Unveiling CRESS DNA Virus Diversity in Oysters by Virome
Abstract
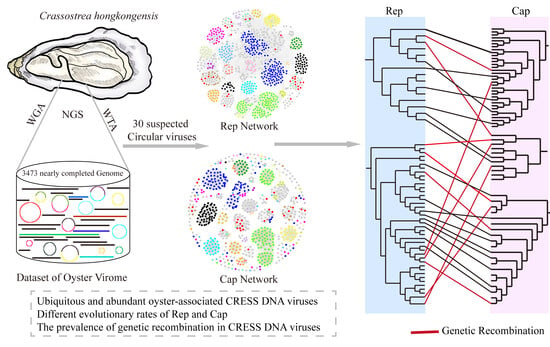
:1. Introduction
2. Materials and Methods
2.1. Sequence Assembling and Virus Discovery
2.2. Open Reading Frame (ORF) Prediction and Annotation
2.3. Similarity Clustering Analysis
2.4. Phylogenetic Tree Construction Based on Rep and Cap
2.5. Analysis of the Abundance of Viruses
3. Results
3.1. Ubiquitous and Abundant Oyster-Associated CRESS DNA Viruses
3.2. Different Evolutionary Rates of Rep and Cap
3.3. The Prevalence of Genetic Recombination in CRESS DNA Viruses
3.4. A Large Number of Unclassified CRESS DNA Viruses in Oysters from Yangjiang, China
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Powell, D.; Subramanian, S.; Suwansa-ard, S.; Zhao, M.; O’Connor, W.; Raftos, D.; Elizur, A. The genome of the oyster Saccostrea offers insight into the environmental resilience of bivalves. DNA Res. 2018, 25, 655–665. [Google Scholar] [CrossRef]
- Olalemi, A.; Baker-Austin, C.; Ebdon, J.; Taylor, H. Bioaccumulation and persistence of faecal bacterial and viral indicators in Mytilus edulis and Crassostrea gigas. Int. J. Hyg. Environ. Health 2016, 219, 592–598. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Fang, X.; Guo, X.; Li, L.; Luo, R.; Xu, F.; Yang, P.; Zhang, L.; Wang, X.; Qi, H.; et al. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 2012, 490, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, Y.; Zhang, Y.; Mao, F.; Xiang, Z.; Xiao, S.; Ma, H.; Yu, Z. The first invertebrate NFIL3 transcription factor with role in immune defense identified from the Hong Kong oyster, Crassostrea hongkongensis. Dev. Comp. Immunol. 2017, 76, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Farley, C.A.; Banfield, W.G.; Kasnic, G., Jr.; Foster, W.S. Oyster herpes-type virus. Science 1972, 178, 759–760. [Google Scholar] [CrossRef]
- Renault, T.; Novoa, B. Viruses infecting bivalve molluscs. Aquat. Living Resour. 2004, 17, 397–409. [Google Scholar] [CrossRef]
- Delmotte, J.; Pelletier, C.; Morga, B.; Galinier, R.; Petton, B.; Lamy, J.B.; Kaltz, O.; Avarre, J.C.; Jacquot, M.; Montagnani, C.; et al. Genetic diversity and connectivity of the Ostreid herpesvirus 1 populations in France: A first attempt to phylogeographic inference for a marine mollusc disease. Virus Evol. 2022, 8, veac039. [Google Scholar] [CrossRef]
- Delmotte, J.; Chaparro, C.; Galinier, R.; de Lorgeril, J.; Petton, B.; Stenger, P.L.; Vidal-Dupiol, J.; Destoumieux-Garzon, D.; Gueguen, Y.; Montagnani, C.; et al. Contribution of Viral Genomic Diversity to Oyster Susceptibility in the Pacific Oyster Mortality Syndrome. Front. Microbiol. 2020, 11, 1579. [Google Scholar] [CrossRef]
- Shi, M.; Lin, X.-D.; Tian, J.-H.; Chen, L.-J.; Chen, X.; Li, C.-X.; Qin, X.-C.; Li, J.; Cao, J.-P.; Eden, J.-S.; et al. Redefining the invertebrate RNA virosphere. Nature 2016, 540, 539–543. [Google Scholar] [CrossRef]
