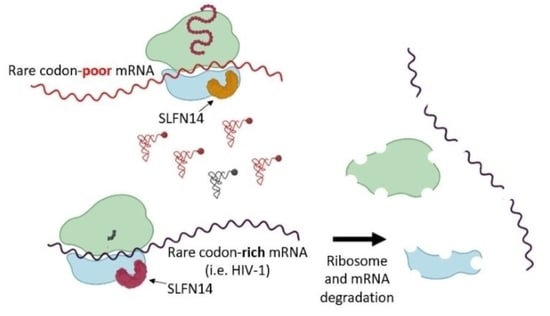
Schlafen14 Impairs HIV-1 Expression in a Codon Usage-Dependent Manner
Abstract
:1. Introduction
2. Materials and Methods
3. Results
3.1. Human and Mouse SLFN14 Preferentially Impairs the Expression of Proteins Enriched in Rare Codons
3.2. Expression of SLFN14 in Cells of the Immune System
3.3. SLFN14 Requires the Endoribonuclease Activity to Impair HIV-1 Expression
3.4. SLFN14 Impairs HIV-1 Protein Expression in CD4+ T Cells and Monocytes
3.5. Effect of SLFN14 on Viral Infection
3.6. SLFN14 Caused Ribosomal RNA Degradation in Cells Co-Expressing Gag Wild Type
3.7. Subcellular Distribution of SLFN14
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Ghaemmaghami, S.; Huh, W.-K.; Bower, K.; Howson, R.W.; Belle, A.; Dephoure, N.; O’Shea, E.K.; Weissman, J.S. Global analysis of protein expression in yeast. Nature 2003, 425, 737–741. [Google Scholar] [CrossRef] [PubMed]
- Brockmann, R.; Beyer, A.; Heinisch, J.J.; Wilhelm, T. Posttranscriptional expression regulation: What determines translation rates? PLoS Comput. Biol. 2007, 3, e57. [Google Scholar] [CrossRef] [PubMed]
- Tuller, T.; Kupiec, M.; Ruppin, E. Determinants of protein abundance and translation efficiency in S. cerevisiae. PLoS Comput. Biol. 2007, 3, e248. [Google Scholar] [CrossRef]
- Ishihama, Y.; Schmidt, T.; Rappsilber, J.; Mann, M.; Hartl, F.U.; Kerner, M.J.; Frishman, D. Protein abundance profiling of the Escherichia coli cytosol. BMC Genom. 2008, 9, 102. [Google Scholar] [CrossRef]
- Tuller, T.; Waldman, Y.Y.; Kupiec, M.; Ruppin, E. Translation efficiency is determined by both codon bias and folding energy. Proc. Natl. Acad. Sci. USA 2010, 107, 3645–3650. [Google Scholar] [CrossRef]
- Hia, F.; Takeuchi, O. The effects of codon bias and optimality on mRNA and protein regulation. Cell Mol. Life Sci. 2021, 78, 1909–1928. [Google Scholar] [CrossRef]
- Hanson, G.; Coller, J. Codon optimality, bias and usage in translation and mRNA decay. Nat. Rev. Mol. Cell Biol. 2018, 19, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Carlini, D.B.; Stephan, W. In vivo introduction of unpreferred synonymous codons into the Drosophila Adh gene results in reduced levels of ADH protein. Genetics 2003, 163, 239–243. [Google Scholar] [CrossRef]
- Pershing, N.L.; Lampson, B.L.; Belsky, J.A.; Kaltenbrun, E.; MacAlpine, D.M.; Counter, C.M. Rare codons capacitate Kras-driven de novo tumorigenesis. J. Clin. Investig. 2015, 125, 222–233. [Google Scholar] [CrossRef]
- Kotsopoulou, E.; Kim, V.N.; Kingsman, A.J.; Kingsman, S.M.; Mitrophanous, K.A. A Rev-independent human immunodeficiency virus type 1 (HIV-1)-based vector that exploits a codon-optimized HIV-1 gag-pol gene. J. Virol. 2000, 74, 4839–4852. [Google Scholar] [CrossRef]
- Peterson, J.; Li, S.; Kaltenbrun, E.; Erdogan, O.; Counter, C.M. Expression of transgenes enriched in rare codons is enhanced by the MAPK pathway. Sci. Rep. 2020, 10, 22166. [Google Scholar] [CrossRef]
