Comparison of the Structure of Soil Microbial Communities of Different Ecosystems Using the Microbiome Sequencing Approach
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site and Soil Sampling
2.2. Climate Conditions
2.3. Soil Agrochemical Analysis
2.4. Quantification of Cultivable Bacteria and Fungi
2.5. Soil DNA Extraction and Microbiomic Analysis
2.6. Bioinformatics Analysis
2.7. Statistical Analysis
3. Results
3.1. Soil Agrochemical Analysis
3.2. Quantification of Cultivable Bacteria and Fungi
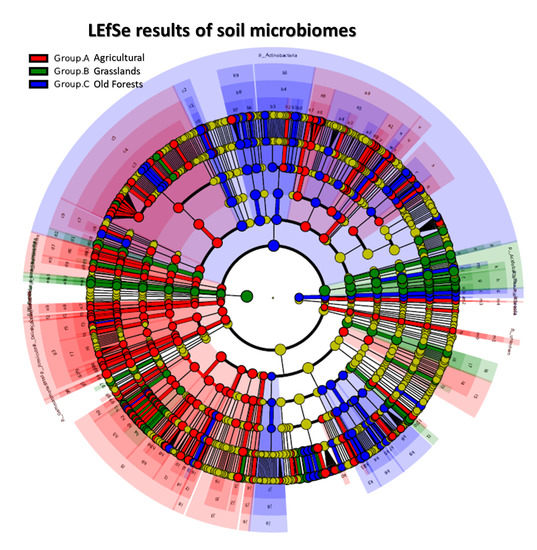
3.3. Soil Microbiomic Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Abatenh, E.; Gizaw, B.; Tsegaye, Z.; Tefera, G. Microbial Function on Climate Change—A Review. Open J. Environ. Biol. 2018, 3, 001–007. [Google Scholar] [CrossRef]
- Cavicchioli, R.; Ripple, W.J.; Timmis, K.N.; Azam, F.; Bakken, L.R.; Baylis, M.; Behrenfeld, M.J.; Boetius, A.; Boyd, P.W.; Classen, A.T.; et al. Scientists’ warning to humanity: Microorganisms and climate change. Nat. Rev. Microbiol. 2019, 17, 569–586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jansson, J.K.; Hofmockel, K.S. Soil microbiomes and climate change. Nat. Rev. Microbio. 2020, 18, 35–46. [Google Scholar] [CrossRef]
- Tiedje, J.M.; Bruns, M.A.; Casadevalli, A.; Criddle, C.S.; Eloe-Fadrosh, E.; Karl, D.M.; Nguyen, N.K.; Zhou, J. Microbes and Climate Change: A Research Prospectus for the Future. mBio 2022, 13, e0080022. [Google Scholar] [CrossRef]
- Balser, T.C.; Gutknecht, J.L.M.; Liang, C. How will climate change impact soil microbial communities? In Soil Microbiology and Sustainable Crop Production; Dixon, G.R., Tilston, E.L., Eds.; Springer Science + Business Media B.V.: Berlin/Heidelberg, Germany, 2010; pp. 373–397. [Google Scholar]
- Deng, J.; Yin, Y.; Luo, J.; Zhu, W.; Zhou, Y. Different revegetation types alter soil physical-chemical characteristics and fungal community in the Baishilazi Nature Reserve. PeerJ 2019, 6, e6251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, R.; Liu, J.; Wang, J.; Han, W.; Shen, Z.; Muraina, T.O.; Chen, J.; Sun, D. Comparison of soil microbial community between reseeding grassland and natural grassland in Songnen Meadow. Sci. Rep. 2020, 10, 16884. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Xu, N. Variations of soil bacterial microbial community and functional structure under different land-uses. Rev. Bras. Cienc. Solo 2022, 46, e0220090. [Google Scholar] [CrossRef]
- Tan, X.; Yanxia Nie, Y.; Xiaomin Ma, X.; Guo, Z.; Liu, Y.; Tian, H.; Megharaj, M.; Shen, W.; He, W. Soil chemical properties rather than the abundance of active and potentially active microorganisms control soil enzyme kinetics. Sci. Total Environ. 2021, 770, 144500. [Google Scholar] [CrossRef]
- He, Z.; Yuan, C.; Chen, P.; Rong, Z.; Peng, T.; Farooq, T.H.; Wang, G.; Yan, W.; Wang, J. Soil Microbial Community Composition and Diversity Analysis under Different Land Use Patterns in Taojia River Basin. Forests 2023, 14, 1004. [Google Scholar] [CrossRef]
