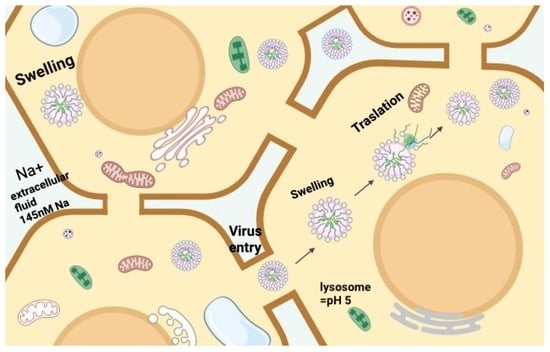
An Observation of a Very High Swelling of Bromovirus Members at Specific Ionic Strengths and pH
Abstract
:1. Introduction
2. Materials and Methods
2.1. Characterization Methods
2.1.1. Dynamic Light Scattering (DLS)
2.1.2. Atomic Force Microscopy
2.1.3. Transmission Electronic Microscopy (TEM)
3. Results and Discussion
4. General Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Konecny, R.; Trylska, J.; Tama, F.; Zhang, D.; Baker, N.A.; Brooks, C.L.; McCammon, J.A. Electrostatic Properties of Cowpea Chlorotic Mottle Virus and Cucumber Mosaic Virus Capsids. Biopolymers 2006, 82, 106–120. [Google Scholar] [CrossRef]
- Chen, C.; Daniel, M.-C.; Quinkert, Z.T.; De, M.; Stein, B.; Bowman, V.D.; Chipman, P.R.; Rotello, V.M.; Kao, C.C.; Dragnea, B. Nanoparticle-templated assembly of viral protein Cages. Nano Lett. 2006, 6, 611–615. [Google Scholar] [CrossRef] [PubMed]
- Lane, L.C. The Bromoviruses. Adv. Virus Res. 1974, 19, 151–220. [Google Scholar] [CrossRef] [PubMed]
- Speir, J.A.; Munshi, S.; Wang, G.; Baker, T.S.; E Johnson, J. Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by X-ray crystallography and cryo-electron microscopy. Structure 1995, 3, 63–78. [Google Scholar] [CrossRef] [PubMed]
- Bancroft, J.B.; Hills, G.J.; Markham, R. A study of the self-assembly process in a small spherical virus formation of organized structures from protein subunits in vitro. Virology 1967, 31, 354–379. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.M.; Wang, G.; Speir, J.A.; Olson, N.H.; Johnson, J.E.; Baker, T.S.; Young, M.J. Comparison of the native CCMV virion within vitroassembled CCMV virions by cryoelectron microscopy and image reconstruction. Virology 1998, 244, 212–218. [Google Scholar] [CrossRef]
- Ahlquist, P. Bromovirus RNA replication and transcription. Curr. Opin. Genet. Dev. 1992, 2, 71–76. [Google Scholar] [CrossRef]
- Sacher, R.; Ahlquist, P. Effects of deletions in the N-terminal basic arm of brome mosaic virus coat protein on RNA packaging and systemic infection. J. Virol. 1989, 63, 4545–4552. [Google Scholar] [CrossRef]
- Caspar, D.L.; Klug, A. Physical principles in the construction of regular viruses. In Cold Spring Harbor Symposia on Quantitative Biology; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1962; Volume 27, pp. 1–24. [Google Scholar] [CrossRef]
- Bancroft, J.B.; Hiebert, E.; Rees, M.W.; Markham, R. Properties of cowpea chlorotic mottle virus, its protein and nucleic acid. Virology 1968, 34, 224–239. [Google Scholar] [CrossRef]
- Incardona, N.L.; Kaesberg, P. A pH-induced structural change in bromegrass mosaic virus. Biophys. J. 1964, 4, 11–21. [Google Scholar] [CrossRef]
- Chauvin, C.; Pfeiffer, P.; Witz, J.; Jacrot, B. Structural polymorphism of bromegrass mosaic virus: A neutron small angle scattering investigation. Virology 1978, 88, 138–148. [Google Scholar] [CrossRef] [PubMed]
- Tama, F.; Brooks, C.L. The mechanism and pathway of pH induced swelling in cowpea chlorotic mottle Virus. J. Mol. Biol. 2002, 318, 733–747. [Google Scholar] [CrossRef] [PubMed]
- Wilts, B.D.; Schaap, I.A.; Schmidt, C.F. Swelling and softening of the cowpea chlorotic mottle virus in response to pH shifts. Biophys. J. 2015, 108, 2541–2549. [Google Scholar] [CrossRef] [PubMed]
- Liepold, L.O.; Revis, J.; Allen, M.; Oltrogge, L.; Young, M.; Douglas, T. Structural transitions in Cowpea chlorotic mottle virus (CCMV). Phys. Biol. 2005, 2, S166–S172. [Google Scholar] [CrossRef]
