right: Example data showing the time course of target word recognition (solid line) and semantic competition: the semantically related competitor (dotted line) was fixated more than the unrelated distractors (dashed line)
Gaze preprocessing requires four steps:
-
(1)
Reading in the data
-
(2)
Eliminating trackloss (out-of-bounds) data
-
(3)
Assigning areas of interest
-
(4)
Samples to bins (optional)
Reading in Gaze Data
In order to process the EDF files generated by the EyeLink system you will need to first install the EDF API provided by SR-Research, which is free of chargeFootnote 2 and required for the ‘edfR’ package (Hubbard & von der Malsburg, 2015), which gazeR uses to read EDF files. In order to read the EDF files, for both pupil and visual world data, two folder paths must be specified: one path where the EDF files are located and one where the raw CSV files should be saved.

Once folder paths are specified, you can call the parse_edf function. This function imports the sample data from the EDF files. The type argument must be specified as either “pupil” or “vwp”, depending on experimental design used. The parse_edf function merges the sample and message data from your raw EDF files and wrangles them into a format suitable for preprocessing with gazeR. Specifically, the function places time in milliseconds, adds participant ids, trials, and sample messages, and computes the mean x and y gaze coordinates and diameter values for a monocular eye variable (the left, right, or mean of both eyes).
The parse_edf function generates a CSV file from each EDF file in a directory specified by the user. The merge_gazer_files function can then aggregate those new CSV data files stored in the directory_csv_from_edf_conversion path specified above. For files that were processed with parse_edf, you need to set the filetype argument to “edf.”

Behavioral Data
When using raw EDF files, relevant behavioral message variables (e.g., conditional variables, RTs , and accuracy) are usually sent outside the sampling frequency of the eye tracker. To display the relevant trial variables in a nice table, you can run the find_messages_edf function, which will produce a CSV file from each EDF file. In order for the function to work properly the user must specify specific variable names (varnames) and the patterns that need to be replaced (patterns). SR prepends a “TRIAL VAR” marker to behavioral variables. After running this function, you can merge each participant’s behavioral report with the merge_gazer_files function. The behavioral report can then be joined to the gaze sample report.

A pre-read version of this data set is included in gazeR to demonstrate what the sample data should look like after merging the message information:

For this example data set the sample gaze data contain eye-tracking variables and experiment-specific values (positions of different objects, trial condition, participant accuracy and response time) that were extracted from the raw EDF files

Trackloss
Once the data are loaded, some researchers might prefer to remove trials with excessive trackloss (instances where the eyes travel outside of the viewing screen). This can be determined by the X and Y coordinates at each sample relative to the size (resolution) of the screen. Trackloss from the EyeLink systems use 1e+08. Theget_trackloss function determines the on/off screen status of each sample, computes the proportion of trackloss by trial and participant, and filters out trials and subjects that pass a user-defined threshold (this filtering can be omitted by setting the threshold to 1.0). The screen_size argument must be supplied as a numeric vector of the X and Y dimensions of the computer screen used during the experiment. In this example, we will not be throwing out data due to trackloss.

Parsing areas of interest
The following preprocessing assumes that the interest areas (locations of objects) were static and that the fixation report includes columns indicating the location of each object for each trial. For this example, the objects were always presented in the four corners of the screen, though which object was in which corner was randomized. The four possible image locations are labeled as image1, image2, image3, and image4. The TargetLoc variable identifies which of those locations was the target object and the CompPort variable identifies which of those locations was the critical semantically related competitor. The gaze position was recorded in terms of (X,Y) coordinates. In order to determine which (if any) of the objects were being fixated, first identify the locations of the target and competitor images, then use gaze coordinates to determine which image location (if any) was being fixated, then compare gaze location to target and competitor locations. If gaze location has already been coded in terms of interest areas (many experiment programs do this dynamically, as the data are being collected), then this step can be skipped.
The assign_aoi function will match gaze positions to numbered areas of interest (AOI) based on screen coordinates (by default, 400x300 rectangles in the corners of the 1024x768 screen), which will need to be matched to image location labels:

Now determine which object was being fixated by matching AOI codes with target and competitor locations:

Gathering Data
The specifics of data organization and aggregation depend on the design and hypotheses of the specific study. For this example, the fixation locations need to be “gathered” from separate columns into a single column (see Supplemental Figure for a demonstration of this) and “NA” values need to be re-coded as no-fixations:

Samples to Bins (Optional)
You can downsample your data, if you choose, into larger time bins using the downsample_gaze function. This function aggregates the set of samples into a time series consisting of standardized time bins with a size specified by the user (default is 50ms). In addition, it drops columns that are no longer necessary. The user needs to specify a list columns (aggvars) that define the aggregation level (e.g., individual trials) and should be kept after the binning is done. If you would like to keep the raw data unbinned, you can skip this part.