- Rosani, U.; Gerdol, M. A bioinformatics approach reveals seven nearly-complete RNA-virus genomes in bivalve RNA-seq data. Virus Res. 2017, 239, 33–42. [Google Scholar] [CrossRef]
- Rosani, U.; Venier, P. Oyster RNA-seq Data Support the Development of Malacoherpesviridae Genomics. Front. Microbiol. 2017, 8, 1515. [Google Scholar] [CrossRef] [PubMed]
- Rosani, U.; Shapiro, M.; Venier, P.; Allam, B. A Needle in A Haystack: Tracing Bivalve-Associated Viruses in High-Throughput Transcriptomic Data. Viruses 2019, 11, 205. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.-Z.; Fang, Y.-F.; Wei, H.-Y.; Zhu, P.; Liu, M.; Yuan, W.-G.; Yang, L.-L.; Guo, Y.-X.; Jin, T.; Shi, M.; et al. A remarkably diverse and well-organized virus community in a filter-feeding oyster. Microbiome 2023, 11, 2. [Google Scholar]
- Zhu, P.; Liu, G.-F.; Liu, C.; Yang, L.-L.; Liu, M.; Xie, K.-M.; Shi, S.-K.; Shi, M.; Jiang, J.-Z. Novel RNA viruses in oysters revealed by virome. iMeta 2022, 1, e65. [Google Scholar] [CrossRef]
- Rosario, K.; Schenck, R.O.; Harbeitner, R.C.; Lawler, S.N.; Breitbart, M. Novel circular single-stranded DNA viruses identified in marine invertebrates reveal high sequence diversity and consistent predicted intrinsic disorder patterns within putative structural proteins. Front. Microbiol. 2015, 6, 696. [Google Scholar] [CrossRef] [PubMed]
- Krupovic, M.; Varsani, A. Naryaviridae, Nenya viridae, and Vilyaviridae: Three new families of single-stranded DNA viruses in the phylum Cressdnaviricota. Arch. Virol. 2022, 167, 2907–2921. [Google Scholar] [CrossRef]
- Krupovic, M.; Varsani, A.; Kazlauskas, D.; Breitbart, M.; Delwart, E.; Rosario, K.; Yutin, N.; Wolf, Y.I.; Harrach, B.; Zerbini, F.M.; et al. Cressdnaviricota: A Virus Phylum Unifying Seven Families of Rep-Encoding Viruses with Single-Stranded, Circular DNA Genomes. J. Virol. 2020, 94, e00582-20. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Rosario, K.; Breitbart, M.; Duffy, S. Eukaryotic Circular Rep-Encoding Single-Stranded DNA (CRESS DNA) Viruses: Ubiquitous Viruses with Small Genomes and a Diverse Host Range. Adv. Virus Res. 2019, 103, 71–133. [Google Scholar]
- Abbas, A.A.; Taylor, L.J.; Dothard, M.I.; Leiby, J.S.; Fitzgerald, A.S.; Khatib, L.A.; Collman, R.G.; Bushman, F.D. Redondoviridae, a Family of Small, Circular DNA Viruses of the Human Oro-Respiratory Tract Associated with Periodontitis and Critical Illness. Cell Host Microbe 2019, 25, 719–729.e714. [Google Scholar] [CrossRef]
- Dayaram, A.; Goldstien, S.; Argüello-Astorga, G.R.; Zawar-Reza, P.; Gomez, C.; Harding, J.S.; Varsani, A. Diverse small circular DNA viruses circulating amongst estuarine molluscs. Infect. Genet. Evol. 2015, 31, 284–295. [Google Scholar] [CrossRef]
- Gezon, N.R.; Haywick, D.W.; Sanders, J.M.; Hewson, I.; Strychar, K.B. Circular Rep encoding single stranded (CRESS) DNA virus-like sequences detected in quagga mussels (Dreissena rostriformis bugensis) and sediments from the central Lake Michigan benthos. J. Great Lakes Res. 2020, 46, 302–310. [Google Scholar] [CrossRef]