- Kavitha, P.G.; Sudha, A.; Ahila Devi, P.; Kumaran, K. A Comparative Study on Forest Soil Microbial Diversity and Biomass in Nilgiri Biosphere of Southern India. Int. J. Curr. Microbiol. Appl. Sci. 2020, 9, 3701–3715. [Google Scholar] [CrossRef]
- Liu, L.; Zhu, K.; Wurzburger, N.; Zhang, J. Relationships between plant diversity and soil microbial diversity vary across taxonomic groups and spatial scales. Ecosphere 2020, 11, e02999. [Google Scholar] [CrossRef] [Green Version]
- Fu, Q.; Shao, Y.; Wang, S.; Liu, F.; Tian, G.; Chen, Y.; Yuan, Z.; Ye, Y. Soil Microbial Distribution Depends on Different Types of Landscape Vegetation in Temperate Urban Forest Ecosystems. Front. Ecol. Evol. 2022, 10, 858254. [Google Scholar] [CrossRef]
- World Bank Data on Agricultural Land. Available online: https://data.worldbank.org/indicator/AG.LND.AGRI.ZS (accessed on 26 March 2023).
- Lanz, B.; Dietz, S.; Swanson, T. The expansion of modern agriculture and global biodiversity decline: An integrated assessment. Ecol. Econ. 2018, 144, 260–277. [Google Scholar] [CrossRef]
- Dai, Z.; Sua, W.; Chenb, H.; Barberánc, A.; Zhaoa, H.; Yua, M.; Yua, L.; Brookesa, P.C.; Schadtb, C.W.; Changd, S.X.; et al. Long-term nitrogen fertilization decreases bacterial diversity and favors the growth of Actinobacteria and Proteobacteria in agro-ecosystems across the globe. Glob. Chang. Biol. 2018, 24, 3452–3461. [Google Scholar] [CrossRef]
- Global Forests Report 2020. Available online: https://www.cdp.net/en/research/global-reports/global-forests-report-2020 (accessed on 23 January 2023).
- Godfray, H.C.; Godfray, J.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; et al. Food security: The challenge of feeding 9 billion people. Science 2010, 327, 812–818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bahram, M.; Hildebrand, F.; Forslund, S.K.; Anderson, J.L.; Soudzilovskaia, N.A.; Bodegom, P.M.; Bengtsson-Palme, J.; Anslan, S.; Coelho, L.P.; Harend, H.; et al. Structure and function of the global topsoil microbiome. Nature 2018, 560, 233–237. [Google Scholar] [CrossRef] [Green Version]
- Maestre, F.T.; Delgado-Baquerizob, M.; Jeffriesb, T.C.; Eldridg, D.J.; Ochoa, V.; Gozalo, B.; Quero, J.L.; García-Gómez, M.; Gallardo, A.; Ulrich, W.; et al. Increasing aridity reduces soil microbial diversity and abundance in global drylands. Proc. Natl. Acad. Sci. USA 2015, 112, 15684–15689. [Google Scholar] [CrossRef]
- Jing, X.; Sanders, N.J.; Shi, Y.; Chu, H.; Classen, A.T.; Zhao, K.; Chen, L.; Shi, Y.; Jiang, Y.; He, J.S. The links between ecosystem multifunctionality and above- and belowground biodiversity are mediated by climate. Nat. Commun. 2015, 6, 8159. [Google Scholar] [CrossRef] [Green Version]
- Lithuanian Hydrometeorological Service Database. Available online: http://www.meteo.lt/en/ (accessed on 23 January 2023).
- ISO 10390:2005; Soil Quality—Determination of pH. ISO: Geneva, Switzerland, 2005. Available online: https://www.iso.org/standard/40879.html (accessed on 30 April 2023).
