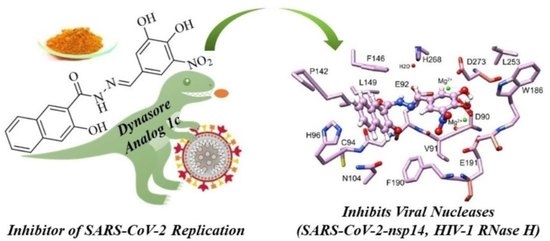
Analogs of the Catechol Derivative Dynasore Inhibit HIV-1 Ribonuclease H, SARS-CoV-2 nsp14 Exoribonuclease, and Virus Replication
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemistry
2.1.1. General Procedures and Materials
2.1.2. (E)-N’-(4-Hydroxy-3-nitrobenzylidene)-3-hydroxy-2-naphthohydrazide (1a)
2.1.3. (E)-N′-(4-Hydroxy-5-methoxy-3-nitrobenzylidene)-3-hydroxy-2-naphthohydrazide (1b)
2.1.4. (E)-N′-(3,4-Dihydroxy-5-nitrobenzylidene)-3-hydroxy-2-naphthohydrazide (1c)
2.1.5. (E)-N’-(2,3-Dihydroxy-4-methoxybenzylidene)-3-hydroxy-2-naphthohydrazide (1e)
2.1.6. (E)-N’-[4-(N-Methylpiperazinyl)benzylidene]-3-hydroxy-2-naphthohydrazide (1l)
2.1.7. (E)-N′-(3,4-Dihydroxy-5-nitrobenzylidene)-2-hydroxybenzohydrazide (2a)
2.1.8. (E)-N′-(3,4-Dihydroxy-5-nitrobenzylidene)-3-fluorobenzohydrazide (2b)
2.1.9. (E)-N′-(3,4-Dihydroxy-5-nitrobenzylidene)-nicotinoylhydrazide (2c)
2.2. Plasmid Construction and Protein Purification
2.3. HIV-1 RNase H Inhibition
2.4. SARS-CoV-2 nsp14 Exonuclease Activity Assays
2.5. SARS-CoV-2 nsp14 N7-Methyltransferase Activity Assays
2.6. Drug Toxicity to Human Adenocarcinoma Lung Cells
2.7. SARS-CoV-2 Replication Inhibition in A549 ACE2+ Cells
2.8. Plaque Reduction Assay
2.9. Molecular Docking
3. Results
3.1. Chemistry
3.2. Inhibition of HIV-1 RNase H
3.3. Inhibition of ExoN Activity of SARS-CoV-2 nsp14/nsp10
3.4. Inhibitor 1c Does Not Interfere with nsp14 MTase Activity
3.5. Inhibitor 1c Docks in the Catalytic Site of nsp14-nsp10 Complex
3.6. Compound 1c Does Not Affect Cell Viability
3.7. Compound 1c Inhibits SARS-CoV-2 Replication
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, B.; Tian, E.-K.; Tian, L.; Han, R.; Wang, S.; Xiang, Q.; Zhang, S.; El Arnaout, T.; Cheng, W. Overview of lethal human coronaviruses. Signal Transduct. Target. Ther. 2020, 5, 89. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.-L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Markov, P.V.; Ghafari, M.; Beer, M.; Lythgoe, K.; Simmonds, P.; Stilianakis, N.I.; Katzourakis, A. The evolution of SARS-CoV-2. Nat. Rev. Microbiol. 2023, 21, 361–379. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Yi, S.V. On the origin and evolution of SARS-CoV-2. Exp. Mol. Med. 2021, 53, 537–547. [Google Scholar] [CrossRef]
- Al-Awwal, N.; Dweik, F.; Mahdi, S.; El-Dweik, M.; Anderson, S.H. A review of SARS-CoV-2 disease (COVID-19): Pandemic in our time. Pathogens 2022, 11, 368. [Google Scholar] [CrossRef]
- Park, S.E. Epidemiology, virology, and clinical features of severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2; Coronavirus Disease-19). Clin. Exp. Pediatr. 2020, 4, 119–124. [Google Scholar] [CrossRef] [Green Version]
- Zhong, L.; Zhao, Z.; Peng, X.; Zou, J.; Yang, S. Recent advances in small-molecule therapeutics for COVID-19. Precis. Clin. Med. 2022, 5, pbac024. [Google Scholar] [CrossRef]