Aggregating Data
In the final stage of preprocessing, the error and practice trials can be removed and the time window can be restricted to make the data ready for aggregation. For this example, we group the trials by Subject, Condition, and Object type to calculate number of valid trials in each cell. We then also group by time point to calculate the number of object fixations and mean fixation proportion at each time point; that is, the time course of fixation. These are the subject-by-condition time courses that would go into an analysis.

Plot fixation time course
After the fixations have been assigned to the object type and converted to time bins, they are ready for visualization and statistical analysis. Below is a plot (see Fig. 2) of the time course of fixation proportions for each target type.

Preprocessing Pupil Data from a Lexical Decision Task
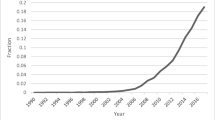
Recent advances in eye-tracking technology have led to a burgeoning interest in cognitive pupillometry (i.e., measurement of changes in pupil size as it relates to higher-level processing). According to a recent PubMed search (see Fig. 3), the number of studies employing pupillometry has grown exponentially since the first modern boom more than a half-century ago. The reason for this is quite simple: pupil size has been shown to be a reliable and valid index of cognitive effort or arousal across many domains, including word recognition (Geller, Still, & Morris, 2016), normal and impaired auditory perception (Zekveld et al., 2018), semantic cognition (Geller, Landrigan, & Mirman, 2019), attention allocation (Endogenous attention: Mathôt, van der Linden, Grainger, and Vitu, 2013), working memory load (Granholm, Asarnow, Sarkin, & Dykes, 1996; Van Gerven, Paas, Van Merriënboer, & Schmidt, 2004), face perception (Goldinger, He, and Papesh, 2009), and general cognitive processing (Murphy et al., 2014). While there are a number of good open-source programs available in R to analyze pupil data (see Forbes, 2019; Tsukahara, 2018), there are not many walk-throughs demonstrating how to go from raw data to fully pre-processed data. A recent methods review by Winn et al. (2018) describes and illustrates general principles such as blink detection, interpolation, and filtering. The gazeR package includes functions for implementing these steps, and here we demonstrate their use.
To demonstrate analysis of pupil data, we will be using an example data set containing data from a lexical decision task. For this walkthrough, participants (N=3) judged the lexicality (i.e. “was this a word or not a word?”) of printed and cursive stimuli while pupil diameter was recorded. Because cursive stimuli are non-segmented and could be ambiguous, it was predicted that recognizing cursive stimuli would require more effort than printed words would (cf., Barnhart & Goldinger, 2010; Geller, Still, Dark, & Carpenter, 2018), resulting in larger pupil dilation.
Preprocessing pupil data requires the following steps:
-
(1)
Read data
-
(2)
De-blinking
-
Extending blinks
-
Smoothing and interpolation
-
-
(3)
Baseline correction
-
(4)
Re-scaling
-
(5)
Artifact Rejection
-
Missing data
-
Unlikely pupil values
-
Median absolute deviation (MAD)
-
-
(6)
Event time alignment
-
(7)
Samples to bins
Reading in Pupil Data
In this example, we consider data from several subjects – the typical experimental scenario. For your own data (that includes many individual files), the function parse_edf with the type argument set to “pupil” will produce the necessary columns needed for gazeR to work. For non-EDF files, you can use the make_gazer function to make your data suitable for gazeR. This returns a data frame with column names changed to: subject, trial, blink, x, y, pupil, time, and message.
Once each EDF file is saved as a CSV with the parse_edf function (explained above), you can call the merge_gazer_files function to aggregate all your pupil sample files.

Behavioral Data (Optional)
For those interested in analyzing behavioral data, the find_messages_edf function can be used to cull the important behavioral data and merge with the sample data.

Due to processing constraints, we use a data file that includes data from a few participants with all behavioral data included. The parse_edf , merge_gazer_files, and find_messages_edf functions can be tested using example data that are available to download from the Open Science Framework (OSF): https://osf.io/fzu38/. While reading in the data is fast (even with many participants), some of the functions performed on the data can be computationally intensive.

Once merged, the behave_data function will cull the important behavioral data from the sample report. The function will return a data frame without errors when omiterrors=TRUE or a data frame with errors for accuracy/error analysis when omiterrors=FALSE. The columns relevant for your experiment need to be specified within the behave_col names argument. This function does not eliminate outliers; you must use your preferred method. Grange’s (2015) trimr package implements multiple standard methods of outlier exclusion (https://github.com/JimGrange/trimr). The overall item and subject accuracy can be merged into the pupil sample report.

In our data, we want to remove subject accuracy lower than 75% and item accuracy below 60%. We can use the data frame generated above to calculate this when argument omiterrors=FALSE. We then merge accuracy by items and subjects into the main pupil file.
We can now restrict preprocessing to valid trials by removing practice blocks, trials with incorrect responses, conditions that are not words, subjects with accuracy below 75%, and items with accuracy below 60%.

