Associations of Leaf Spectra with Genetic and Phylogenetic Variation in Oaks: Prospects for Remote Detection of Biodiversity
Abstract
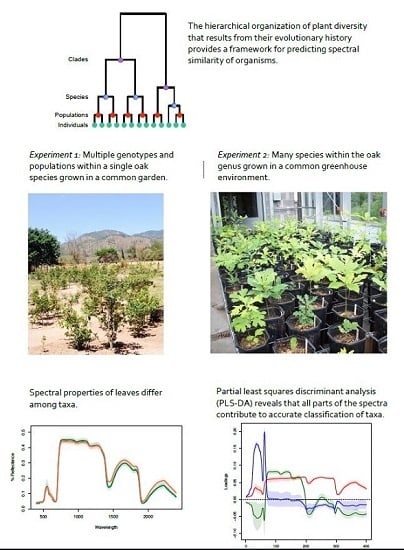
:1. Introduction
- Do closely related species tend to have similar spectral profiles?
- Can leaf spectra be used to detect variation: (i) among populations within a single species; (ii) among species within clades; and (iii) among clades? Where in the biological hierarchy are spectral signals best able to differentiate taxa?
- Does statistical comparison of high dimensional spectral data allow greater accuracy in detection of phylogenetic differences among taxa than estimates of functional traits derived from spectra?
2. Materials and Methods
2.1. Experimental System 1: Population-Level Variation within a Single Species (Tropical Live Oak, Quercus oleoides) in a Field Experiment in Zamorano, Honduras
2.2. Experimental System 2: Greenhouse Experiment with 28 Oak Species from a Range of Geographic Regions and Climatic Zones across North and Central America Grown in a Controlled Environment
2.3. Phylogenetic Information
2.4. Statistical Analyses
3. Results
3.1. Detection of Population-Level Variation within a Single Species
3.2. Detection of Variation among Species at the Phylogenetic Level
3.3. Comparison of Model Accuracy for Traits and Spectra across the Biological Hierarchy of Diversity
3.4. Most Informative Regions of the Spectra and Traits
4. Discussion
Caveats and Limits
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Cavender-Bares, J.; Heffernan, J.; King, E.; Polasky, S.; Balvanera, P.; Clark, W.C. Sustainability and biodiversity. In Encyclopedia of Biodiversity, 2nd ed.; Levin, S.E., Ed.; Academic Press: Amsterdam, The Netherlands, 2013; pp. 71–84. [Google Scholar]
- Díaz, S.; Demissew, S.; Carabias, J.; Joly, C.; Lonsdale, M.; Ash, N.; Larigauderie, A.; Adhikari, J.R.; Arico, S.; Báldi, A.; Bartuska, A.; et al. The IPBES conceptual framework—Connecting nature and people. Curr. Opin. Environ. Sustain. 2015, 14, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Balvanera, P.; Pfisterer, A.B.; Buchmann, N.; He, J.-S.; Nakashizuka, T.; Raffaelli, D.; Schmid, B. Quantifying the evidence for biodiversity effects on ecosystem functioning and services. Ecol. Lett. 2006, 9, 1146–1156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cavender-Bares, J.; Balvanera, P.; King, E.; Polasky, S. Ecosystem service trade-offs across global contexts and scales. Ecol. Soc. 2015, 20, 22. [Google Scholar] [CrossRef]
- Jetz, W.; Cavender-Bares, J.; Pavlick, R.; Schimel, D.; Davis, F.W.; Asner, G.P.; Guralnick, R.; Kattge, J.; Latimer, A.M.; Moorcroft, P.; Schaepman, M.E.; et al. Monitoring plant functional diversity from space. Nat. Plants 2016, 2. [Google Scholar] [CrossRef]
- Turner, W. Sensing biodiversity. Science 2014, 346, 301–302. [Google Scholar] [CrossRef] [PubMed]
- Skidmore, A.K.; Pettorelli, N.; Coops, N.C.; Geller, G.N.; Hansen, M.; Lucas, R.; Mücher, C.A.; O’Connor, B.; Paganini, M.; Pereira, H.M.; et al. Environmental science: Agree on biodiversity metrics to track from space. Nature 2015, 523, 403–405. [Google Scholar] [CrossRef] [PubMed]
- Schimel, D.; Pavlick, R.; Fisher, J.B.; Asnor, G.; Saatchi, S.; Townsend, P.; Miller, C.; Frankenberg, C.; Hibbard, K.; Cox, P. Observing terrestrial ecosystems and the carbon cycle from space. Glob. Chang. Biol. 2015, 21, 1762–1776. [Google Scholar] [CrossRef] [PubMed]