- Egnér, H.; Riehm, H.; Domingo, W.R. Investigations on soil chemical analysis as a basis of the evaluation of plant nutrient status of soils II. Chemical extraction methods for phosphorus and potassium determination. Lantbr. Ann. 1960, 26, 199–215. [Google Scholar]
- ISO 14256-2:2005; Soil Quality—Determination of Nitrate, Nitrite and Ammonium in Field-Moist Soils by Extraction with Potassium Chloride Solution. ISO: Geneva, Switzerland, 2005. Available online: https://www.iso.org/obp/ui/#iso:std:iso:14256:-2:ed-1:v1:en (accessed on 30 April 2023).
- ISO 10694:1995; Soil Quality—Determination of Organic and Total Carbon after Dry Combustion (Elementary Analysis). ISO: Geneva, Switzerland, 1995. Available online: https://www.iso.org/standard/18782.html (accessed on 30 April 2023).
- Küster, E. Outline of a comparative study of criteria used in characterization of the actinomycetes. Int. Bull. Bacteriol. Nomencl. Taxon. 1959, 9, 98–104. [Google Scholar] [CrossRef] [Green Version]
- Aquilanti, L.; Favilli, F.; Clemeti, F. Comparison of different strategies for isolation and preliminary identification of Azotobacter from soil samples. Soil Biol. Biochem. 2004, 36, 1475–1483. [Google Scholar] [CrossRef]
- Carter, M.R.; Gregorich, E.G. Soil Sampling and Methods of Analysis; CRC Press: Boka Raton, FL, USA, 2007; pp. 342–351. [Google Scholar]
- Nelson, P.E.; Toussoun, T.A.; Marasas, W.F.O. Fusarium Species: An Illustrated Manual for Identification; Penn State University Press: University Park, PA, USA, 1990. [Google Scholar]
- Watanabe, T. Pictorial Atlas of Soil and Seed Fungi/Morphologies of Cultured Fungi and Key to Species; CRC Press: Boca Raton, FL, USA, 2002. [Google Scholar]
- Domsch, K.H.; Gams, W.; Anderson, T.H. Compendium of Soil Fungi; IHW-Verlag: Eching, Germany, 2007; p. 672. [Google Scholar]
- Samson, R.A.; Visagie, C.M.; Houbraken, J.; Hong, S.B.; Hubka, V.; Klaassen, C.H.W.; Perrone, G.; Seifert, K.A.; Susca, A.; Tanney, J.B.; et al. Phylogeny, identification and nomenclature of the genus Aspergillus. Stud. Mycol. 2014, 78, 141–173. [Google Scholar] [CrossRef] [Green Version]
- Visagie, C.M.; Houbraken, J.; Frisvad, J.C.; Hong, S.B.; Klaassen, C.H.W.; Perrone, G.; Seifert, K.A.; Varga, J.; Yaguchi, T.; Samsom, R.A. Identification and nomenclature of the genus Penicillium. Stud. Mycol. 2014, 78, 343–371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Kim, D.; Song, L.; Breitwieser, F.P.; Salzberg, S.L. Centrifuge: Rapid and sensitive classification of metagenomic sequences. Genome Res. 2016, 12, 1721–1729. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mandal, A.; Neenu, S. Impact of climate change on soil biodiversity—A review. Agri. Rev. 2012, 33, 283–292. [Google Scholar]
- Wasak, K.; Drewnik, M. Land use effects on soil organic carbon sequestration in calcareous Leptosols in former pastureland—A case study from the Tatra Mountains (Poland). Solid Earth 2015, 6, 1103–1115. [Google Scholar] [CrossRef] [Green Version]
- Martens, D.A.; Reedy, T.R.; Lewis, D.T. Soil organic carbon content and composition of 130-year crop, pasture and forest land-use managements. Glob. Chang. Biol. 2004, 10, 65–78. [Google Scholar] [CrossRef] [Green Version]
- McKinley, D.C.; Rice, C.R.; Blair, J.M. Conversion of grassland to coniferous woodland has limited effects on soil nitrogen cycle processes. Soil Biol. Biochem. 2008, 40, 2627–2633. [Google Scholar] [CrossRef]