Pupil Preprocessing is now ready to begin!
De-blinking
An important step in preprocessing pupil data is de-blinking. A major artifact in pupil data comes from blinking. When the eye blinks, the pupil momentarily becomes smaller as it is occluded more and more by the eyelids, making it difficult to compute the center of the pupil. Eye-trackers interpret this as a fast shift in pupil position and might erroneously classify it as a saccade. Additionally, the estimate of pupil size will rapidly decrease as the pupil occupies less of the camera image. This process happens in reverse (albeit a bit more slowly) as the eye is opening, so blinks are always flanked by an artifact that includes a false saccade and/or false change in pupil size. Occasionally there will be some additional artifacts, such as short fixations preceding or following the blink. It is thus advisable to de-blink the data, which involves identifying blinks, removing data during the blink, removing data slightly before and after the blink, and then interpolating data during the period that was removed. In gazeR, blinks are identified automatically when importing raw EDFs using the saccades package (von der Malsburg, 2019). For data in another format, the detect_blink function can be used to identify blinks. Blink detection from the saccades package uses a velocity-based algorithm taken from Engbert & Kliegl (2003). Blink events are identified as anything that looks like a fixation but has much lower dispersion than the typical fixation. In the saccades package, a blink is an event with a dispersion that is smaller than the median dispersion minus four times the median absolute deviation of the dispersion, and only if this is the case for horizontal and vertical dispersion.Footnote 3
A less trivial matter in pre-processing pupil data is deciding how many data points to remove before and after the blink. It has generally been recommended that data 100 ms before and after the blink should be eliminated. There are several ways one can deal with blinks (see Hershman, Henik, & Cohen, 2018). One method is to eliminate all blinks from a trial. This is generally not recommended as it can eliminate too much data, resulting in a loss of power. A more acceptable approach, and the one implemented in gazeR, is to extend the time window around the blinks so the interpolation starts 100-200 ms before the blink and continues after the blink (Nyström, Hooge, & Andersson, 2016; Satterthwaite et al., 2007). Extending the time window around the blinks eliminates spurious samples caused by the closing and opening of the eyelids. The extend_blinks function implements this method, with the fillback argument specifying how far blinks should extend back in time (before the blink) and the fillforward argument specifying how far blinks should extend forward in time (after the blink). This function is robust to different sampling rates as long as the eye-tracker sampling rate is specified in the hz argument. For this experiment, the tracker sampled at 250Hz (once every 4 ms) and blinks were extended 100 ms forward and backward in time.

Smoothing/Filtering and Interpolation
Pupil data can be extremely noisy. To remove some of this noise, filtering and interpolation are commonly done. In gazeR this is done in one step using the smooth_interpolate_pupil function. With the step.first argument users can choose to either smooth the data first with an n-moving average, and then interpolate (step.first = “smooth”) or vice versa (step.first = “interp”). Depending on which methods are selected, the order of the steps can have negligible or substantial effects (see Figs. 4 and 5); if applying cubic-spline interpolation, smoothing before interpolation is generally advisable. The gazeR package currently implements two common ways to smooth pupil data: n-point moving average and Hanning window (we plan to include more smoothing options in future updates to the package). To smooth the data, you must specify the column that contains the pupil information and the size (in samples) of the moving average window. In this example, we use a 5-point moving average (n=5) which, at a sampling rate of 250 Hz, corresponds to a 1250 ms moving average.
Missing data stemming from blinks or failure of the eye tracker need to be interpolated. The smooth_interpolate_pupil function searches the data and reconstructs the smoothed pupil size for each trial from the relevant samples using either linear interpolation (Bradley, Miccoli, Escrig, & Lang, 2008; Cohen et al., 2015; Siegle, Steinhauer, Carter, Ramel, & Thase, 2003) or cubic-spline interpolation (Mathôt, 2018). Considering the short duration of blinks and the relatively low speed of blinks, the choice of linear versus cubic interpolation will ultimately have negligible effect. If extendblinks= FALSE, samples with blinks are turned into NAs and are then interpolated. This function returns a tibble object with a column called pup_interp, which contains interpolated values from the moving averaged pupillary data. There are also options to denote the max number of gaps to interpolate over (the maxgap argument is set to Inf by default). This requires the sampling rate (hz) of the eye tracker be specified. As an important note, if the Data Viewer was used to extend blinks, the extendblinks argument should be set to FALSE. If the extend_blinks function was used, the extendblink argument should be set to TRUE. It is important to note that SR only extends the blink column and does not set pupil size estimates during blinks to “NA” in the Sample Report. For this example, we will set extendblinks to TRUE and use linear interpolation. You can use cubic interpolation by changing type to “cubic.”