- Yang, L.-L.; Guo, Y.-X.; Wei, H.-Y.; Wang, M.; Fang, Y.-F.; Zhu, P.; Jiang, J.-Z. Identification of a new type of oyster-related circovirus genome Comparative genome analysis of oyster-related circoviruses. South China Fish. Sci. 2021, 18, 65–75. [Google Scholar]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Li, D.; Luo, R.; Liu, C.M.; Leung, C.M.; Ting, H.F.; Sadakane, K.; Yamashita, H.; Lam, T.W. MEGAHIT v1.0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods 2016, 102, 3–11. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.M.; Luo, R.; Sadakane, K.; Lam, T.W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Huson, D.H.; Beier, S.; Flade, I.; Gorska, A.; El-Hadidi, M.; Mitra, S.; Ruscheweyh, H.J.; Tappu, R. MEGAN Community Edition—Interactive Exploration and Analysis of Large-Scale Microbiome Sequencing Data. PLoS Comput. Biol. 2016, 12, e1004957. [Google Scholar] [CrossRef] [PubMed]
- Hyatt, D.; Chen, G.L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Kazlauskas, D.; Varsani, A.; Krupovic, M. Pervasive Chimerism in the Replication-Associated Proteins of Uncultured Single-Stranded DNA Viruses. Viruses 2018, 10, 187. [Google Scholar] [CrossRef]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An Open Source Software for Exploring and Manipulating Networks. Proc. Int. AAAI Conf. Web Soc. Media 2009, 3, 361–362. [Google Scholar] [CrossRef]
- Muhire, B.M.; Varsani, A.; Martin, D.P. SDT: A Virus Classification Tool Based on Pairwise Sequence Alignment and Identity Calculation. PLoS ONE 2014, 9, e108277. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Capella-Gutierrez, S.; Silla-Martinez, J.M.; Gabaldon, T. TrimAl: A Tool for Automated Alignment Trimming in Large-Scale Phylogenetic Analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed]
- Varsani, A.; Krupovic, M. Smacoviridae: A new family of animal-associated single-stranded DNA viruses. Arch. Virol. 2018, 163, 2005–2015. [Google Scholar] [CrossRef]
- Krupovic, M.; Varsani, A. A 2021 taxonomy update for the family Smacoviridae. Arch. Virol. 2021, 166, 3245–3253. [Google Scholar] [CrossRef]
- Breitbart, M.; Delwart, E.; Rosario, K.; Segalés, J.; Varsani, A.; ICTV Report Consortium. ICTV Virus Taxonomy Profile: Circoviridae. J. Gen. Virol. 2017, 98, 1997–1998. [Google Scholar] [CrossRef]
- Desingu, P.A.; Nagarajan, K. Genetic Diversity and Characterization of Circular Replication (Rep)-Encoding Single-Stranded (CRESS) DNA Viruses. Microbiol. Spectr. 2022, 10, e0105722. [Google Scholar] [CrossRef]
- Cheng, R.; Li, X.; Jiang, L.; Gong, L.; Geslin, C.; Shao, Z. Virus diversity and interactions with hosts in deep-sea hydrothermal vents. Microbiome 2022, 10, 235. [Google Scholar] [CrossRef]