- National Research Council. Earth Science and Applications from Space: National Imperatives for the Next Decade and Beyond; The National Academies Press: Washington, DC, USA, 2007. [Google Scholar]
- Reich, P.B.; Wright, I.J.; Cavender-Bares, J.; Craine, J.M.; Oleksyn, J.; Westoby, M.; Walters, M.B. The evolution of plant functional variation: Traits, spectra, and strategies. Int. J. Plant Sci. 2003, 164, S143–S164. [Google Scholar] [CrossRef]
- Reich, P.B. The world-wide “fast–slow” plant economics spectrum: A traits manifesto. J. Ecol. 2014, 102, 275–301. [Google Scholar] [CrossRef]
- Wright, I.J.; Reich, P.B.; Westoby, M.; Ackerly, D.D.; Baruch, Z.; Bongers, F.; Cavender-Bares, J.; Chapin, T.; Cornelissen, J.H.C.; Diemer, M.; et al. The worldwide leaf economics spectrum. Nature 2004, 428, 821–827. [Google Scholar] [CrossRef] [PubMed]
- Cavender-Bares, J.; Kitajima, K.; Bazzaz, F.A. Multiple trait associations in relation to habitat differentiation among 17 Floridian oak species. Ecol. Monogr. 2004, 74, 635–662. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E.; Carranza-Jimenez, L.; Sinca, F.; Tupayachi, R.; Anderson, C.B.; Martinez, P. Functional and biological diversity of foliar spectra in tree canopies throughout the Andes to Amazon region. New Phytol. 2014, 204, 127–139. [Google Scholar] [CrossRef] [PubMed]
- Gamon, J.; Surfus, J. Assessing leaf pigment content and activity with a reflectometer. New Phytol. 1999, 143, 105–117. [Google Scholar] [CrossRef]
- Couture, J.; Serbin, S.; Townsend, P. Spectroscopic sensitivity of real-time, rapidly induced phytochemical change in response to damage. New Phytol. 2013, 198, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Townsend, P.; Serbin, S.; Kruger, E.; Gamon, J. Disentangling the contribution of biological and physical properties of leaves and canopies in imaging spectroscopy data. Proc. Natl. Acad. Sci. USA 2013, 110, E1074–E1074. [Google Scholar] [CrossRef] [PubMed]
- Serbin, S.P.; Singh, A.; McNeil, B.E.; Kingdon, C.C.; Townsend, P.A. Spectroscopic determination of leaf morphological and biochemical traits for northern temperate and boreal tree species. Ecol. Appl. 2014, 7, 1651–1669. [Google Scholar] [CrossRef]
- Ustin, S.L.; Gitelson, A.A.; Jacquemoud, S.; Schaepman, M.E.; Asner, G.P.; Gamon, J.A.; Zarco-Tejada, P. Retrieval of foliar information about plant pigment systems from high resolution spectroscopy. Remote Sens. Environ. 2009, 113, S67–S77. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.; Serbin, S.P.; McNeil, B.E.; Kingdon, C.C.; Townsend, P.A. Imaging spectroscopy algorithms for mapping canopy foliar chemical and morphological traits and their uncertainties. Ecol. Appl. 2015. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E. Airborne spectranomics: Mapping canopy chemical and taxonomic diversity in tropical forests. Front. Ecol. Environ. 2009, 7. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E.; Tupayachi, R.; Anderson, C.B.; Sinca, F.; Carranza-Jiménez, L.; Martinez, P. Amazonian functional diversity from forest canopy chemical assembly. Proc. Natl. Acad. Sci. USA 2014, 11, 5604–5609. [Google Scholar] [CrossRef] [PubMed]
- Féret, J.B.; Asner, G.P. Microtopographic controls on lowland Amazonian canopy diversity from imaging spectroscopy. Ecol. Appl. 2014, 24, 1297–1310. [Google Scholar] [CrossRef]