Baseline correction
To control for variability in overall pupil size arising from non-task related (tonic) state of arousal, baseline correction is commonly used (but see Attard-Johnson, Ó Ciardha, & Bindemann, 2019). The two most popular types of baseline correction to identify task-evoked dilation are subtractive (pupil size - baseline) and divisive (change in pupil size - baseline / baseline). Subtractive baseline correction is more common in the literature (cf., Beatty, 1982; Laeng et al., 2012; Zekveld, Koelewijn, & Kramer, 2018), and this practice has been argued by Reilly, Kelly, Kim, Jett, and Zuckerman (2018) to be reflective of the linearity of the pupil response independent of baseline size, although the basis of that argument has been questionedFootnote 4. The baseline_correction_pupil function finds the median pupil size during a specified baseline period for each trial and performs a subtraction baseline correction by default (see Mathôt et al., 2018, for argument that baseline correction should be done using the median, and not the mean, baseline value). By changing the baseline_method argument to “div”, you will get proportion change from baseline. In this example, subtractive baseline correction is applied to pupil size in arbitrary units (pupil_colname = "pup_interp"), though the same can be done for pupil size in mm or z-score. The baseline time window can be set with numerical values using the baseline_window argument from the baseline_correction_pupil function if events are fixed or static.

If events in your experiment occur dynamically, the baseline_correction_pupil_msg function can be used. This function takes a user-specified baseline period immediately preceding a stimulus onset message. In the below example, we set the baseline_dur argument to 100 ms and the event argument to “target.” This will calculate the median baseline value for 100 ms preceding target onset. The choice of baseline duration appears to be largely inconsequential (Winn et al., 2018).

Re-Scaling
So far, the analysis steps have used arbitrary pupil units. It is advised that these be transformed into a standardized unit in order to make comparisons between individuals. Numerous options have been used in prior studies: z-scores (see Cohen, Moyal, & Henik, 2015; Einhauser, Stout, Koch, & Carter, 2008; Kang & Wheatley, 2015), absolute changes in mm (e.g., Beatty, 1982; Geller, Landrigan, & Mirman, 2019; Geller et al., 2016), proportional change relative to baseline (Winn, 2016), proportional change relative to peak (Winn, 2016; Winn & Moore, 2018), and absolute change relative to dynamic range of pupil reactivity elicited by the light reflex (Piquado, Isaacowitz, & Wingfield, 2010). To convert arbitrary pupil size to mm, we measured the scaling factor by running a short experiment with an artificial pupil (5 mm in size) and calculated the average pupil size in arbitrary units. At a fixed camera-to-pupil distance of 90 cm, the 5mm pupil was coded as 5570 arbitrary pixel units. This information was entered into the equation below to convert arbitrary units to mm. Specifically, the smoothed pupil size value is multiplied by 5/5570 to re-scale the values to mm.

Alternatively, the arbitrary pupil units can be converted to a z-score using the scale function.

For the rest of the walkthrough, arbitrary pupil size will be used.
Artifact Rejection
Missingness
The count_missing_pupil function will remove subjects and items that have a large amount of missing data – the threshold for “a large amount” is specified by the researcher. It has been recommended by Winn et al. (2018) that a reasonable threshold is 20%, but that the exact importance of missing data might be weighted by specific timing landmarks in the experiment trials. For this example, we have set the missingthresh argument to .2 and the pupil argument to the raw, non-interpolated, pupil dilation column in our dataset. The count_missing_pupil function returns the percentage of subjects and trials that have been excluded for reporting.

Spurious pupil values
Unlikely pupil values that are too small and too large should be removed from the data (Mathôt et al., 2018; Winn et al., 2018). Mathôt (2018) recommended against removing data based on a subject-independent fixed criterion (e.g., above or below an SD cut-off or a specified lower and upper pupil boundary). This is due to the inherent heterogeneity of pupil sizes across experiments. Instead, Mathôt (2018) recommend visual inspection to determine unlikely pupil values. This can be done using a simple histogram to plot the pupillometric data. Based on the histogram below, it seems reasonable to remove pupil sizes less than 2500 and greater than 5000. Fig 6


Median absolute deviation (MAD)
After interpolation, it is a good idea to perform a second pass to make sure that the data are not contaminated by rapid pupil size disturbances. These artifacts can be detected using the median absolute deviation, which is a more robust variability metric than traditional measures of variability (e.g., standard deviation; Hampel, 1974; Kret & Sjak-Shie, 2018). For each time point, the speed_pupil function calculates the normalized dilation speed, which is the absolute change in pupil size between samples divided by the temporal separation between them. To detect outliers, the MAD is calculated from the dilation speed variable and multiplied by a constant (in this case 16, which is the value used in Kret & Sjak-Shie, 2018). The constant controls the sensitivity threshold: The higher the constant, the more extreme a value needs to be in order to be marked for removal. The MAD is added to the median dilation speed variable using the calc_mad function; values above this threshold are then removed.

Event Time Alignment
In most psychological experiments, each trial includes several events. In the example experiment, each trial began with a fixation screen (small cross in the center of the screen) and the stimulus of interest appeared on screen 1s after trial onset. These events are documented in the data file: the onset of the target is denoted by the trial message “target.” We can use this information to align the data so that time=0 corresponds to stimulus onset (i.e., the analysis window of interest) rather than trial onset. The onset_pupil function performs this alignment using three arguments: time column, sample message column, and the event of interest (“target” in our example). In the output below, we can see that our experiment now starts at zero, when the target was displayed on screen.