- Yang, Q.; Yu, C.-H.; Zhao, F.; Dang, Y.; Wu, C.; Xie, P.; Sachs, M.S.; Liu, Y. eRF1 mediates codon usage effects on mRNA translation efficiency through premature termination at rare codons. Nucleic Acids Res. 2019, 47, 9243–9258. [Google Scholar] [CrossRef]
- Wu, C.C.; Zinshteyn, B.; Wehner, K.A.; Green, R. High-Resolution Ribosome Profiling Defines Discrete Ribosome Elongation States and Translational Regulation during Cellular Stress. Mol. Cell 2019, 73, 959–970.e5. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, D.E.; Shah, P.; Eichhorn, S.W.; Hussmann, J.A.; Plotkin, J.B.; Bartel, D.P. Improved Ribosome-Footprint and mRNA Measurements Provide Insights into Dynamics and Regulation of Yeast Translation. Cell Rep. 2016, 14, 1787–1799. [Google Scholar] [CrossRef] [PubMed]
- Hussmann, J.A.; Patchett, S.; Johnson, A.; Sawyer, S.; Press, W.H. Understanding Biases in Ribosome Profiling Experiments Reveals Signatures of Translation Dynamics in Yeast. PLoS Genet. 2015, 11, e1005732. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Hoek, T.A.; Vale, R.D.; Tanenbaum, M.E. Dynamics of Translation of Single mRNA Molecules In Vivo. Cell 2016, 165, 976–989. [Google Scholar] [CrossRef] [PubMed]
- Darnell, A.M.; Subramaniam, A.R.; O’Shea, E.K. Translational Control through Differential Ribosome Pausing during Amino Acid Limitation in Mammalian Cells. Mol. Cell 2018, 71, 229–243.e11. [Google Scholar] [CrossRef] [PubMed]
- Joazeiro, C.A.P. Mechanisms and functions of ribosome-associated protein quality control. Nat. Rev. Mol. Cell Biol. 2019, 20, 368–383. [Google Scholar] [CrossRef] [PubMed]
- De, S.; Muhlemann, O. A comprehensive coverage insurance for cells: Revealing links between ribosome collisions, stress responses and mRNA surveillance. RNA Biol. 2022, 19, 609–621. [Google Scholar] [CrossRef] [PubMed]
- Presnyak, V.; Alhusaini, N.; Chen, Y.-H.; Martin, S.; Morris, N.; Kline, N.; Olson, S.; Weinberg, D.; Baker, K.E.; Graveley, B.R.; et al. Codon optimality is a major determinant of mRNA stability. Cell 2015, 160, 1111–1124. [Google Scholar] [CrossRef]
- Letzring, D.P.; Dean, K.M.; Grayhack, E.J. Control of translation efficiency in yeast by codon-anticodon interactions. RNA 2010, 16, 2516–2528. [Google Scholar] [CrossRef]
- Chu, D.; Kazana, E.; Bellanger, N.; Singh, T.; Tuite, M.F.; von der Haar, T. Translation elongation can control translation initiation on eukaryotic mRNAs. EMBO J. 2014, 33, 21–34. [Google Scholar] [CrossRef]
- Torrent, M.; Chalancon, G.; de Groot, N.S.; Wuster, A.; Madan Babu, M. Cells alter their tRNA abundance to selectively regulate protein synthesis during stress conditions. Sci. Signal. 2018, 11, eaat6409. [Google Scholar] [CrossRef]
- Pang, Y.L.; Abo, R.; Levine, S.S.; Dedon, P.C. Diverse cell stresses induce unique patterns of tRNA up- and down-regulation: tRNA-seq for quantifying changes in tRNA copy number. Nucleic Acids Res. 2014, 42, e170. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.L.; Chen, G.; Wilke, C.O.; Krug, R.M. Avian Influenza Virus PB1 Gene in H3N2 Viruses Evolved in Humans To Reduce Interferon Inhibition by Skewing Codon Usage toward Interferon-Altered tRNA Pools. mBio 2018, 9, e01222-18. [Google Scholar] [CrossRef] [PubMed]