- Féret, J.B.; Asner, G.P. Mapping tropical forest canopy diversity using high-fidelity imaging spectroscopy. Ecol. Appl. 2014, 24, 1289–1296. [Google Scholar] [CrossRef]
- White, J.C.; Gómez, C.; Wulder, M.A.; Coops, N.C. Characterizing temperate forest structural and spectral diversity with Hyperion EO-1 data. Remote Sens. Environ. 2010, 114, 1576–1589. [Google Scholar] [CrossRef]
- Hulshof, C.M.; Swenson, N.G. Variation in leaf functional trait values within and across individuals and species: An example from a Costa Rican dry forest. Funct. Ecol. 2010, 24, 217–223. [Google Scholar] [CrossRef]
- Koehler, K.; Center, A.; Cavender-Bares, J. Evidence for a freezing tolerance—Growth rate trade-off in the live oaks (Quercus series Virentes) across the tropical-temperate divide. New Phytol. 2012, 193, 730–744. [Google Scholar] [CrossRef] [PubMed]
- Silvertown, J.; Dodd, M.; Gowing, D.; Lawson, C.; McConway, K. Phylogeny and the hierarchical organization of plant diversity. Ecology 2006, 87, S39–S49. [Google Scholar] [CrossRef]
- Donoghue, M.J. A phylogenetic perspective on the distribution of plant diversity. Proc. Natl. Acad. Sci. USA 2008, 105 (Suppl. 1), 11549–11555. [Google Scholar] [CrossRef] [PubMed]
- Wiens, J.J.; Ackerly, D.D.; Allen, A.P.; Anacker, B.L.; Buckley, L.B.; Cornell, H.V.; Damschen, E.I.; Davies, T.J.; Grytnes, J.-A.; Harrison, S.P.; et al. Niche conservatism as an emerging principle in ecology and conservation biology. Ecol. Lett. 2010, 13, 1310–1324. [Google Scholar] [CrossRef] [PubMed]
- Butler, M.; King, A.A. Phylogenetic comparative analysis: A modeling approach for adaptive evolution. Am. Nat. 2004, 164, 683–695. [Google Scholar] [CrossRef]
- Ackerly, D.D. Evolution, origin and age of lineages in the Californian and Mediterranean floras. J. Biogeogr. 2009, 36, 1221–1233. [Google Scholar] [CrossRef]
- Cavender-Bares, J.; Ackerly, D.D.; Baum, D.A.; Bazzaz, F.A. Phylogenetic overdispersion in Floridian oak communities. Am. Nat. 2004, 163, 823–43. [Google Scholar] [CrossRef] [PubMed]
- Zanne, A.E.; Tank, D.C.; Cornwell, W.K.; Eastman, J.M.; Smith, S.A.; FitzJohn, R.G.; McGlinn, D.J.; O’Meara, B.C.; Moles, A.T.; Reich, P.B.; et al. Three keys to the radiation of angiosperms into freezing environments. Nature 2014, 506, 89–92. [Google Scholar] [CrossRef] [PubMed]
- Cavender-Bares, J.; Gonzalez-Rodriguez, A.; Pahlich, A.; Koehler, K.; Deacon, N. Phylogeography and climatic niche evolution in live oaks (Quercus series Virentes) from the tropics to the temperate zone. J. Biogeogr. 2011, 38, 962–981. [Google Scholar] [CrossRef]
- Cavender-Bares, J.; Gonzalez-Rodriguez, A.; Eaton, D.A.R.; Hipp, A.A.L.; Beulke, A.; Manos, P.S. Phylogeny and biogeography of the American live oaks (Quercus subsection Virentes): A genomic and population genetics approach. Mol. Ecol. 2015, 24, 3668–3687. [Google Scholar] [CrossRef] [PubMed]
- Klemens, J.A.; Deacon, N.J.; Cavender-Bares, J. Pasture recolonization by a tropical oak and the regeneration ecology of seasonally dry tropical forests. In Seasonally Dry Tropical Forests; Island Press/Center for Resource Economics: Washington, DC, USA, 2011; pp. 221–237. [Google Scholar]
- Serbin, S. Spectroscopic Determination of Leaf Nutritional, Morphological, and Metabolic Traits. Ph.D. Thesis, The University of Wisconsin, Madison, WI, USA, 2012. [Google Scholar]
