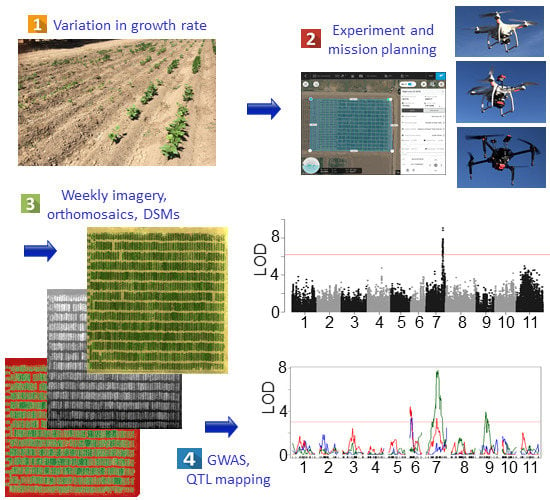
Determining the Genetic Control of Common Bean Early-Growth Rate Using Unmanned Aerial Vehicles
Abstract
:1. Introduction
2. Material and Methods
2.1. Plant Materials
2.2. Phenotyping
2.3. Genotyping and Genetic Mapping
3. Results
3.1. Growth Measurements
3.2. Genome-Wide Association Studies
3.3. Biparental Population Phenotyping
3.4. Linkage Map Development and QTL Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Oerke, E.C. Crop losses to pests. J. Agric. Sci. 2005, 144, 31–43. [Google Scholar] [CrossRef]
- Lammerts van Bueren, E.T.; Jones, S.S.; Tamm, L.; Murphy, K.M.; Myers, J.R.; Leifert, C.; Messmer, M.M. The need to breed crop varieties suitable for organic farming, using wheat, tomato and broccoli as examples: A review. NJAS Wagening. J. Life Sci. 2011, 58, 193–205. [Google Scholar] [CrossRef]
- Place, G.T.; Reberg-Horton, S.C.; Carter, T.E.; Brinton, S.R.; Smith, A.N. Screening tactics for identifying competitive soybean genotypes. Commun. Soil. Sci. Plan. 2011, 42, 2654–2665. [Google Scholar] [CrossRef]
- Manalil, S.; Coast, O.; Werth, J.; Chauhan, B.S. Weed management in cotton (Gossypium hirsutum L.) through weed-crop competition: A review. Crop Prot. 2017, 95, 53–59. [Google Scholar] [CrossRef] [Green Version]
- Dass, A.; Shekhawat, K.; Choudhary, A.K.; Sepat, S.; Rathore, S.S.; Mahajan, G.; Chauhan, B.S. Weed management in rice using crop competition—A review. Crop Prot. 2017, 95, 45–52. [Google Scholar] [CrossRef]
- Andrew, I.K.S.; Storkey, J.; Sparkes, D.L. A review of the potential for competitive cereal cultivars as a tool in integrated weed management. Weed Res. 2015, 55, 239–248. [Google Scholar] [CrossRef]
- Hansen, T.; Sørensen, M.I.; Eriksen, M.L.R. How the interplay between consumer motivations and values influences organic food identity and behavior. Food Policy 2018, 74, 39–52. [Google Scholar] [CrossRef] [Green Version]
- Willer, H.; Lernoud, J. Research Institute of Organic Agriculture FiBL and IFOAM Organics International. In The World of Organic Agriculture. Statistics and Emerging Trends 2019; Research Institute of Organic Agriculture FiBL and IFOAM Organics International: Frick, Switzerland, 2019; pp. 1–336. [Google Scholar]
- Varanasi, A.; Prasad, P.V.; Jugulam, M. Impact of climate change factors on weeds and herbicide efficacy. In Advances in Agronomy; Academic Press: Cambridge, MA, USA, 2016; Volume 135, pp. 107–146. [Google Scholar]
- Guan, H.; Wilson, J.L. A hybrid dual-source model for potential evaporation and transpiration partitioning. J. Hydrol. 2009, 377, 405–416. [Google Scholar] [CrossRef]
- Sivasakthi, K.; Thudi, M.; Tharanya, M.; Kale, S.M.; Kholová, J.; Halime, M.H.; Jaganathan, D.; Baddam, R.; Thirunalasundari, T.; Gaur, P.M.; et al. Plant vigour QTLs co-map with an earlier reported QTL hotspot for drought tolerance while water saving QTLs map in other regions of the chickpea genome. BMC Plant Biol. 2018, 18, 29. [Google Scholar] [CrossRef]
- Sherwood, S.; Fu, Q. A drier future? Science 2014, 343, 737–739. [Google Scholar] [CrossRef]