- Saikia, M.; Wang, X.; Mao, Y.; Wan, J.; Pan, T.; Qian, S.B. Codon optimality controls differential mRNA translation during amino acid starvation. RNA 2016, 22, 1719–1727. [Google Scholar] [CrossRef]
- Yue, T.; Zhan, X.; Zhang, D.; Jain, R.; Wang, K.-W.; Choi, J.H.; Misawa, T.; Su, L.; Quan, J.; Hildebrand, S.; et al. SLFN2 protection of tRNAs from stress-induced cleavage is essential for T cell-mediated immunity. Science 2021, 372, eaba4220. [Google Scholar] [CrossRef]
- Chan, C.T.; Dyavaiah, M.; DeMott, M.S.; Taghizadeh, K.; Dedon, P.C.; Begley, T.J. A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet. 2010, 6, e1001247. [Google Scholar] [CrossRef]
- Chionh, Y.H.; McBee, M.; Babu, I.R.; Hia, F.; Lin, W.; Zhao, W.; Cao, J.; Dziergowska, A.; Malkiewicz, A.; Begley, T.J.; et al. tRNA-mediated codon-biased translation in mycobacterial hypoxic persistence. Nat. Commun. 2016, 7, 13302. [Google Scholar] [CrossRef]
- Patil, A.; Dyavaiah, M.; Joseph, F.; Rooney, J.P.; Chan, C.T.Y.; Dedon, P.C.; Begley, T.J. Increased tRNA modification and gene-specific codon usage regulate cell cycle progression during the DNA damage response. Cell Cycle 2012, 11, 3656–3665. [Google Scholar] [CrossRef]
- Li, M.; Kao, E.; Gao, X.; Sandig, H.; Limmer, K.; Pavon-Eternod, M.; Jones, T.E.; Landry, S.; Pan, T.; Weitzman, M.D.; et al. Codon-usage-based inhibition of HIV protein synthesis by human schlafen 11. Nature 2012, 491, 125–128. [Google Scholar] [CrossRef]
- Valdez, F.; Salvador, J.; Palermo, P.M.; Mohl, J.E.; Hanley, K.A.; Watts, D.; Llano, M. Schlafen 11 Restricts Flavivirus Replication. J. Virol. 2019, 93, e00104-19. [Google Scholar] [CrossRef]
- Pavon-Eternod, M.; Wei, M.; Pan, T.; Kleiman, L. Profiling non-lysyl tRNAs in HIV-1. RNA 2010, 16, 267–273. [Google Scholar] [CrossRef]
- van Weringh, A.; Ragonnet-Cronin, M.; Pranckeviciene, E.; Pavon-Eternod, M.; Kleiman, L.; Xia, X. HIV-1 modulates the tRNA pool to improve translation efficiency. Mol. Biol. Evol. 2011, 28, 1827–1834. [Google Scholar] [CrossRef] [PubMed]
- Jungfleisch, J.; Böttcher, R.; Talló-Parra, M.; Pérez-Vilaró, G.; Merits, A.; Novoa, E.M.; Díez, J. CHIKV infection reprograms codon optimality to favor viral RNA translation by altering the tRNA epitranscriptome. Nat. Commun. 2022, 13, 4725. [Google Scholar] [CrossRef] [PubMed]
- Stabell, A.C.; Hawkins, J.; Li, M.; Gao, X.; David, M.; Press, W.H.; Sawyer, S.L. Non-human Primate Schlafen11 Inhibits Production of Both Host and Viral Proteins. PLoS Pathog. 2016, 12, e1006066. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Kao, E.; Malone, D.; Gao, X.; Wang, J.Y.J.; David, M. DNA damage-induced cell death relies on SLFN11-dependent cleavage of distinct type II tRNAs. Nat. Struct. Mol. Biol. 2018, 25, 1047–1058. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.-Y.; Deng, X.-Y.; Li, Y.-S.; Ma, X.-C.; Feng, J.-X.; Yu, B.; Chen, Y.; Luo, Y.-L.; Wang, X.; Chen, M.-L.; et al. Structure of Schlafen13 reveals a new class of tRNA/rRNA- targeting RNase engaged in translational control. Nat. Commun. 2018, 9, 1165. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Hoyt, S.; Wu, X.; Garvie, C.; McGaunn, J.; Shekhar, M.; Tötzl, M.; Rees, M.G.; Cherniack, A.D.; Meyerson, M.; et al. Velcrin-induced selective cleavage of tRNA(Leu)(TAA) by SLFN12 causes cancer cell death. Nat. Chem. Biol. 2023, 19, 301–310. [Google Scholar] [CrossRef]
- Metzner, F.J.; Huber, E.; Hopfner, K.P.; Lammens, K. Structural and biochemical characterization of human Schlafen 5. Nucleic Acids Res. 2022, 50, 1147–1161. [Google Scholar] [CrossRef]
- Fletcher, S.J.; Pisareva, V.P.; Khan, A.O.; Tcherepanov, A.; Morgan, N.V.; Pisarev, A.V. Role of the novel endoribonuclease SLFN14 and its disease-causing mutations in ribosomal degradation. RNA 2018, 24, 939–949. [Google Scholar] [CrossRef] [PubMed]
- Pisareva, V.P.; Muslimov, I.A.; Tcherepanov, A.; Pisarev, A.V. Characterization of Novel Ribosome-Associated Endoribonuclease SLFN14 from Rabbit Reticulocytes. Biochemistry 2015, 54, 3286–3301. [Google Scholar] [CrossRef] [PubMed]
- Stapley, R.J.; Smith, C.W.; Haining, E.J.; Bacon, A.; Lax, S.; Pisareva, V.P.; Pisarev, A.V.; Watson, S.P.; Khan, A.O.; Morgan, N.V. Heterozygous mutation SLFN14 K208N in mice mediates species-specific differences in platelet and erythroid lineage commitment. Blood Adv. 2021, 5, 377–390. [Google Scholar] [CrossRef]
- Mao, Y.; Ding, J.W.; Chen, M.S.; Cen, S.; Li, X.Y. SLFN14 inhibits LINE-1 transposition activity. Yi Chuan 2020, 42, 669–679. [Google Scholar] [CrossRef] [PubMed]
- Seong, R.-K.; Seo, S.-W.; Kim, J.-A.; Fletcher, S.J.; Morgan, N.V.; Kumar, M.; Choi, Y.-K.; Shin, O.S. Schlafen 14 (SLFN14) is a novel antiviral factor involved in the control of viral replication. Immunobiology 2017, 222, 979–988. [Google Scholar] [CrossRef]
- Sharp, P.M.; Li, W.H. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987, 15, 1281–1295. [Google Scholar] [CrossRef]
- Pandit, A.; Sinha, S. Differential trends in the codon usage patterns in HIV-1 genes. PLoS ONE 2011, 6, e28889. [Google Scholar] [CrossRef]
- Martrus, G.; Nevot, M.; Andres, C.; Clotet, B.; Martinez, M.A. Changes in codon-pair bias of human immunodeficiency virus type 1 have profound effects on virus replication in cell culture. Retrovirology 2013, 10, 78. [Google Scholar] [CrossRef]
- Vidyavijayan, K.K.; Hassan, S.; Precilla, L.K.; Ashokkumar, M.; Chandrasekeran, P.; Swaminathan, S.; Hanna, L.E. Biased Nucleotide Composition and Differential Codon Usage Pattern in HIV-1 and HIV-2. AIDS Res. Hum. Retroviruses 2017, 33, 298–307. [Google Scholar] [CrossRef]
- Llano, M.; Saenz, D.T.; Meehan, A.; Wongthida, P.; Peretz, M.; Walker, W.H.; Teo, W.; Poeschla, E.M. An essential role for LEDGF/p75 in HIV integration. Science 2006, 314, 461–464. [Google Scholar] [CrossRef]