- Gottlieb, R.L.; Vaca, C.E.; Paredes, R.; Mera, J.; Webb, B.J.; Perez, G.; Oguchi, G.; Ryan, P.; Nielsen, B.U.; Brown, M.; et al. Early remdesivir to prevent progression to severe Covid-19 in outpatients. N. Engl. J. Med. 2022, 386, 305–315. [Google Scholar] [CrossRef]
- Dal-Ré, R.; Becker, S.L.; Bottieau, E.; Holm, S. Availability of oral antivirals against SARS-CoV-2 infection and the requirement for an ethical prescribing approach. Lancet Infect. Dis. 2022, 22, e231–e238. [Google Scholar] [CrossRef]
- Kabinger, F.; Stiller, C.; Schmitzová, J.; Dienemann, C.; Kokic, G.; Hillen, H.S.; Höbartner, C.; Cramer, P. Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis. Nat. Struct. Mol. Biol. 2021, 28, 740–746. [Google Scholar] [CrossRef]
- Yang, L.; Wang, Z. Bench-to-bedside: Innovation of small molecule anti-SARS-CoV-2 drugs in China. Eur. J. Med. Chem. 2023, 257, 115503. [Google Scholar] [CrossRef] [PubMed]
- Kumari, M.; Lu, R.-M.; Li, M.-C.; Huang, J.-L.; Hsu, F.-F.; Ko, S.-H.; Ke, F.-Y.; Su, S.-C.; Liang, K.-H.; Yuan, J.P.-Y.; et al. A critical overview of current progress for COVID-19: Development of vaccines, antiviral drugs, and therapeutic antibodies. J. Biomed. Sci. 2022, 29, 68. [Google Scholar] [CrossRef] [PubMed]
- Lei, S.; Chen, X.; Wu, J.; Duan, X.; Men, K. Small molecules in the treatment of COVID-19. Signal Transduct. Target. Ther. 2022, 7, 387. [Google Scholar] [CrossRef] [PubMed]
- Perry, J.K.; Appleby, T.C.; Bilello, J.P.; Feng, J.Y.; Schmitz, U.; Campbell, E.A. An atomistic model of the coronavirus replication-transcription complex as a hexamer assembled around nsp15. J. Biol. Chem. 2021, 297, 101218. [Google Scholar] [CrossRef]
- Ortiz-Alcantara, J.; Bhardwaj, K.; Pananinathan, S.; Frieman, M.; Baric, R.S.; Kao, C.C. Small molecule inhibitors of the SARS-CoV Nsp15 endoribonuclease. Virus Adapt. Treat. 2010, 2, 125–133. [Google Scholar]
- Minskaia, E.; Hertzig, T.; Gorbalenya, A.E.; Campanacci, V.; Cambillau, C.; Canard, B.; Ziebuhr, J. Discovery of an RNA virus 3’-5’ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. USA 2006, 103, 5108–5113. [Google Scholar] [CrossRef]
- Ma, Y.; Wu, L.; Shaw, N.; Gao, Y.; Wang, J.; Sun, Y.; Lou, Z.; Yan, L.; Zhang, R.; Rao, Z. Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex. Proc. Natl. Acad. Sci. USA 2015, 112, 9436–9441. [Google Scholar] [CrossRef]
- Canal, B.; McClure, A.W.; Curran, J.F.; Wu, M.; Ulferts, R.; Weissmann, F.; Zeng, J.; Bertolin, A.P.; Milligan, J.C.; Basu, S.; et al. Identifying SARS-CoV-2 antiviral compounds by screening for small molecule inhibitors of nsp14/nsp10 exoribonuclease. Biochem. J. 2021, 478, 2445–2464. [Google Scholar] [CrossRef]
- Baddock, H.T.; Brolih, S.; Yosaatmadja, Y.; Ratnaweera, M.; Bielinski, M.; Swift, L.P.; Cruz-Migoni, A.; Fan, H.; Keown, J.R.; Walker, A.P.; et al. Characterization of the SARS-CoV-2 ExoN (nsp14ExoN-nsp10) complex: Implications for its role in viral genome stability and inhibitor identification. Nucleic Acids Res. 2022, 50, 1484–1500. [Google Scholar] [CrossRef]