Samples to Bins
If the data are recorded at a relatively high sampling frequency (e.g., 250Hz in this example), it may be useful to aggregate the data into time bins that are somewhat larger than the sample rate (users can specify a time bin size to use). The downsample_gaze function will aggregate the set of samples into a time series consisting of standardized time bins with a size specified by the user for pupil data when the type argument is set to “pupil”. In addition, it will drop columns that are no longer necessary. The user needs to specify a vector of column names (aggvars) that define the aggregation level (e.g., individual trials) and should be kept after the binning is done. This produces an average baseline-corrected pupil diameter for each subject, condition, and timebin. If you would like to keep the raw data unbinned, you can skip this part. This function returns a tibble with an added column called timebins, which can be used for aggregation (e.g., calculating the mean pupil size in each time bin).

Pupillary Data Visualization
After baseline-correction and artifact rejection, the data are ready for visualization and statistical analysis (see Fig. 7). The pre-processed data produced by gazeR are highly flexible and compatible with different visualization strategies.

Data Analysis
The gazeR package is deliberately agnostic to how researchers should analyze their data, leaving the data in a format that is flexible and compatible with a variety of statistical modeling strategies. For instance, various curve-fitting strategies such as growth curve analysis, general additive models and/or functional data analysis (Jackson & Sirois, 2009; Mirman, 2014; Seedorff, Oleson, and McMurray, 2018; van Rij et al., 2019). In the absence of a field-standard statistical approach, we leave it up to the researcher to choose what statistical analysis to use.
For those interested in growth curve modeling, the gazeR package does contain a helper function (code_poly) to facilitate growth curve analysis (GCA) using orthogonal polynomials (Mirman, 2014). The code_poly function takes your time column and adds polynomial transformations of time up to a user-specified order (see Fig. 8). The polynomial transformation can be natural or orthogonal; the orthogonal version can be useful because it renders the time terms uncorrelated and scales them to the same range (for a discussion of natural vs. orthogonal polynomials see Mirman, 2014).