- Podwika, M.; Solek-Podwika, K.; Ciarkowska, K. Changes in the properties of grassland soils as a result of afforestation. IForest 2018, 11, 600–608. [Google Scholar] [CrossRef] [Green Version]
- Furtak, K.; Gajda, A.M. Activity and Variety of Soil Microorganisms Depending on the Diversity of the Soil Tillage System. Sustainability of Agroecosystems. In Sustainability of Agroecosystems; de Oliveira, A.B., Ed.; IntechOpen: London, UK, 2018. [Google Scholar]
- Maraha, N.; Backman, A.; Jansson, J.K. Monitoring physiological status of GFP-tagged Pseudomonas fluorescens SBW25 under different nutrient conditions and in soil by flow cytometry. FEMS Microbiol. Ecol. 2004, 51, 123–132. [Google Scholar] [CrossRef] [Green Version]
- Caracciolo, A.B.; Fajardo, C.; Grenni, P.; Saccà, M.L.; Amalfitano, S.; Ciccoli, R.; Martin, M.; Gibello, A. The role of a groundwater bacterial community in the degradation of the herbicide terbuthylazine. FEMS Microbiol. Ecol. 2009, 71, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Busse, M.D.; Sanchez, F.G.; Ratcliff, A.W.; Butnor, J.R.; Carter, E.A.; Powers, R.F. Soil carbon sequestration and changes in fungal and bacterial biomass following incorporation of forest residues. Soil Biol. Biochem. 2009, 41, 220–227. [Google Scholar] [CrossRef] [Green Version]
- Blagodatskaya, E.; Kuzyakov, Y. Active microorganisms in soil: Critical review of estimation criteria and approaches. Soil Biol. Biochem. 2013, 67, 192–211. [Google Scholar] [CrossRef]
- Bakšienė, E.; Nedzinskienė, T.L.; Ražukas, A.; Salina, O.; Repečkienė, J. Efficiency of various farming systems on an infertile soil. Zemdirb. Agric. 2009, 96, 47–61. [Google Scholar]
- Mikkelson, K.M.; Bearup, L.A.; Maxwell, R.M.; Stednick, J.D.; McCray, J.E.; Sharp, J.O. Bark beetle infestation impacts on nutrient cycling, water quality and interdependent hydrological effects. Biogeochemistry 2013, 115, 1–21. [Google Scholar] [CrossRef]
- Kacergius, A.; Sivojiene, D. Microbial diversity and abundance in loamy sandy soil under renaturalization of former arable land. PeerJ 2023, 11, e14761. [Google Scholar] [CrossRef]
- Ren, C.; Sun, P.; Kang, D.; Zhao, F.; Feng, Y.; Ren, G.; Han, X.; Yang, G. Responsiveness of soil nitrogen fractions and bacterial communities to afforestation in the Loess Hilly Region (LHR) of China. Sci. Rep. 2016, 6, 28469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.; Zhang, Y.; Tang, Z.; Shangguan, Z.; Chang, F.; Jia, F.; Chen, Y.; He, X.; Shi, W.; Deng, L. Effects of grassland afforestation on structure and function of soil bacterial and fungal communities. Sci. Total Environ. 2019, 676, 396–406. [Google Scholar] [CrossRef] [PubMed]
- García-Orenes, F.; Morugán-Coronado, A.; Zornoza, R.; Scow, K. Changes in Soil Microbial Community Structure Influenced by Agricultural Management Practices in a Mediterranean Agro-Ecosystem. PLoS ONE 2013, 8, e80522. [Google Scholar] [CrossRef] [PubMed]








| Group | Sample | Description |
|---|---|---|
| Agricultural | SG-1/22 | Agricultural field (used as the control (without fertilizer—N0P0P0). Sample from the field of a long-term experiment. Soil type: Sandy Loam Haplic Luvisol. (54.609902, 25.124873). |
| SG-7/22 | Agricultural field fertilized with granulated poultry manure (N170) + biological additive Trichoderma spp. The rate of organic fertilizer was calculated based on 170 kg ha−1 of nitrogen-active substance. Soil type: Sandy Loam Haplic Luvisol. (54.609879, 25.124819). | |