- Rona, G.; Zeke, A.; Miwatani-Minter, B.; de Vries, M.; Kaur, R.; Schinlever, A.; Garcia, S.F.; Goldberg, H.V.; Wang, H.; Hinds, T.R.; et al. The NSP14/NSP10 RNA repair complex as a pan-coronavirus therapeutic target. Cell Death Differ. 2022, 29, 285–292. [Google Scholar] [CrossRef]
- Otava, T.; Sála, M.; Li, F.; Fanfrlík, J.; Devkota, K.; Perveen, S.; Chau, I.; Pakarian, P.; Hobza, P.; Vedadi, M.; et al. The structure-based design of SARS-CoV-2 nsp14 methyltransferase ligands yields nanomolar inhibitors. ACS Infect. Dis. 2021, 7, 2214–2220. [Google Scholar] [CrossRef]
- Ogando, N.S.; Zevenhoven-Dobbe, J.C.; van der Meer, Y.; Bredenbeek, P.J.; Posthuma, C.C.; Snijder, E.J. The enzymatic activity of the nsp14 exoribonuclease is critical for replication of MERS-CoV and SARS-CoV-2. J. Virol. 2020, 94, e01246-20. [Google Scholar] [CrossRef]
- Saramago, M.; Bárria, C.; Costa, V.G.; Souza, C.S.; Viegas, S.C.; Domingues, S.; Lousa, D.; Soares, C.M.; Arraiano, C.M.; Matos, R.G. New targets for drug design: Importance of nsp14/nsp10 complex formation for the 3’-5’ exoribonucleolytic activity of SARS-CoV-2. FEBS J. 2021, 288, 5130–5147. [Google Scholar] [CrossRef] [PubMed]
- Beilhartz, G.L.; Götte, M. HIV-1 ribonuclease H: Structure, catalytic mechanism and inhibitors. Viruses 2010, 2, 900–926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuo, Y.; Deutscher, M.P. Exoribonuclease superfamilies: Structural analysis and phylogenetic distribution. Nucleic Acids Res. 2001, 29, 1017–1026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tun, M.M.G.; Morita, K.; Ishikawa, T.; Urata, S. The antiviral effect of the chemical compounds targeting DED/Edh motifs of the viral proteins on lymphocytic choriomeningitis virus and SARS-CoV-2. Viruses 2021, 13, 1220. [Google Scholar]
- Schneider, A.; Corona, A.; Spöring, I.; Jordan, M.; Buchholz, B.; Maccioni, E.; Di Santo, R.; Bodem, J.; Tramontano, E.; Wöhrl, B.M. Biochemical characterization of a multi-drug resistant HIV-1 subtype AG reverse transcriptase: Antagonism of AZT discrimination and excision pathways and sensitivity to RNase H inhibitors. Nucleic Acids Res. 2016, 44, 2310–2322. [Google Scholar] [CrossRef] [Green Version]
- Corona, A.; Seibt, S.; Schaller, D.; Schobert, R.; Volkamer, A.; Biersack, B.; Tramontano, E. Garcinol from Garcinia indica inhibits HIV-1 reverse transcriptase-associated ribonuclease H. Arch. Pharm. 2021, 354, 2100123. [Google Scholar] [CrossRef]
- Macia, E.; Ehrlich, M.; Massol, R.; Boucrot, E.; Brunner, C.; Kirchhausen, T. Dynasore, a cell-permeable inhibitor of dynamin. Dev. Cell 2006, 10, 839–850. [Google Scholar] [CrossRef] [Green Version]
- Preta, G.; Cronin, J.G.; Sheldon, M. Dynasore—Not just a dynamin inhibitor. Cell Commun. Signal. 2015, 13, 24. [Google Scholar] [CrossRef] [Green Version]
- Carro, A.C.; Piccini, L.E.; Damonte, E.B. Blockade of dengue virus entry into myeloid cells by endocytic inhibitors in the presence or absence of antibodies. PLoS Negl. Trop. Dis. 2018, 12, e0006685. [Google Scholar] [CrossRef] [Green Version]