- Gao, B. NDWI–a normalized difference water index for remote sensing of vegetation liquid water from space. Remote Sens. Environ. 1996, 58, 257–266. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Merzlyak, M.N. Spectral reflectance changes associate with autumn senescence of Aesculus hippocastanum L. and Acer platanoides L. leaves. Spectral features and relation to chlorophyll estimation. J. Plant Physiol. 1994, 143, 286–292. [Google Scholar] [CrossRef]
- Gamon, J.A.; Penuelas, J.; Field, C.B. A narrow-waveband spectral index that tracks diurnal changes in photosynthetic efficiency. Remote Sens. Environ. 1992, 41, 35–44. [Google Scholar] [CrossRef]
- Cavender-Bares, J.; Cortes, P.; Rambal, S.; Joffre, R.; Miles, B.; Rocheteau, A. Summer and winter sensitivity of leaves and xylem to minimum freezing temperatures: a comparison of cooccurring Mediterranean oaks that differ in leaf lifespan. New Phytol. 2005, 168, 597–612. [Google Scholar] [CrossRef] [PubMed]
- Cavender-Bares, J.; Holbrook, N.M. Hydraulic properties and freezing-induced cavitation in sympatric evergreen and deciduous oaks with, contrasting habitats. Plant Cell Environ. 2001, 24, 1243–1256. [Google Scholar] [CrossRef]
- Hipp, A.L.; Eaton, D.A.; Cavender-Bares, J.; Fitzek, E.; Nipper, R.; Manos, P.S. A framework phylogeny of the american oak clade based on sequenced RAD data. PLoS ONE 2014, 9, e93975. [Google Scholar]
- Aldrich, P.R.; Cavender-Bares, J. Quercus. In Wild Crop Relatives: Genomic and Breeding Resources; Springer: Berlin, Germany, 2011; pp. 89–129. [Google Scholar]
- Manos, P.S.; Zhou, Z.K.; Cannon, C.H. Systematics of fagaceae: Phylogenetic tests of reproductive trait evolution. Int. J. Plant Sci. 2001, 162, 1361–1379. [Google Scholar] [CrossRef]
- Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Blomberg, S.P.; Garland, T.; Ives, A.R. Testing for phylogenetic signal in comparative data: Behavioral traits are more labile. Evolution 2003, 57, 717–745. [Google Scholar] [CrossRef] [PubMed]
- Revell, L.J. Phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 2012, 3, 217–223. [Google Scholar] [CrossRef]
- Chevallier, S.; Bertrand, D.; Kohler, A.; Courcoux, P. Application of PLS-DA in multivariate image analysis. J. Chemom. 2006, 20, 221–229. [Google Scholar] [CrossRef]
- Castillo, R.; Otto, M.; Freer, J.; Valenzuela, S. Multivariate strategies for classification of Eucalyptus globulus genotypes using carbohydrates content and NIR spectra for evaluation of their cold resistance. J. Chemom. 2008, 22, 268–280. [Google Scholar] [CrossRef]
- Williams, P.; Geladi, P.; Britz, T.; Manley, M. Near-infrared (NIR) hyperspectral imaging and multivariate image analysis to study growth characteristics and differences between species and strains of members of the genus Fusarium. Anal. Bioanal. Chem. 2012, 404, 1759–1769. [Google Scholar] [CrossRef] [PubMed]
- Madritch, M.D.; Kingdon, C.C.; Singh, A.; Mock, K.E.; Lindroth, R.L.; Townsend, P.A. Imaging spectroscopy links aspen genotype with below-ground processes at landscape scales. Philos. Trans. R. S. Lond. B Biol. Sci. 2014, 369, 20130194. [Google Scholar] [CrossRef] [PubMed]
- Wiklund, S.; Johansson, E.; Sjoestroem, L.; Mellerowicz, E.J.; Edlund, U.; Shockcor, J.P.; Gottfries, J.; Moritz, T.; Trygg, J. Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Anal. Chem. 2008, 80, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Gavaghan, C.L.; Wilson, I.D.; Nicholson, J.K. Physiological variation in metabolic phenotyping and functional genomic studies: use of orthogonal signal correction and PLS-DA. FEBS Lett. 2002, 530, 191–196. [Google Scholar] [CrossRef]