- Jannink, J.L.; Orf, J.H.; Jordan, N.R.; Shaw, R.G. Index selection for weed suppressive ability in soybean. Crop Sci. 2000, 40, 1087–1094. [Google Scholar] [CrossRef]
- Bennett, A.C.; Shaw, D.R. Effect of Glycine max cultivar and weed control on weed seed characteristics. Weed Sci. 2017, 48, 431–435. [Google Scholar]
- Shilling, D.G.; Brecke, B.J.; Hiebsch, C.; MacDonald, G. Effect of soybean (Glycine max) cultivar, tillage, and rye (Secale cereale) mulch on sicklepod (Senna obtusifolia). Weed Technol. 1995, 9, 339–342. [Google Scholar] [CrossRef]
- Jordan, N. Differential interference between soybean (Glycine max) varieties and common cocklebur (Xanthium strumarium): A path analysis. Weed Sci. 1992, 40, 614–620. [Google Scholar] [CrossRef]
- Xavier, A.; Hall, B.; Hearst, A.A.; Cherkauer, K.A.; Rainey, K.M. Genetic architecture of phenomic-enabled canopy coverage in Glycine max. Genetics 2017, 206, 1081–1089. [Google Scholar] [CrossRef] [Green Version]
- Moreira, F.F.; Hearst, A.A.; Cherkauer, K.A.; Rainey, K.M. Improving the efficiency of soybean breeding with high-throughput canopy phenotyping. Plant Methods 2019, 15, 139. [Google Scholar] [CrossRef]
- Sanchez-Valdez, I.; Acosta-Gallegos, J.A.; Ibarra-Perez, F.J.; Rosales-Serna, R.; Singh, S.P. Registration of ‘Pinto Saltillo’ common bean. Crop Sci. 2004, 44, 1865–1866. [Google Scholar] [CrossRef]
- Sexton, P.J.; Peterson, C.M.; Boote, K.J.; White, J.W. Early-season growth in relation to region of domestication, seed size, and leaf traits in common bean. Field Crops Res. 1997, 52, 69–78. [Google Scholar] [CrossRef]
- Sankaran, S.; Khot, L.R.; Espinoza, C.Z.; Jarolmasjed, S.; Sathuvalli, V.R.; Vandemark, G.J.; Miklas, P.N.; Carter, A.H.; Pumphrey, M.O.; Knowles, N.R.; et al. Low-altitude, high-resolution aerial imaging systems for row and field crop phenotyping: A review. Eur. J. Agron. 2015, 70, 112–123. [Google Scholar] [CrossRef]
- Tian, Z.; Wang, X.; Lee, R.; Li, Y.; Specht, J.E.; Nelson, R.L.; McClean, P.E.; Qiu, L.; Ma, J. Artificial selection for determinate growth habit in soybean. Proc. Natl. Acad. Sci. USA 2010, 107, 8563–8568. [Google Scholar] [CrossRef] [Green Version]
- Gepts, P.; Aragão, F.J.L.; Barros, E.; Blair, M.W.; Brondani, R.; Broughton, W.; Galasso, I.; Hernández, G.; Kami, J.; Lariguet, P.; et al. Genomics of Phaseolus beans, a major source of dietary protein and micronutrients in the Tropics. In Genomics of Tropical Crop Plants; Moore, P., Ming, R., Eds.; Springer: Berlin, Germany, 2008; pp. 113–143. [Google Scholar]
- Singh, S.P. Common Bean Improvement in the Twenty-First Century; Springer-Science and Business Media: Berlin, Germany, 1999. [Google Scholar]
- USDA Economic Research Service. Table 7. Certified organic beans. In Acres of Soybeans, Dry Beans, Dry Peas/Lentils by State 1997 and 2000-11; USDA Economic Research Service: Washington, DC, USA, 2013. Available online: https://www.ers.usda.gov/data-products/organic-production/ (accessed on 26 May 2020).