- Abban, C.Y.; Bradbury, N.A.; Meneses, P.I. HPV16 and BPV1 infection can be blocked by the dynamin inhibitor dynasore. Am. J. Ther. 2008, 15, 304–311. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Felipe, L.; Villar, E.; Munoz-Barroso, I. Entry of Newcastle Disease Virus into the host cell: Role of acidic pH and endocytosis. Biochim. Biophys. Acta 2014, 1838, 300–309. [Google Scholar] [CrossRef] [Green Version]
- Mues, M.B.; Cheshenko, N.; Wilson, D.W.; Gunther-Cummins, L.; Herold, B.C. Dynasore disrupts trafficking of herpes simplex virus proteins. J. Virol. 2015, 89, 6673–6684. [Google Scholar] [CrossRef] [Green Version]
- Shi, B.J.; Liu, C.C.; Zhou, J.; Wang, S.Q.; Gao, Z.C.; Zhang, X.M.; Zhou, B.; Chen, P.Y. Entry of classical swine fever virus into PK-15 cells via a pH-, dynamin-, and cholesterol-dependent, clathrin-mediated endocytotic pathway that requires Rab5 and Rab7. J. Virol. 2016, 90, 9194–9208. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.H.R.; Hussain, K.M.; Chu, J.J.H. Macropinocytosis dependent entry of Chikungunya virus into human muscle cells. PLoS Negl. Trop. Dis. 2019, 13, e0007610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Xiao, D.; Zhao, Y.; Zhang, L.; Chen, R.; Liu, W.; Wen, Y.; Liao, Y.; Wen, Y.; Wu, R.; et al. Porcine deltacoronavirus (PDCoV) entry into PK-15 cells by caveolae-mediated endocytosis. Viruses 2022, 14, 496. [Google Scholar] [CrossRef]
- Ailenberg, M.; Di Ciano-Oliveira, C.; Szaszi, K.; Dan, Q.; Rozycki, M.; Kapus, A.; Rotstein, O.D. Dynasore enhances the formation of mitochondrial antiviral signaling aggregates and endocytosis-independent NF-κB activation. Br. J. Pharmacol. 2015, 172, 3748–3763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarabia, I.; Novis, C.L.; Macedo, A.B.; Takata, H.; Nell, R.; Kakazu, J.C.; Furler, R.L.; Shakya, B.; Schubert, H.L.; Hill, C.P.; et al. Activation of the anti-oxidative stress response reactivates latent HIV-1 through the mitochondrial antiviral signaling protein isoform miniMAVS. Front. Immunol. 2021, 12, 682182. [Google Scholar] [CrossRef]
- Nitsche, C.; Steuer, C.; Klein, C.D. Acrylcyanoacrylamides as inhibitors of the Dengue and West Nile virus proteases. Bioorg. Med. Chem. 2011, 19, 7318–7337. [Google Scholar] [CrossRef] [PubMed]
- Kaisalo, L.; Latvala, A.; Hase, T. Selective demethylations in 2,3,4-trimethoxyaryl carbonyl compounds. Synth. Commun. 1986, 16, 645–648. [Google Scholar] [CrossRef]
- Saglik, B.N.; Ilgin, S.; Özkay, Y. Synthesis of new donepezil analogues and investigation of their effects on cholinesterase enzymes. Eur. J. Med. Chem. 2016, 124, 1026–1040. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Jung, K.-Y.; Park, J.; Cho, J.-H.; Kim, Y.-C.; Chang, S. Synthesis of potent chemical inhibitors of dynamin GTPase. Bioorg. Med. Chem. Lett. 2010, 20, 4858–4864. [Google Scholar] [CrossRef]