- Haas, J.; Park, E.C.; Seed, B. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr. Biol. 1996, 6, 315–324. [Google Scholar] [CrossRef]
- Kutsch, O.; Benveniste, E.N.; Shaw, G.M.; Levy, D.N. Direct and quantitative single-cell analysis of human immunodeficiency virus type 1 reactivation from latency. J. Virol. 2002, 76, 8776–8786. [Google Scholar] [CrossRef]
- Bueno, M.T.D.; Reyes, D.; Llano, M. LEDGF/p75 Deficiency Increases Deletions at the HIV-1 cDNA Ends. Viruses 2017, 9, 259. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Rivera, J.A.; Bueno, M.T.; Morales, E.; Kugelman, J.R.; Rodriguez, D.F.; Llano, M. Implication of serine residues 271, 273, and 275 in the human immunodeficiency virus type 1 cofactor activity of lens epithelium-derived growth factor/p75. J. Virol. 2010, 84, 740–752. [Google Scholar] [CrossRef]
- Llano, M.; Vanegas, M.; Hutchins, N.; Thompson, D.; Delgado, S.; Poeschla, E.M. Identification and characterization of the chromatin-binding domains of the HIV-1 integrase interactor LEDGF/p75. J. Mol. Biol. 2006, 360, 760–773. [Google Scholar] [CrossRef]
- Gelderblom, H.C.; Vatakis, D.N.; Burke, S.A.; Lawrie, S.D.; Bristol, G.C.; Levy, D.N. Viral complementation allows HIV-1 replication without integration. Retrovirology 2008, 5, 60. [Google Scholar] [CrossRef]
- Lee, S.; Trivedi, U.; Johnson, C.; Farquharson, C.; Bergkvist, G.T. Optimised isolation method for RNA extraction suitable for RNA sequencing from feline teeth collected in a clinical setting and at post mortem. Vet. Res. Commun. 2019, 43, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Puigbo, P.; Bravo, I.G.; Garcia-Vallve, S. CAIcal: A combined set of tools to assess codon usage adaptation. Biol. Direct. 2008, 3, 38. [Google Scholar] [CrossRef]
- Jeon, J.S.; Won, Y.H.; Kim, I.K.; Ahn, J.H.; Shin, O.S.; Kim, J.H.; Lee, C.H. Analysis of single nucleotide polymorphism among Varicella-Zoster Virus and identification of vaccine-specific sites. Virology 2016, 496, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Yamasoba, D.; Sato, K.; Ichinose, T.; Imamura, T.; Koepke, L.; Joas, S.; Reith, E.; Hotter, D.; Misawa, N.; Akaki, K.; et al. N4BP1 restricts HIV-1 and its inactivation by MALT1 promotes viral reactivation. Nat. Microbiol. 2019, 4, 1532–1544. [Google Scholar] [CrossRef]
- Mavrommatis, E.; Fish, E.N.; Platanias, L.C. The schlafen family of proteins and their regulation by interferons. J. Interferon. Cytokine Res. 2013, 33, 206–210. [Google Scholar] [CrossRef]
- Chiu, Y.H.; Macmillan, J.B.; Chen, Z.J. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 2009, 138, 576–591. [Google Scholar] [CrossRef]
- Ran, Y.; Li, D.; Xiong, M.-G.; Liu, H.-N.; Feng, T.; Shi, Z.-W.; Li, Y.-H.; Wu, H.-N.; Wang, S.-Y.; Zheng, H.-X.; et al. African swine fever virus I267L acts as an important virulence factor by inhibiting RNA polymerase III-RIG-I-mediated innate immunity. PLoS Pathog. 2022, 18, e1010270. [Google Scholar] [CrossRef]