- Klonsky, K. Comparison of production costs and resource use for organic and conventional production systems. Am. J Agric. Econ. 2011, 94, 314–321. [Google Scholar] [CrossRef] [Green Version]
- Moghaddam, S.M.; Mamidi, S.; Osorno, J.M.; Lee, R.; Brick, M.; Kelly, J.; Miklas, P.; Urrea, C.; Song, Q.; Cregan, P.; et al. Genome-wide association study identifies candidate loci underlying agronomic traits in a Middle American diversity panel of common bean. Plant Genome USA 2016, 9, 1–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwak, M.; Kami, J.A.; Gepts, P. The putative Mesoamerican domestication center of Phaseolus vulgaris is located in the Lerma-Santiago basin of Mexico. Crop Sci. 2009, 49, 554–563. [Google Scholar] [CrossRef] [Green Version]
- Bitocchi, E.; Bellucci, E.; Giardini, A.; Rau, D.; Rodriguez, M.; Biagetti, E.; Santilocchi, R.; Spagnoletti Zeuli, P.; Gioia, T.; Logozzo, G.; et al. Molecular analysis of the parallel domestication of the common bean (Phaseolus vulgaris) in Mesoamerica and the Andes. New Phytol. 2013, 197, 300–313. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.P. A key for identification of different growth habits of Phaseolus vulgaris L. Annu. Rep. Bean Improv. Coop. 1982, 25, 92–95. [Google Scholar]
- Repinski, S.L.; Kwak, M.; Gepts, P. The common bean growth habit gene PvTFL1y is a functional homolog of Arabidopsis TFL1. Theor. Appl. Genet. 2012, 124, 1539–1547. [Google Scholar] [CrossRef]
- Kwak, M.; Toro, O.; Debouck, D.; Gepts, P. Multiple origins of the determinate growth habit in domesticated common bean (Phaseolus vulgaris L.). Ann. Bot. 2012, 110, 1573–1580. [Google Scholar] [CrossRef] [Green Version]
- Federer, W.T.; Raghavarao, D. On augmented designs. Biometrics 1975, 29–35. [Google Scholar] [CrossRef] [Green Version]
- Hang, A.N.; Silbernagel, M.J.; Miklas, P.N. Registration of ‘Orca’ black-and-white mottled Anasazi-type dry bean. Crop Sci. 2003, 43, 1882. [Google Scholar] [CrossRef]
- Rouse, J.W.; Haas, R.H.; Schell, J.A.; Deering, D.W. Monitoring vegetation systems in the Great Plains with ERTS. In Third Earth Resources Technology Satellite-1 Symposium; NASA Special Publication; Technical Presentations, Section, A; Goddard Space Flight Center: Washington, DC, USA, 1974; Volume I, pp. 309–318. [Google Scholar]
- QGIS Development Team. QGIS Geographic Information System. Open Source Geospatial Foundation Project. Available online: http://qgis.osgeo.org (accessed on 16 May 2018).
- Woebbecke, D.M.; Meyer, G.E.; Von Bargen, K.; Mortensen, D.A. Color indices for weed identification under various soil, residue, and lighting conditions. Trans. ASAE 1995, 38, 259–269. [Google Scholar] [CrossRef]
- Cleveland, W.S.; Devlin, S.J. Locally weighted regression: An approach to regression analysis by local fitting. J. Am. Stat. Assoc. 1988, 83, 596–610. [Google Scholar] [CrossRef]
- BeanCAP (Bean Coordinated Agricultural Project) data. Available online: arsftfbean.uprm.edu/beancap/research (accessed on 26 May 2020).