- McCluskey, A.; Daniel, J.A.; Hadzic, G.; Chau, N.; Clayton, E.L.; Mariana, A.; Whiting, A.; Gorgani, N.N.; Lloyd, J.; Quan, A.; et al. Building a better dynasore: The dyngo compounds potently inhibit dynamin and endocytosis. Traffic 2013, 14, 1272–1289. [Google Scholar] [CrossRef]
- Caboni, L.; Egan, B.; Kelly, B.; Blanco, F.; Fayne, D.; Meegan, M.J.; Lloyd, D.G. Structure-activity relationships in non-ligand binding pocket (non-LBP) diarylhydrazide antiandrogens. J. Chem. Inf. Model. 2013, 53, 2116–2130. [Google Scholar] [CrossRef] [PubMed]
- Buu-Hoi, N.P.; Xuong, N.D.; Nam, N.H.; Binon, F.; Royer, R. Tuberculostatic hydrazides and their derivatives. J. Chem. Soc. 1953, 1358–1364. [Google Scholar] [CrossRef]
- Caboni, L.; Gálvez-Llompart, M.; Gálvez, J.; Blanco, F.; Rubio-Martinez, J.; Fayne, D.; Lloyd, D.G. Molecular topology applied to the discovery of 1-benzyl-2-(3-fluorophenyl)-4-hydroxy-3-(3-phenylpropanoyl)-2H-pyrrole-5-one as a non-ligand-binding-pocket antiandrogen. J. Chem. Inf. Model. 2014, 54, 2953–2966. [Google Scholar] [CrossRef]
- Asthana, A.; Gaughan, C.; Dong, B.; Weiss, S.R.; Silverman, R.H. Specificity and mechanism of coronavirus, rotavirus, and mammalian two-histidine phosphodiesterases that antagonize antiviral innate immunity. mBio 2021, 12, e0178121. [Google Scholar] [CrossRef]
- Corona, A.; Meleddu, R.; Esposito, F.; Distinto, S.; Bianco, G.; Masaoka, T.; Maccioni, E.; Menéndez-Arias, L.; Alcaro, S.; Le Grice, S.F.; et al. Ribonuclease H/DNA polymerase HIV-1 reverse transcriptase dual inhibitor: Mechanistic studies on the allosteric mode of action of isatin-based compound RMNC6. PLoS ONE 2016, 11, e0147225. [Google Scholar] [CrossRef] [Green Version]
- Corona, A.; Schneider, A.; Schweimer, K.; Rösch, P.; Wöhrl, B.M.; Tramontano, E. Inhibition of foamy virus reverse transcriptase by human immunodeficiency virus type 1 RNase H inhibitors. Antimicrob. Agents Chemother. 2014, 58, 4086–4093. [Google Scholar] [CrossRef] [Green Version]
- Ogando, N.S.; El Kazzi, P.; Zevenhoven-Dobbe, J.C.; Bontes, B.W.; Decombe, A.; Posthuma, C.C.; Thiel, V.; Canard, B.; Ferron, F.; Decroly, E.; et al. Structure-function analysis of the nsp14 N7-guanine methyltransferase reveals an essential role in betacoronavirus replication. Proc. Natl. Acad. Sci. USA 2021, 118, e2108709118. [Google Scholar] [CrossRef]
- Wang, W.; Shin, W.-J.; Zhang, B.; Choi, Y.; Yoo, J.-S.; Zimmerman, M.I.; Frederick, T.E.; Bowman, G.R.; Gross, M.L.; Leung, D.W.; et al. The cap-snatching SFTSV endonuclease domain is an antiviral target. Cell Rep. 2020, 30, 153–163.e5. [Google Scholar] [CrossRef] [Green Version]
- Shin, W.-J.; Nam, K.-Y.; Kim, N.-D.; Kim, S.-H.; No, K.-T.; Seong, B.-L. Identification of a small benzamide inhibitor of influenza virus using a cell-based screening. Chemotherapy 2016, 61, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Hasipek, M.; Jiang, D.; Tiwari, A.D.; Grabowski, D.R.; Pagliuca, S.; Kongkiatkamon, S.; Patel, B.; Singh, S.; Parker, Y.; et al. Eltrombobag inhibits TET dioxygenase to contribute to hematopoietic stem cell expansion in aplastic anemia. J. Clin. Investig. 2022, 132, e149856. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Tiwari, A.D.; Phillips, J.G.; Hasipek, M.; Grabowski, D.R.; Pagliuca, S.; Gopal, P.; Kerr, C.M.; Adema, V.; Radivoyevitch, T.; et al. A therapeutic strategy for preferential targeting of TET2 mutant and TET-dioxygenase deficient cells in myeloid neoplasms. Blood Cancer Discov. 2021, 2, 146–161. [Google Scholar] [CrossRef] [PubMed]