The gazeR package also includes a function (get_ranef) for extracting random effects from a growth curve model in a format that is convenient for quantifying individual differences (see Mirman, 2014, Chapter 7) and a function for estimating p-values for linear mixed effects models using the normal and/or Kenward-Roger approximations (get_pvalues).
Discussion
While there are a number of viable solutions available to process eye-tracking data, not all are suitable for research purposes:
-
An all-graphical interface seldom provides information about the underlying data analysis and can be difficult to reproduce.
-
File formats are sometimes proprietary and undocumented, lacking detailed annotation necessary for replicability.
-
Source code and description of the algorithms are not always accessible to the user.
-
Some implementations rely on expensive proprietary software.
The research community needs solutions that are completely open, with the possibility of directly manipulating and annotating the code, data, and parameters so that others may replicate or critique the methods. This article summarized and demonstrated the functionality of gazeR, a free, open-source package written in R. We walked through important functions needed to pre-process both gaze position and pupil size data and make them suitable for analysis. This provides a generalized, replicable, and transparent method for preprocessing raw eye-tracking data.
How does gazeR compare to other existing solutions?
While there exist several packages in R to analyze pupillometry data or visual world paradigm data (see Tables 1 and 2), gazeR has several advantages over existing solutions. First, gazeR is currently the only package that allows one to analyze pupillometic and gaze data all in one package. Second, many of the R packages rely on proprietary software and algorithms for key analysis steps (such as fixation/saccade parsing and blink detection). GazeR avoids this by allowing users to read EDF files directly and by using open-source event detection algorithms. Third, when deciding on a package to use, it is important the package be well documented and supported. Many of the packages listed in the table were developed and updated several years ago, and it is unclear if the package developers are still servicing the packages. Most crucially, the source code and descriptions for many of the packages are not well explained. Lastly, a significant problem for the standardization of analyses, especially with the implementation of a package, is that with constantly changing software environments, results likely differ. For example, different R versions could change the results, as could differences between platforms (Windows, macOS, or Linux). To this end, a containerized version of gazeR is available via Binder on Github, which allows users to follow along with this walkthrough in the exact environment in which it was created, thus supporting reproducibility. Our hope is this encapsulated environment will help facilitate use of our pipeline to users’ own data. In the future we hope to host gazer on Docker or Code Ocean,, similar to other preprocessing pipelines (e.g., fmriprep; Esteban et al., 2019). Overall, we feel that this walkthrough, end-to-end implementation in the free, cross-platform environment of R, and the fact that gazeR conforms to best practices for pupillometry laid out in recent reviews (Winn et al., 2018; Mathot et al., 2018) makes gazeR a valuable tool for analyzing your pupillometry and gaze data.
Limitations
The gazeR pre-possessing pipeline is not exhaustive. The implemented set of functions should suffice for researchers to pre-process their gaze and pupil data based on recent recommendations, but there are factors that are not yet included. For example, gaze position is known to influence pupil size (Brisson et al., 2013; Gagl, Hawelka, & Hutzler, 2011), called the pupil foreshortening effect. This effect occurs when rotations of the eyes change the angle at which the camera records the pupil, and therefore also the pupil’s apparent size. As such, this manifestation of gaze position in pupil size should ideally be controlled or corrected for. A simple way to do this would be to include X and Y gaze coordinates into the analysis model as a co-variate. If reading EDF files, the X-Y gaze coordinates are included and can easily be included in this analysis. Additionally, various aspects of pupil dilation might be more or less important to the analysis, which might benefit from examination of additional features such as onset and offset slopes (c.f., Winn & Moore, 2018). Because the gazeR package is open-source, modifications can always be made to incorporate additional functionality. Suggestions and contributions from users are encouraged and can be submitted through the package github page: https://github.com/dmirman/gazer.
Finally, the current instantiation of gazeR has only been tested with data files from the SR EyeLink (i.e., raw EDFs and sample reports). Much of the gazeR functionality is easily compatible with other eye-trackers with the addition of functions for reading data and renaming columns (variables) to match the gazeR conventions. Future updates to the package will include added support for Tobii and Gazepoint eye trackers.
To summarize, the gazeR package provides general, open-source tools for replicable and transparent processing of gaze and pupillometry data. GazeR grew out of in-house preprocessing code in several research groups and is already being used by several additional research groups. It is our hope that more researchers will use it and will contribute to its improvement.
Notes
Although not necessary, some EyeLink users nevertheless find it convenient to use the Fixation Reports or Sample Reports generated by DataViewer. A walkthrough for Sample and Fixation reports can be found here: https://psyarxiv.com/gvcxb/