- van der Kuyl, A.C.; Berkhout, B. The biased nucleotide composition of the HIV genome: A constant factor in a highly variable virus. Retrovirology 2012, 9, 92. [Google Scholar] [CrossRef] [PubMed]
- Chesebro, B.; Wehrly, K.; Metcalf, J.; Griffin, D.E. Use of a new CD4-positive HeLa cell clone for direct quantitation of infectious human immunodeficiency virus from blood cells of AIDS patients. J. Infect. Dis. 1991, 163, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Maddon, P.J.; Dalgleish, A.G.; McDougal, J.S.; Clapham, P.R.; Weiss, R.A.; Axel, R. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell 1986, 47, 333–348. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.; Shi, J.; Márquez, C.L.; Lau, D.; Walsh, J.; Faysal, K.M.R.; Byeon, C.H.; Byeon, I.-J.L.; Aiken, C.; Böcking, T. Functional analysis of the secondary HIV-1 capsid binding site in the host protein cyclophilin A. Retrovirology 2019, 16, 10. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, A.; Mueller, O.; Stocker, S.; Salowsky, R.; Leiber, M.; Gassmann, M.; Lightfoot, S.; Menzel, W.; Granzow, M.; Ragg, T. The RIN: An RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 2006, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Bustos, O.; Naik, S.; Ayers, G.; Casola, C.; Perez-Lamigueiro, M.A.; Chippindale, P.T.; Pritham, E.J.; de la Casa-Esperón, E. Evolution of the Schlafen genes, a gene family associated with embryonic lethality, meiotic drive, immune processes and orthopoxvirus virulence. Gene 2009, 447, 1–11. [Google Scholar] [CrossRef] [PubMed]
- de la Casa-Esperon, E. From mammals to viruses: The Schlafen genes in developmental, proliferative and immune processes. Biomol. Concepts 2011, 2, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi-Ishihara, M.; Smutná, K.F.; Alonso, F.E.; Argilaguet, J.; Esteve-Codina, A.; Geiger, K.; Genescà, M.; Grau-Expósito, J.; Duran-Castells, C.; Rogenmoser, S.; et al. Schlafen 12 restricts HIV-1 latency reversal by a codon-usage dependent post-transcriptional block in CD4+ T cells. Commun. Biol. 2023, 6, 487. [Google Scholar] [CrossRef] [PubMed]
- Frenkel-Morgenstern, M.; Danon, T.; Christian, T.; Igarashi, T.; Cohen, L.; Hou, Y.; Jensen, L.J. Genes adopt non-optimal codon usage to generate cell cycle-dependent oscillations in protein levels. Mol. Syst. Biol. 2012, 8, 572. [Google Scholar] [CrossRef] [PubMed]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valenzuela, C.; Saucedo, S.; Llano, M. Schlafen14 Impairs HIV-1 Expression in a Codon Usage-Dependent Manner. Viruses 2024, 16, 502. https://doi.org/10.3390/v16040502
Valenzuela C, Saucedo S, Llano M. Schlafen14 Impairs HIV-1 Expression in a Codon Usage-Dependent Manner. Viruses. 2024; 16(4):502. https://doi.org/10.3390/v16040502
Chicago/Turabian StyleValenzuela, Carlos, Sergio Saucedo, and Manuel Llano. 2024. "Schlafen14 Impairs HIV-1 Expression in a Codon Usage-Dependent Manner" Viruses 16, no. 4: 502. https://doi.org/10.3390/v16040502