- Imprachim, N.; Yosaatmadja, Y.; Newman, J.A. Crystal structures and fragment screening of SARS-CoV-2 NSP14 reveal details of exoribonuclease activation and mRNA capping and provide starting points for antiviral drug development. Nucleic Acids Res. 2023, 51, 475–487. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Shi, W.; Becker, S.T.; Schatz, D.G.; Liu, B.; Yang, Y. Structural basis of mismatch recognition by a SARS-CoV-2 proofreading enzyme. Science 2021, 373, 1142–1146. [Google Scholar] [CrossRef]
- Chen, Y.; Cai, H.; Pan, J.; Xiang, N.; Tien, P.; Ahola, T.; Guo, D. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc. Natl. Acad. Sci. USA 2009, 106, 3484–3489. [Google Scholar] [CrossRef]
- Masaoka, T.; Chung, S.; Caboni, P.; Rausch, J.W.; Wilson, J.A.; Taskent-Sezgin, H.; Beutler, J.A.; Tocco, G.; Le Grice, S.F.J. Exploiting drug-resistant enzymes as tools to identify thienopyrimidinone inhibitors of human immunodeficiency virus reverse transcriptase-associated ribonuclease H. J. Med. Chem. 2013, 56, 5436–5445. [Google Scholar] [CrossRef]
- Martín-Alonso, S.; Kang, D.; del Río, J.M.; Luczkowiak, J.; Frutos-Beltrán, E.; Zhang, L.; Cheng, X.; Liu, X.; Zhan, P.; Menéndez-Arias, L. Novel RNase H inhibitors blocking RNA-directed strand displacement DNA synthesis by HIV-1 reverse transcriptase. J. Mol. Biol. 2022, 434, 167507. [Google Scholar] [CrossRef]
- Zhang, L.; Wei, F.; Borrego, D.; Zhao, F.; del Río, J.M.; Frutos-Beltrán, E.; Zhang, J.; Xu, S.; López-Carrobles, N.; Gao, S.; et al. Design, synthesis, and biological evaluation of novel double-winged galloyl derivatives as HIV-1 RNase H inhibitors. Eur. J. Med. Chem. 2022, 240, 114563. [Google Scholar] [CrossRef]
- Kang, D.; Urhan, C.; Wei, F.; Frutos-Beltrán, E.; Sun, L.; Álvarez, M.; Feng, D.; Tao, Y.; Pannecouque, C.; De Clercq, E.; et al. Discovery, optimization, and target identification of novel coumarin derivatives as HIV-1 reverse transcriptase-associated ribonuclease H inhibitors. Eur. J. Med. Chem. 2021, 225, 113769. [Google Scholar] [CrossRef]
- Malet, H.; Williams, H.M.; Cusack, S.; Rosenthal, M. The mechanisms of genome replication and transcription in bunyaviruses. PLoS Pathog. 2023, 19, e1011060. [Google Scholar] [CrossRef]
- Bohn, P.; Waßmann, I.; Wendt, L.; Leske, A.; Hoenen, T.; Tews, B.A.; Groseth, A. A dsRNA-binding mutant reveals only a minor role of exonuclease activity in interferon antagonism by the arenavirus nucleoprotein. PLoS Pathog. 2023, 19, e1011049. [Google Scholar] [CrossRef]