A comparison of results using the blink detection algorithm in the saccades package and the SR-EyeLink algorithm resulted in negligible differences, at least on grand averaged data. However, more extensive testing should be done.
Reilly et al. varied luminance in order to elicit different baseline sizes, but that is not the typical source of baseline pupil size differences. Tonic baseline pupil size differences due to arousal, age, or other variables may affect the range of dilation reactivity in ways that differ from changes that are elicited by changes in luminance. Additionally, Wang et al. (2018) suggested that brighter lighting condition elicit larger dilations, on account of suppression of the parasympathetic suppressive influence on dilations. These factors can be used to motivate divisive baseline correction.
References
Attard-Johnson, J., CÓ Ciardha, & Bindemann, M. (2019). Comparing methods for the analysis of pupillary response. Behavior Research Methods, 51(1), 83–95. https://doi.org/10.3758/s13428-018-1108-6
Barnhart, A. S., & Goldinger, S. D. (2010). Interpreting chicken-scratch: Lexical access for handwritten words. Journal of Experimental Psychology: Human Perception and Performance, 36(4), 906–923. https://doi.org/10.1037/a0019258
Beatty, J. (1982). Task-evoked pupillary responses, processing load, and the structure of processing resources. Psychological Bulletin, 91(2), 276–292. https://doi.org/10.1037/0033-2909.91.2.276
Bradley, M. M., Miccoli, L., Escrig, M. A., & Lang, P. J. (2008). The pupil as a measure of emotional arousal and autonomic activation. Psychophysiology, 45(4), 602–607. https://doi.org/10.1111/j.1469-8986.2008.00654.x
Brisson, J., Mainville, M., Mailloux, D., Beaulieu, C., Serres, J., & Sirois, S. (2013). Pupil diameter measurement errors as a function of gaze direction in corneal reflection eyetrackers. Behavior Research Methods, 45(4), 1322–1331. https://doi.org/10.3758/s13428-013-0327-0
Cohen, N., Moyal, N., & Henik, A. (2015). Executive control suppresses pupillary responses to aversive stimuli. Biological Psychology, 112, 1–11. https://doi.org/10.1016/j.biopsycho.2015.09.006
Cooper, R. M. (1974). The control of eye fixation by the meaning of spoken language: A new methodology for the real-time investigation of speech perception, memory, and language processing. Cognitive Psychology, 6(1), 84–107. https://doi.org/10.1016/0010-0285(74)90005-X
Einhauser, W., Stout, J., Koch, C., & Carter, O. (2008). Pupil dilation reflects perceptual selection and predicts subsequent stability in perceptual rivalry. Proceedings of the National Academy of Sciences, 105(5), 1704–1709. https://doi.org/10.1073/pnas.0707727105
Engbert, R., & Kliegl, R. (2003). Microsaccades uncover the orientation of covert attention. Vision Research, 43(9), 1035–1045. https://doi.org/10.1016/S0042-6989(03)00084-1
Esteban, O., Markiewicz, C. J., Blair, R. W., Moodie, C. A., Isik, A. I., Erramuzpe, A., … Gorgolewski, K. J. (2019). fMRIPrep: a robust preprocessing pipeline for functional MRI. Nature Methods, 16(1), 111–116. https://doi.org/10.1038/s41592-018-0235-4
Forbes, S.H. (2019). pupillometryR: An R package for preparing and analysing pupillometry data. Retrieved from https://github.com/samhforbes/PupillometryR
Gagl, B., Hawelka, S., & Hutzler, F. (2011). Systematic influence of gaze position on pupil size measurement: analysis and correction. Behavior Research Methods, 43(4), 1171–1181. https://doi.org/10.3758/s13428-011-0109-5
Geller, J., Still, M. L., & Morris, A. L. (2016). Eyes wide open: Pupil size as a proxy for inhibition in the masked-priming paradigm. Memory & Cognition, 44(4), 554–564. https://doi.org/10.3758/s13421-015-0577-4
Geller, J., Still, M. L., Dark, V. J., & Carpenter, S. K. (2018). Would disfluency by any other name still be disfluent? Examining the disfluency effect with cursive handwriting. Memory & Cognition, 46(7), 1109–1126. https://doi.org/10.3758/s13421-018-0824-6
Geller, J., Landrigan, J.-F., & Mirman, D. (2019). A Pupillometric Examination of Cognitive Control in Taxonomic and Thematic Semantic Memory. Journal of Cognition, 2(1). https://doi.org/10.5334/joc.56
Goldinger, S. D., He, Y., & Papesh, M. H. (2009). Deficits in cross-race face learning: Insights from eye movements and pupillometry. Journal of Experimental Psychology: Learning, Memory, and Cognition, 35(5), 1105–1122. https://doi.org/10.1037/a0016548
Grange, J.A. (2015). trimr: An implementation of common response time trimming methods. R package version 1.0.1. https://cran.r-project.org/web/packages/trimr/index.html
Granholm, E., Asarnow, R. F., Sarkin, A. J., & Dykes, K. L. (1996). Pupillary responses index cognitive resource limitations. Psychophysiology, 33(4), 457–461. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/8753946
Hampel, F. R. (1974). The influence curve and its role in robust estimation. Journal of the American Statistical Association, 69(346), 383–393. https://doi.org/10.1080/01621459.1974.10482962
Hershman, R., Henik, A., & Cohen, N. (2018). A novel blink detection method based on pupillometry noise. Behavior Research Methods, 50(1), 107–114. https://doi.org/10.3758/s13428-017-1008-1
Hubbard, J., & von der Malsburg, T. (2015). edfR: Basic functions for importing SR-Research Eyelink EDF files. R package version 0.1
Jackson, I., & Sirois, S. (2009). Infant cognition: going full factorial with pupil dilation. Developmental Science, 12(4), 670–679. https://doi.org/10.1111/j.1467-7687.2008.00805.x
Kang, O., & Wheatley, T. (2015). Pupil dilation patterns reflect the contents of consciousness. Consciousness and Cognition, 35, 128–135. https://doi.org/10.1016/j.concog.2015.05.001
Karatekin, C., Couperus, J. W., & Marcus, D. J. (2004). Attention allocation in the dual-task paradigm as measured through behavioral and psychophysiological responses. Psychophysiology, 41(2), 175–185. https://doi.org/10.1111/j.1469-8986.2004.00147.x