- Center, A. Physiological and fitness consequences of seasonal rainfall variation in neotropical live oak seedlings (Quercus oleoides): implications for global change. In Department of Plant Biology; University of Minnesota: Minneapolis, MN, USA, 2015; p. 116. [Google Scholar]
- Knyazikhin, Y.; Schull, M.A.; Stenberg, P.; Mõttus, M.; Rautiainen, M.; Yang, Y.; Marshak, A.; Carmona, P.L.; Kaufmann, R.K.; Lewis, P.; et al. Hyperspectral remote sensing of foliar nitrogen content. Proc. Natl. Acad. Sci. USA 2013, 110, E185–E192. [Google Scholar] [CrossRef] [PubMed]
- Ustin, S.L.; Gamon, J.A. Remote sensing of plant functional types. New Phytol. 2010, 186, 795–816. [Google Scholar] [CrossRef] [PubMed]
- Baret, F. Use of reflectance spectral variation to retrieve canopy biophysical characteristics. In Advances in Environmental Remote Sensing; Danson, F., Plummer, S., Eds.; John Wiley: Chichester, UK, 1994; pp. 33–51. [Google Scholar]
- Harmon, L.J.; Losos, J.B.; Jonathan Davies, T.; Gillespie, R.G.; Gittleman, J.L.; Bryan Jennings, W.; Kozak, K.H.; McPeek, M.A.; Moreno-Roark, F.; Near, T.J.; et al. Early bursts of body size and shape evolution are rare in comparative data. Evolution 2010, 64, 2385–2396. [Google Scholar] [CrossRef] [PubMed]
- Kozak, K.H.; Wiens, J.J. Accelerated rates of climatic-niche evolution underlie rapid species diversification. Ecol. Lett. 2010, 13, 1378–1389. [Google Scholar] [CrossRef] [PubMed]
- Uyeda, J.C.; Caetano, D.S.; Pennell, M.W. Comparative analysis of principal components can be misleading. Syst. Biol. 2015, 64, 677–689. [Google Scholar] [CrossRef] [PubMed]
- Cybis, G.B.; Sinsheimer, J.S.; Bedford, T.; Mather, A.E.; Lemey, P.; Suchard, M.A. Assessing phenotypic correlation through the multivariate phylogenetic latent liability model. Ann. Appl. Stat. 2015, 9, 969–991. [Google Scholar] [CrossRef]





| Classification Unit | Spectra | Traits | ||
|---|---|---|---|---|
| Kappa: Mean (SD) | # Components | Kappa: Mean (SD) | # Components | |
| Population | 0.344 (0.042) | 29 | 0.222 (0.033) | 7 |
| Species | 0.61 (0.027) | 60 | 0.271 (0.021) | 8 |
| Clade | 0.813 (0.025) | 35 | 0.516 (0.028) | 5 |
| Leaf type | 0.365 (0.046) | 44 | 0.13 (0.044) | 8 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cavender-Bares, J.; Meireles, J.E.; Couture, J.J.; Kaproth, M.A.; Kingdon, C.C.; Singh, A.; Serbin, S.P.; Center, A.; Zuniga, E.; Pilz, G.; et al. Associations of Leaf Spectra with Genetic and Phylogenetic Variation in Oaks: Prospects for Remote Detection of Biodiversity. Remote Sens. 2016, 8, 221. https://doi.org/10.3390/rs8030221
Cavender-Bares J, Meireles JE, Couture JJ, Kaproth MA, Kingdon CC, Singh A, Serbin SP, Center A, Zuniga E, Pilz G, et al. Associations of Leaf Spectra with Genetic and Phylogenetic Variation in Oaks: Prospects for Remote Detection of Biodiversity. Remote Sensing. 2016; 8(3):221. https://doi.org/10.3390/rs8030221
Chicago/Turabian StyleCavender-Bares, Jeannine, Jose Eduardo Meireles, John J. Couture, Matthew A Kaproth, Clayton C. Kingdon, Aditya Singh, Shawn P. Serbin, Alyson Center, Esau Zuniga, George Pilz, and et al. 2016. "Associations of Leaf Spectra with Genetic and Phylogenetic Variation in Oaks: Prospects for Remote Detection of Biodiversity" Remote Sensing 8, no. 3: 221. https://doi.org/10.3390/rs8030221