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Dereeper, A.; Nicolas, S.; Le Cunff, L.; Bacilieri, R.; Doligez, A.; Peros, J.P.; Ruiz, M.; This, P. SNiPlay: A web-based tool for detection, management and analysis of SNPs. Application to grapevine diversity projects. BMC Bioinform. 2011, 12, 134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, S. qqman: An R package for visualizing GWAS results using Q-Q and manhattan plots. J. Open Source Softw. 2018, 3, 731. [Google Scholar] [CrossRef] [Green Version]
- Song, Q.; Jia, G.; Hyten, D.L.; Jenkins, J.; Hwang, E.Y.; Schroeder, S.G.; Osorno, J.M.; Schmutz, J.; Jackson, S.A.; McClean, P.E.; et al. SNP assay development for linkage map construction, anchoring whole-genome sequence, and other genetic and genomic applications in common bean. G3-Genes Genom. Genet. 2015, 5, 2285–2290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, J.; Butler, D. R Package ASMap: Efficient genetic linkage map construction and diagnosis. J. Stat Softw. 2017, 79, 1–29. [Google Scholar] [CrossRef] [Green Version]
- Lander, E.S.; Botstein, D. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 1989, 121, 185–199. [Google Scholar]
- Broman, K.W.; Wu, H.; Sen, Ś.; Churchill, G.A. R/qtl: QTL mapping in experimental crosses. Bioinformatics 2003, 19, 889–890. [Google Scholar] [CrossRef] [Green Version]
- Freyre, R.; Skroch, P.W.; Geffroy, V.; Adam-Blondon, A.F.; Shirmohamadali, A.; Johnson, W.C.; Llaca, V.; Nodari, R.O.; Pereira, P.A.; Tsai, S.M.; et al. Towards an integrated linkage map of common bean. 4. Development of a core linkage map and alignment of RFLP maps. Theor. Appl. Genet. 1998, 97, 847–856. [Google Scholar] [CrossRef]
- Pedrosa-Harand, A.; Porch, T.; Gepts, P. Standard nomenclature for common bean chromosomes and linkage groups. Annu. Rep. Bean Improv. Coop. 2008, 51, 106–107. [Google Scholar]
- Clouse, S.D. Brassinosteroid signal transduction: Clarifying the pathway from ligand perception to gene expression. Mol. Cell 2002, 10, 973–982. [Google Scholar] [CrossRef]
- Kwak, M.; Velasco, D.M.; Gepts, P. Mapping homologous sequences for determinacy and photoperiod sensitivity in common bean (Phaseolus vulgaris). J. Hered. 2008, 99, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, M.; Nascimento, A.C.C.; Silva, F.F.; Barili, L.D.; Vale, N.M.; Carneiro, J.E.; Cruz, C.D.; Carneiro, P.C.S.; Serão, N.V.L. Quantile regression for genome-wide association study of flowering time-related traits in common bean. PLoS ONE 2018, 13, e0190303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McClean, P.E.; Bett, K.E.; Stonehouse, R.; Lee, R.; Pflieger, S.; Moghaddam, S.M.; Geffroy, V.; Miklas, P.; Mamidi, S. White seed color in common bean (Phaseolus vulgaris) results from convergent evolution in the P (pigment) gene. New Phytol. 2018, 219, 1112–1123. [Google Scholar] [CrossRef] [Green Version]
- Weller, J.L.; Vander Schoor, J.K.; Perez-Wright, E.C.; Hecht, V.; González, A.M.; Capel, C.; Yuste-Lisbona, F.J.; Lozano, R.; Santalla, M. Parallel origins of photoperiod adaptation following dual domestications of common bean. J. Exp. Bot. 2019, 70, 1209–1219. [Google Scholar] [CrossRef]
- Parker, T.A.; Berny Mier y Teran, J.C.; Palkovic, A.; Jernstedt, J.; Gepts, P. Pod indehiscence is a domestication and aridity resilience trait in common bean. New Phytol. 2020, 225, 558–570. [Google Scholar] [CrossRef] [Green Version]