Kret, M. E., & Sjak-Shie, E. E. (2018). Preprocessing pupil size data: Guidelines and code. Behavior Research Methods, 1–7. https://doi.org/10.3758/s13428-018-1075-y
Laeng, B., Sirois, S., & Gredebäck, G. (2012). Pupillometry. Perspectives on Psychological Science, 7(1), 18–27. https://doi.org/10.1177/1745691611427305
Mathôt, S. (2018). Pupillometry: Psychology, Physiology, and Function. Journal of Cognition, 1(1). https://doi.org/10.5334/joc.18
Mathôt, S., van der Linden, L., Grainger, J., & Vitu, F. (2013). The Pupillary Light Response Reveals the Focus of Covert Visual Attention. PLoS ONE, 8(10), e78168. https://doi.org/10.1371/journal.pone.0078168
Mathôt, S., Fabius, J., Van Heusden, E., & Van der Stigchel, S. (2018). Safe and sensible preprocessing and baseline correction of pupil-size data. Behavior Research Methods, 50(1), 94–106. https://doi.org/10.3758/s13428-017-1007-2
Mirman, D. (2014). Growth Curve Analysis and Visualization Using R. Boca Raton, FL: Chapman and Hall/CRC Press.
Mirman, D., & Graziano, K. M. (2012). Individual differences in the strength of taxonomic versus thematic relations. Journal of Experimental Psychology: General, 141(4), 601–609. https://doi.org/10.1037/a0026451
Murphy, P. R., O'connell, R. G., O'sullivan, M., Robertson, I. H., & Balsters, J. H. (2014). Pupil diameter covaries with BOLD activity in human locus coeruleus. Human Brain Mapping, 35(8), 4140-4154.
Nyström, M., Hooge, I., & Andersson, R. (2016). Pupil size influences the eye-tracker signal during saccades. Vision Research, 121, 95–103. https://doi.org/10.1016/J.VISRES.2016.01.009
Piquado, T., Isaacowitz, D., & Wingfield, A. (2010). Pupillometry as a measure of cognitive effort in younger and older adults. Psychophysiology, 47(3), 560–569. https://doi.org/10.1111/j.1469-8986.2009.00947.x
R Core Team. (2019). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL http://www.R-project.org/.
Reilly, J., Kelly, A., Kim, S. H., Jett, S., & Zuckerman, B. (2018). The human task-evoked pupillary response function is linear: Implications for baseline response scaling in pupillometry. Behavior Research Methods. https://doi.org/10.3758/s13428-018-1134-4
Salverda, A. P., & Tanenhaus, M. K. (2018). The visual world paradigm. In Annette M. B. de Groot and Peter Hagoort (Eds) Research methods in psycholinguistics and the neurobiology of language: A practical guide, pp. 89-110. Wiley Blackwell.
Satterthwaite, T. D., Green, L., Myerson, J., Parker, J., Ramaratnam, M., & Buckner, R. L. (2007). Dissociable but inter-related systems of cognitive control and reward during decision making: Evidence from pupillometry and event-related fMRI. NeuroImage, 37(3), 1017–1031. https://doi.org/10.1016/j.neuroimage.2007.04.066
Seedorff, M., Oleson, J., & McMurray, B. (2018). Detecting when timeseries differ: Using the Bootstrapped Differences of Timeseries (BDOTS) to analyze Visual World Paradigm data (and more). Journal of Memory and Language, 102, 55–67. https://doi.org/10.1016/J.JML.2018.05.004
Siegle, G. J., Steinhauer, S. R., Carter, C. S., Ramel, W., & Thase, M. E. (2003). Do the Seconds Turn Into Hours? Relationships between Sustained Pupil Dilation in Response to Emotional Information and Self-Reported Rumination. Cognitive Therapy and Research, 27(3), 365–382. https://doi.org/10.1023/A:1023974602357
Tanenhaus, M. K., Spivey-Knowlton, M. J., Eberhard, K. M., & Sedivy, J. C. (1995). Integration of visual and linguistic information in spoken language comprehension. Science (New York, N.Y.), 268(5217), 1632–1634. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/7777863
Tsukahara, J.S. (2018). pupillometry: An R Package to Preprocess Pupil Data. Retrieved from https://dr-jt.github.io/pupillometry
Van Gerven, P. W. M., Paas, F., Van Merriënboer, J. J. G., & Schmidt, H. G. (2004). Memory load and the cognitive pupillary response in aging. Psychophysiology, 41(2), 167–174. https://doi.org/10.1111/j.1469-8986.2003.00148.x
van Rij, J., Hendriks, P., van Rijn, H., Baayen, R. H., & Wood, S. N. (2019). Analyzing the Time Course of Pupillometric Data. Trends in Hearing, 23, 233121651983248. https://doi.org/10.1177/2331216519832483
von der Malsburg, T. (2019). Saccades: Detection of fixations in eye-tracking data. Retrieved from https://github.com/tmalsburg/saccades
Wang, Y., Kramer, S. E., Wendt, D., Naylor, G., Lunner, T., & Zekveld, A. A. (2018). The pupil dilation response during speech perception in dark and light: The involvement of the parasympathetic nervous system in listening effort. Trends in Hearing, 22. https://doi.org/10.1177/2331216518816603
Winn, M.B., (2016). Rapid release from listening effort resulting from semantic context, and effects of spectral degradation and cochlear implants. Trends in Hearing, 20, 1–17. https://doi.org/10.1177/2331216516669723
Winn, M., Moore, A. (2018). Pupillometry reveals that context benefit in speech perception can be disrupted by later-occurring sounds, especially in listeners with cochlear implants. Trends in Hearing, 22:2331216518808962. https://doi.org/10.1177/2331216518808962
Winn, M. B., Wendt, D., Koelewijn, T., & Kuchinsky, S. E. (2018). Best Practices and Advice for Using Pupillometry to Measure Listening Effort: An Introduction for Those Who Want to Get Started. Trends in Hearing, 22, 2331216518800869. https://doi.org/10.1177/2331216518800869
Zekveld, A. A., Koelewijn, T., & Kramer, S. E. (2018). The pupil dilation response to auditory stimuli: current state of knowledge. Trends in Hearing, 22. https://doi.org/10.1177/2331216518777174
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 48 kb)
Rights and permissions
About this article
Cite this article
Geller, J., Winn, M.B., Mahr, T. et al. GazeR: A Package for Processing Gaze Position and Pupil Size Data. Behav Res 52, 2232–2255 (2020). https://doi.org/10.3758/s13428-020-01374-8
Published:
Issue Date:
DOI: https://doi.org/10.3758/s13428-020-01374-8