- Lucas, R.W.; Kuznetsov, Y.G.; Larson, S.B.; McPherson, A. Crystallization of brome mosaic virus and T = 1 brome mosaic virus particles following a structural transition. Virology 2001, 286, 290–303. [Google Scholar] [CrossRef]
- Yamazaki, H.; Kaesberg, P. Degradation of bromegrass mosaic virus with calcium chloride and the isolation of its protein and nucleic acid. J. Mol. Biol. 1963, 7, 760–762. [Google Scholar] [CrossRef]
- Lavelle, L.; Michel, J.-P.; Gingery, M. The disassembly, reassembly and stability of CCMV protein capsids. J. Virol. Methods 2007, 146, 311–316. [Google Scholar] [CrossRef]
- Pfeiffer, P.; Hirth, L. Aggregation states of Brome mosaic virus protein. Virology 1974, 61, 160–167. [Google Scholar] [CrossRef]
- Bancroft, J.B.; Hiebert, E.; Bracker, C.E. The effects of various polyanions on shell formation of some spherical viruses. Virology 1969, 39, 924–930. [Google Scholar] [CrossRef]
- Lavelle, L.; Gingery, M.; Phillips, M.; Gelbart, W.M.; Knobler, C.M.; Cadena-Nava, R.D.; Vega-Acosta, J.R.; Pinedo-Torres, L.A.; Ruiz-Garcia, J. Phase diagram of self-assembled viral capsid protein polymorphs. J. Phys. Chem. B 2009, 113, 3813–3819. [Google Scholar] [CrossRef]
- Ramos-Carreño, S.; Giffard-Mena, I.; Zamudio-Ocadiz, J.N.; Nuñez-Rivera, A.; Valencia-Yañez, R.; Ruiz-Garcia, J.; Viana, M.T.; Cadena-Nava, R.D. Antiviral therapy in shrimp through plant virus VLP containing VP28 dsRNA against WSSV. Beilstein J. Org. Chem. 2021, 17, 1360–1373. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.B.; Knobler, C.M.; Gelbart, W.M.; Mason, T.G. Curvature dependence of viral protein structures on encapsidated nanoemulsion droplets. ACS Nano 2008, 2, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Durán-Meza, A.L.; Escamilla-Ruiz, M.I.; Segovia-González, X.F.; Villagrana-Escareño, M.V.; Vega-Acosta, J.R.; Ruiz-Garcia, J. Encapsidation of Different Plasmonic Gold Nanoparticles by the CCMV CP. Molecules 2020, 25, 2628. [Google Scholar] [CrossRef]
- Lucas, R.W.; Larson, S.B.; McPherson, A. The crystallographic structure of brome mosaic virus. J. Mol. Biol. 2002, 317, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Uchida, M.; Klem, M.T.; Allen, M.; Suci, P.; Flenniken, M.; Gillitzer, E.; Varpness, Z.; Liepold, L.O.; Young, M.; Douglas, T. Biological containers: Protein cages as multifunctional nanoplatforms. Adv. Mater. 2007, 19, 1025–1042. [Google Scholar] [CrossRef]
- Llauró, A.; Coppari, E.; Imperatori, F.; Bizzarri, A.R.; Castón, J.R.; Santi, L.; Cannistraro, S.; de Pablo, P.J. Calcium ions modulate the mechanics of tomato bushy stunt virus. Biophys. J. 2015, 109, 390–397. [Google Scholar] [CrossRef] [PubMed]
- Loredo-Tovias, M.; Duran-Meza, A.L.; Villagrana-Escareño, M.V.; Vega-Acosta, R.; Reynaga-Hernández, E.; Flores-Tandy, L.M.; Valdes-Resendiz, O.E.; Cadena-Nava, R.D.; Alvizo-Paez, E.R.; Ruiz-Garcia, J. Encapsidated ultrasmall nanolipospheres as novel nanocarriers for highly hydrophobic anticancer drugs. Nanoscale 2017, 9, 11625–11631. [Google Scholar] [CrossRef]
- Michel, J.-P.; Gingery, M.; Lavelle, L. Efficient purification of bromoviruses by ultrafiltration. J. Virol. Methods 2004, 122, 195–198. [Google Scholar] [CrossRef]
- Stoll, V.S.; Blanchard, J.S. Buffers: Principles and practice. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 2009; Volume 463, pp. 43–56. [Google Scholar] [CrossRef]