- Yoshida, M.; Takaki, Y.; Eitoku, M.; Nunoura, T.; Takai, K. Metagenomic analysis of viral communities in (hado)pelagic sediments. PLoS ONE 2013, 8, e57271. [Google Scholar] [CrossRef]
- McDaniel, L.D.; Rosario, K.; Breitbart, M.; Paul, J.H. Comparative metagenomics: Natural populations of induced prophages demonstrate highly unique, lower diversity viral sequences. Environ. Microbiol. 2014, 16, 570–585. [Google Scholar] [CrossRef]
- Lőrincz, M.; Dán, A.; Láng, M.; Csaba, G.; Tóth, A.G.; Székely, C.; Cságola, A.; Tuboly, T. Novel circovirus in European catfish (Silurus glanis). Arch. Virol. 2012, 157, 1173–1176. [Google Scholar] [CrossRef]
- Doszpoly, A.; Tarján, Z.L.; Glávits, R.; Müller, T.; Benkő, M. Full genome sequence of a novel circo-like virus detected in an adult European eel Anguilla anguilla showing signs of cauliflower disease. Dis. Aquat. Organ. 2014, 109, 107–115. [Google Scholar] [CrossRef]
- Lorincz, M.; Csagola, A.; Farkas, S.L.; Szekely, C.; Tuboly, T. First detection and analysis of a fish circovirus. J. Gen. Virol. 2011, 92, 1817–1821. [Google Scholar] [CrossRef]
- Zhang, Y.P.; Qin, Z.C.; Li, T.T.; Zhu, X.D. Carbon dioxide uptake overrides methane emission at the air-water interface of algae-shellfish mariculture ponds: Evidence from eddy covariance observations. Sci. Total Environ. 2022, 815, 152867. [Google Scholar] [CrossRef]
- Li, L.; Kapoor, A.; Slikas, B.; Bamidele, O.S.; Wang, C.; Shaukat, S.; Masroor, M.A.; Wilson, M.L.; Ndjango, J.B.; Peeters, M.; et al. Multiple diverse circoviruses infect farm animals and are commonly found in human and chimpanzee feces. J. Virol. 2010, 84, 1674–1682. [Google Scholar] [CrossRef]
- Kim, K.I.; Kwon, W.J.; Kim, Y.C.; Kim, M.-S.; Hong, S.; Jeong, H.D. Surveillance of Aquatic Animal Viruses in Seawater and Shellfish in Korea. Aquaculture 2016, 461, 17–24. [Google Scholar] [CrossRef]
- Martin, D.P.; Biagini, P.; Lefeuvre, P.; Golden, M.; Roumagnac, P.; Varsani, A. Recombination in eukaryotic single stranded DNA viruses. Viruses 2011, 3, 1699–1738. [Google Scholar] [CrossRef]
- Li, L.; Shan, T.; Soji, O.B.; Alam, M.M.; Kunz, T.H.; Zaidi, S.Z.; Delwart, E. Possible cross-species transmission of circoviruses and cycloviruses among farm animals. J. Gen. Virol. 2011, 92, 768–772. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, P.; Liu, C.; Liu, G.-F.; Liu, H.; Xie, K.-M.; Zhang, H.-S.; Xu, X.; Xiao, J.; Jiang, J.-Z. Unveiling CRESS DNA Virus Diversity in Oysters by Virome. Viruses 2024, 16, 228. https://doi.org/10.3390/v16020228
Zhu P, Liu C, Liu G-F, Liu H, Xie K-M, Zhang H-S, Xu X, Xiao J, Jiang J-Z. Unveiling CRESS DNA Virus Diversity in Oysters by Virome. Viruses. 2024; 16(2):228. https://doi.org/10.3390/v16020228
Chicago/Turabian StyleZhu, Peng, Chang Liu, Guang-Feng Liu, Hong Liu, Ke-Ming Xie, Hong-Sai Zhang, Xin Xu, Jian Xiao, and Jing-Zhe Jiang. 2024. "Unveiling CRESS DNA Virus Diversity in Oysters by Virome" Viruses 16, no. 2: 228. https://doi.org/10.3390/v16020228