- Cruz-González, A.; Munoz-Verlasco, I.; Cottom-Salas, W.; Becerra, A.; Campillo-Balderas, J.A.; Hernández-Morales, R.; Vázquez-Salazar, A.; Jácome, R.; Lazcano, A. Structural analysis of viral ExoN domains reveals polyphyletic hijacking events. PLoS ONE 2021, 16, e0246981. [Google Scholar] [CrossRef]
- Huang, K.-W.; Hsu, K.-C.; Chu, L.-Y.; Yang, J.-M.; Yuan, H.S.; Hsiao, Y.-Y. Identification of inhibitors for the DEDDh family of exonucleases and a unique inhibition mechanism by crystal structure analysis of CRN-4 bound with 2-morpholin-4-ylethanesulfonate (MES). J. Med. Chem. 2016, 59, 8019–8029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, M.; Guan, X. Aurintricarboxylic acid suppresses hepatitis B virus replication by inhibition of RNase H activity. Front. Virol. 2022, 2, 861494. [Google Scholar] [CrossRef]
- Thota, S.; Rodrigues, D.A.; de Sena Murteira Pinheiro, P.; Lima, L.M.; Fraga, C.A.M.; Barreiro, E.J. N-Acylhydrazones as drugs. Bioorg. Med. Chem. Lett. 2018, 28, 2797–2806. [Google Scholar] [CrossRef] [PubMed]
- Socea, L.-I.; Barbuceanu, S.-F.; Pahontu, E.M.; Dumitru, A.-C.; Nitulescu, G.M.; Sfetea, R.C.; Apostol, T.-V. Acylhydrazones and Their Biological Activity: A Review. Molecules 2022, 27, 8719. [Google Scholar] [CrossRef]
- Holm, K.J.; Spencer, C.M. Entacapone—A review of its use in Parkinson’s disease. Drugs 1999, 58, 159–177. [Google Scholar] [CrossRef]








| Compound | HIV-1 RNase H | Compound | HIV-1 RNase H |
|---|---|---|---|
| Dynasore | 3.10 ± 0.52 | 1i | 27.54 ± 2.65 |
| 1a | 69.56 ± 8.46 | 1j | >100 (91%) 2 |
| 1b | 7.16 ± 2.35 | 1k | 7.94 ± 2.17 |
| 1c | 0.57 ± 0.04 | 1l | >100 (70%) 2 |
| 1d | 35.95 ± 8.60 | 2a | 13.99 ± 0.97 |
| 1e | 1.02 ± 0.15 | 2b | 15.23 ± 1.35 |
| 1f | 0.93 ± 0.24 | 2c | 24.68 ± 3.89 |
| 1g | >100 (84%) 2 | RDS1643 | 10.8 ± 5.2 |
| 1h | 36.38 ± 10.46 |
| Virus | EC50 1 (μM) | |||
|---|---|---|---|---|
| 1c | 2a | 2b | 2c | |
| SARS-CoV-2 WA1/2020 | 10.23 ± 0.01 | 43.97 ± 0.51 | 35.30 ± 0.59 | >50 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Asthana, A.; Corona, A.; Shin, W.-J.; Kwak, M.-J.; Gaughan, C.; Tramontano, E.; Jung, J.U.; Schobert, R.; Jha, B.K.; Silverman, R.H.; et al. Analogs of the Catechol Derivative Dynasore Inhibit HIV-1 Ribonuclease H, SARS-CoV-2 nsp14 Exoribonuclease, and Virus Replication. Viruses 2023, 15, 1539. https://doi.org/10.3390/v15071539
Asthana A, Corona A, Shin W-J, Kwak M-J, Gaughan C, Tramontano E, Jung JU, Schobert R, Jha BK, Silverman RH, et al. Analogs of the Catechol Derivative Dynasore Inhibit HIV-1 Ribonuclease H, SARS-CoV-2 nsp14 Exoribonuclease, and Virus Replication. Viruses. 2023; 15(7):1539. https://doi.org/10.3390/v15071539
Chicago/Turabian StyleAsthana, Abhishek, Angela Corona, Woo-Jin Shin, Mi-Jeong Kwak, Christina Gaughan, Enzo Tramontano, Jae U. Jung, Rainer Schobert, Babal Kant Jha, Robert H. Silverman, and et al. 2023. "Analogs of the Catechol Derivative Dynasore Inhibit HIV-1 Ribonuclease H, SARS-CoV-2 nsp14 Exoribonuclease, and Virus Replication" Viruses 15, no. 7: 1539. https://doi.org/10.3390/v15071539