- Dey, P.; Bergmann, T.; Cuellar-Camacho, J.L.; Ehrmann, S.; Chowdhury, M.S.; Zhang, M.; Dahmani, I.; Haag, R.; Azab, W. Multivalent flexible nanogels exhibit broad-spectrum antiviral activity by blocking virus entry. ACS Nano 2018, 12, 6429–6442. [Google Scholar] [CrossRef]
- Cao, B.; Xu, H.; Mao, C. Transmission electron microscopy as a tool to image bioinorganic nanohybrids: The case of phage-gold nanocomposites. Microsc. Res. Tech. 2011, 74, 627–635. [Google Scholar] [CrossRef]
- Krupovic, M.; Koonin, E.V. Multiple origins of viral capsid proteins from cellular ancestors. Proc. Natl. Acad. Sci. USA 2017, 114, E2401–E2410. [Google Scholar] [CrossRef] [PubMed]
- Duran-Meza, A.L.; Villagrana-Escareño, M.V.; Ruiz-García, J.; Knobler, C.M.; Gelbart, W.M. Controlling the surface charge of simple viruses. PLoS ONE 2021, 16, e0255820. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Johnson, J.E.; Ortoleva, P.J. All-atom multiscale simulation of cowpea chlorotic mottle virus capsid swelling. J. Phys. Chem. B 2010, 114, 11181–11195. [Google Scholar] [CrossRef] [PubMed]
- Dietzgen, R.G.; Mann, K.S.; Johnson, K.N. Plant Virus–insect vector interactions: Current and potential future research directions. Viruses 2016, 8, 303. [Google Scholar] [CrossRef] [PubMed]
- Colebatch, G.; East, P.; Cooper, P. Preliminary characterisation of digestive proteases of the green mirid, Creontiades dilutus (Hemiptera: Miridae). Insect Biochem. Mol. Biol. 2001, 31, 415–423. [Google Scholar] [CrossRef]
- Kader, M.A.; Lindberg, S. Cytosolic calcium and pH signaling in plants under salinity stress. Plant Signal. Behav. 2010, 5, 233–238. [Google Scholar] [CrossRef]
- Minocha, S.C. pH of the medium and the growth and metabolism of cells in culture. In Cell and Tissue Culture in Forestry; Springer: Dordrecht, The Netherlands, 1987; pp. 125–141. [Google Scholar] [CrossRef]
- Wilson, T.M.A. Cotranslational disassembly of tobacco mosaic virus in vitro. Virology 1984, 137, 255–265. [Google Scholar] [CrossRef]
- Roenhorst, J.W.; Verduin, B.J.M.; Goldbach, R.W. Virus-ribosome complexes from cell-free translation systems supplemented with cowpea chlorotic mottle virus particles. Virology 1989, 168, 138–146. [Google Scholar] [CrossRef]
- Sorger, P.K.; Stockley, P.G.; Harrison, S.C. Structure and assembly of turnip crinkle virus: II. Mechanism of reassembly in vitro. J. Mol. Biol. 1986, 191, 639–658. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Segovia-González, X.F.; Villagrana-Escareño, M.V.; Ríos-Ramírez, M.; de la Cruz, V.S.; Mejía-Hernández, J.N.; Cuellar-Camacho, J.L.; Patrón-Soberano, A.; Sportsman, R.; Ruiz-García, J. An Observation of a Very High Swelling of Bromovirus Members at Specific Ionic Strengths and pH. Viruses 2023, 15, 2046. https://doi.org/10.3390/v15102046
Segovia-González XF, Villagrana-Escareño MV, Ríos-Ramírez M, de la Cruz VS, Mejía-Hernández JN, Cuellar-Camacho JL, Patrón-Soberano A, Sportsman R, Ruiz-García J. An Observation of a Very High Swelling of Bromovirus Members at Specific Ionic Strengths and pH. Viruses. 2023; 15(10):2046. https://doi.org/10.3390/v15102046
Chicago/Turabian StyleSegovia-González, Xochitl Fabiola, Maria Veronica Villagrana-Escareño, Maricarmen Ríos-Ramírez, Vianey Santiago de la Cruz, Jessica Nathaly Mejía-Hernández, Jose Luis Cuellar-Camacho, Araceli Patrón-Soberano, Richard Sportsman, and Jaime Ruiz-García. 2023. "An Observation of a Very High Swelling of Bromovirus Members at Specific Ionic Strengths and pH" Viruses 15, no. 10: 2046. https://doi.org/10.3390/v15102046