| SG-13/22 | Agricultural field fertilized with granulated poultry manure (N85) + mineral fertilizers (N60 *). The rate of organic fertilizer was calculated based on 85 kg ha−1 of nitrogen-active substance. Soil type: Sandy Loam Haplic Luvisol. (54.609857, 25.124779). | |
| SG-14/22 | Agricultural field fertilized with granulated cattle manure (N85) + mineral fertilizers (N60 *). The rate of organic fertilizer was calculated based on 85 kg ha−1 of nitrogen-active substance. Soil type: Sandy Loam Haplic Luvisol. (54.609840, 25.124760). | |
| Grasslands | SG-17/22 | Natural grassland nearby forest in Neris Regional Reserve Park. Natura 2000 area 6120 Grassland of carbonate sands. Soil type: Sandy Haplic Arenosol. (54.848614, 24.883169). |
| SG-18/22 | Natural grassland in Neris Regional Reserve Park. Natura 2000 area 6210 Steppe grassland. Soil type: Sandy Eutric Regosol. (54.846232, 24.878042). | |
| SG-20/22 | Natural grassland (not flooded) near to the Minija River. Soil type: Sandy Eutric Fluvisol. (55.727205, 21.414129). | |
| SG-21/22 | Natural grassland (not flooded) near to the Dubysa River. Soil type: Sandy loam Haplic Arenosol. (55.237039, 23.517158). | |
| Old Forests | SG-15/22 | Old-growth coniferous forest in Neris Regional Reserve Park. Natura 2000 area 9060 Coniferous Forest on fluvioglacials. Soil type: Sandy Haplic Arenosol. (54.861738, 24.874491). |
| SG-16/22 | Old-growth coniferous forest in Neris Regional Reserve Park. Natura 2000 area 9010 Western Taiga. Soil type: Sandy Eutric Regosol. (54.844590, 24.871234). | |
| SG-19/22 | Old-growth forest in the Curonian Spit Parabolic dunes area. Soil type: Sandy Haplic Arenosol. (55.515580, 21.113550). | |
| SG-22/22 | Old growth mixed forest nearby glacial geological formation, “Devil’s pit”—a funnel-shaped pit in width ~200 m, depth 30–40 m. Soil type: Sandy loam Haplic Arenosol. (54.612031, 24.515918). |
| Group | Sample | pHKCl | Mobile Phosphorus (P2O5; mg kg−1) | Mobile Potassium (K2O; mg kg−1) | Organic Carbon (Corg; %) | Mineral Nitrogen (Nmin, mg kg−1) |
|---|---|---|---|---|---|---|
| Agricultural | SG-1/22 | 5.9 | 237 | 106 | 1.24 | 3.13 |
| SG-7/22 | 5.8 | 294 | 151 | 1.69 | 6.00 | |
| SG-13/22 | 5.7 | 271 | 139 | 1.38 | 5.54 | |
| SG-14/22 | 5.7 | 268 | 149 | 1.36 | 4.91 | |
| Grasslands | SG-17/22 | 5.7 | 163 | 143 | 1.65 | 6.13 |
| SG-18/22 | 6.7 | 481 | 127 | 1.39 | 12.08 | |
| SG-20/22 | 7.2 | 58 | 129 | 2.19 | 9.16 | |
| SG-21/22 | 6.7 | 70 | 123 | 3.50 | 5.09 | |
| Old Forests | SG-15/22 | 4.0 | 164 | 16 | 0.84 | 3.17 |
| SG-16/22 | 3.3 | 41 | 25 | 0.99 | 2.93 | |
| SG-19/22 | 4.0 | 101 | 24 | 0.68 | 1.65 | |
| SG-22/22 | 4.6 | 63 | 53 | 2.15 | 4.18 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kačergius, A.; Sivojienė, D.; Gudiukaitė, R.; Bakšienė, E.; Masevičienė, A.; Žičkienė, L. Comparison of the Structure of Soil Microbial Communities of Different Ecosystems Using the Microbiome Sequencing Approach. Soil Syst. 2023, 7, 70. https://doi.org/10.3390/soilsystems7030070
Kačergius A, Sivojienė D, Gudiukaitė R, Bakšienė E, Masevičienė A, Žičkienė L. Comparison of the Structure of Soil Microbial Communities of Different Ecosystems Using the Microbiome Sequencing Approach. Soil Systems. 2023; 7(3):70. https://doi.org/10.3390/soilsystems7030070
Chicago/Turabian StyleKačergius, Audrius, Diana Sivojienė, Renata Gudiukaitė, Eugenija Bakšienė, Aistė Masevičienė, and Lina Žičkienė. 2023. "Comparison of the Structure of Soil Microbial Communities of Different Ecosystems Using the Microbiome Sequencing Approach" Soil Systems 7, no. 3: 70. https://doi.org/10.3390/soilsystems7030070