- Rau, D.; Murgia, M.L.; Rodriguez, M.; Bitocchi, E.; Bellucci, E.; Fois, D.; Albani, D.; Nanni, L.; Gioia, T.; Santo, D.; et al. Genomic dissection of pod shattering in common bean: Mutations at non-orthologous loci at the basis of convergent phenotypic evolution under domestication of leguminous species. Plant J. 2019, 97, 693–714. [Google Scholar] [CrossRef]
- Di Vittori, V.; Bitocchi, E.; Rodriguez, M.; Alseekh, S.; Bellucci, E.; Nanni, L.; Gioia, T.; Marzario, S.; Logozzo, G.; Rossato, M.; et al. Indehiscence in common bean is associated to the fine regulation of PvMYB26 and a non-functional abscission layer. bioRxiv 2020. [Google Scholar] [CrossRef]
- Resende, R.T.; de Resende, M.D.V.; Azevedo, C.F.; Fonseca e Silva, F.; Melo, L.C.; Pereira, H.S.; Souza, T.L.P.O.; Valdisser, P.A.M.R.; Brondani, C.; Vianello, R.P. Genome-wide association and regional heritability mapping of plant architecture, lodging and productivity in Phaseolus vulgaris. G3-Genes Genom. Genet. 2018, 8, 2841–2854. [Google Scholar] [CrossRef] [Green Version]
- Trapp, J.J.; Urrea, C.A.; Zhou, J.; Khot, L.R.; Sankaran, S.; Miklas, P.N. Selective phenotyping traits related to multiple stress and drought response in dry bean. Crop Sci. 2016, 56, 1460–1472. [Google Scholar] [CrossRef] [Green Version]
- Sankaran, S.; Zhou, J.; Khot, L.R.; Trapp, J.J.; Mndolwa, E.; Miklas, P.N. High-throughput field phenotyping in dry bean using small unmanned aerial vehicle based multispectral imagery. Comput. Electron. Agric. 2018, 151, 84–92. [Google Scholar] [CrossRef]
- Zhou, J.; Khot, L.R.; Boydston, R.A.; Miklas, P.N.; Porter, L. Low altitude remote sensing technologies for crop stress monitoring: A case study on spatial and temporal monitoring of irrigated pinto bean. Precis. Agric. 2018, 19, 555–569. [Google Scholar] [CrossRef]
- Sankaran, S.; Quirós, J.J.; Miklas, P.N. Unmanned aerial system and satellite-based high resolution imagery for high-throughput phenotyping in dry bean. Comput. Electron. Agric. 2019, 165, 104965. [Google Scholar] [CrossRef]







| Year of Trial | Population Evaluated | Aircraft | Primary Camera | Camera Type | Altitude | Mission Planning Application | GSD |
|---|---|---|---|---|---|---|---|
| 2016 | MDP | DJI Phantom 3 Professional | Stock (Sony EXMOR 1/2.3 sensor) | RGB | 17 m | Map Pilot | 0.7 cm/px |
| 2017 * | MDP, BxO | DJI Phantom 3 Professional | Parrot Sequoia | Multi-spectral | 20 m | Atlas Flight | 1.9 cm/px |
| 2018 | MDP, BxO | DJI Matrice 100 | Micasense RedEdge-M | Multi-spectral | 20 m | DJI GS Pro | 1.4 cm/px |
| 2019 | BxO | DJI Matrice 100 | Micasense RedEdge-M | Multi-spectral | 20 m | DJI GS Pro | 1.4 cm/px |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parker, T.A.; Palkovic, A.; Gepts, P. Determining the Genetic Control of Common Bean Early-Growth Rate Using Unmanned Aerial Vehicles. Remote Sens. 2020, 12, 1748. https://doi.org/10.3390/rs12111748
Parker TA, Palkovic A, Gepts P. Determining the Genetic Control of Common Bean Early-Growth Rate Using Unmanned Aerial Vehicles. Remote Sensing. 2020; 12(11):1748. https://doi.org/10.3390/rs12111748
Chicago/Turabian StyleParker, Travis A., Antonia Palkovic, and Paul Gepts. 2020. "Determining the Genetic Control of Common Bean Early-Growth Rate Using Unmanned Aerial Vehicles" Remote Sensing 12, no. 11: 1748. https://doi.org/10.3390/rs12111748