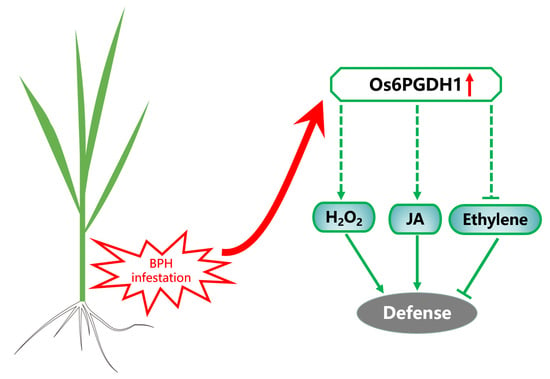
Overexpression of a Cytosolic 6-Phosphogluconate Dehydrogenase Gene Enhances the Resistance of Rice to Nilaparvata lugens
Abstract
:1. Introduction
2. Results
2.1. cDNA Cloning and Phylogenetic Analysis of the Os6PGDH1 Sequence
2.2. Expression of Os6PGDH1
2.3. Overexpressing of Os6PGDH1
2.4. Overexpression of Os6PGDH1 Enhances Levels of BPH-Induced JA, JA-Ile, and H2O2, but Suppresses Ethylene
2.5. Os6PGDH1 Positively Regulates the Resistance of Rice to BPH
3. Discussion
4. Materials and Methods
4.1. Plant Growth
4.2. Insects
4.3. Isolation and Characterization of Os6PGDH1
4.4. Sequence and Phylogenetic Analysis of Os6PGDH1
4.5. Generation and Characterization of Transgenic Plants
4.6. Plant Treatments
4.7. Quantitative Real-Time PCR
4.8. Os6PGDH1 mRNA Expression Analysis
4.9. Measurement of Plant Growth Parameters
4.10. JA, JA-Ile, and SA Analysis
4.11. Hydrogen Oxide Analysis
4.12. Ethylene Analysis
4.13. BPH Bioassays
4.14. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Furstenberg-Hagg, J.; Zagrobelny, M.; Bak, S. Plant defense against insect herbivores. Int. J. Mol. Sci. 2013, 14, 10242–10297. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Baldwin, I.T. New insights into plant responses to the attack from insect herbivores. Annu. Rev. Genet. 2010, 44, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Howe, G.A.; Jander, G. Plant immunity to insect herbivores. Annu. Rev. Plant. Biol. 2008, 59, 41–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erb, M.; Reymond, P. Molecular interactions between plants and insect herbivores. Annu. Rev. Plant. Biol. 2019, 70, 527–557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kruger, N.J.; von Schaewen, A. The oxidative pentose phosphate pathway: Structure and organisation. Curr. Opin. Plant. Biol. 2003, 6, 236–246. [Google Scholar] [CrossRef]
- Stincone, A.; Prigione, A.; Cramer, T.; Wamelink, M.M.; Campbell, K.; Cheung, E.; Olin-Sandoval, V.; Gruning, N.M.; Kruger, A.; Tauqeer Alam, M.; et al. The return of metabolism: Biochemistry and physiology of the pentose phosphate pathway. Biol. Rev. Camb. Philos. Soc. 2015, 90, 927–963. [Google Scholar] [CrossRef] [Green Version]
- Lansing, H.; Doering, L.; Fischer, K.; Baune, M.C.; von Schaewen, A. Analysis of potential redundancy among Arabidopsis 6-phosphogluconolactonase isoforms in peroxisomes. J. Exp. Bot. 2020, 71, 823–836. [Google Scholar] [CrossRef]
- Esposito, S. Nitrogen assimilation, abiotic stress and glucose 6-phosphate dehydrogenase: The full circle of reductants. Plants 2016, 5, 24. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.Z.; Wang, X.M.; Wang, X.Y.; Wu, K.L.; Li, P.; Chang, N.; Wang, J.F.; Wang, F.; Li, J.L.; Bi, Y.R. Glucose-6-phosphate dehydrogenase and alternative oxidase are involved in the cross tolerance of highland barley to salt stress and UV-B radiation. J. Plant. Physiol. 2015, 181, 83–95. [Google Scholar] [CrossRef]
- Landi, S.; Nurcato, R.; De Lillo, A.; Lentini, M.; Grillo, S.; Esposito, S. Glucose-6-phosphate dehydrogenase plays a central role in the response of tomato (Solanum lycopersicum) plants to short and long-term drought. Plant. Physiol. Biochem. 2016, 105, 79–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Y.Z.; Lin, S.Z.; Guo, H.; Zhang, Z.Y.; Chen, X.Y. Functional analysis of PsG6PDH, a cytosolic glucose-6-phosphate dehydrogenase gene from Populus suaveolens, and its contribution to cold tolerance improvement in tobacco plants. Biotechnol. Lett. 2013, 35, 1509–1518. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.H.; Hou, J.J.; Li, Y.; Zhang, Y.Y.; Huang, J.J.; Liang, W.H. Nitric oxide-mediated cytosolic glucose-6-phosphate dehydrogenase is involved in aluminum toxicity of soybean under high aluminum concentration. Plant. Soil 2017, 416, 39–52. [Google Scholar] [CrossRef]
- Hu, Y.F.; You, J.; Li, J.S.; Wang, C.L. Loss of cytosolic glucose-6-phosphate dehydrogenase increases the susceptibility of Arabidopsis thaliana to root-knot nematode infection. Ann. Bot. 2019, 123, 37–46. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Cui, Y.F.; Huang, S.Y.; Yu, J.Y.; Wang, X.Y.; Xin, D.W.; Li, X.; Liu, Y.H.; Dai, Y.X.; Qi, Z.M.; et al. Genome-wide analysis of the glucose-6-phosphate dehydrogenase family in soybean and functional identification of GmG6PDH2 involvement in salt stress. Front. Plant. Sci. 2020, 11, 214. [Google Scholar] [CrossRef] [Green Version]
- Scharte, J.; Schon, H.; Tjaden, Z.; Weis, E.; von Schaewen, A. Isoenzyme replacement of glucose-6-phosphate dehydrogenase in the cytosol improves stress tolerance in plants. Proc. Natl. Acad. Sci. USA 2009, 106, 8061–8066. [Google Scholar] [CrossRef] [Green Version]
- Debnam, P.M.; Emes, M.J. Subcellular distribution of enzymes of the oxidative pentose phosphate pathway in root and leaf tissues. J. Exp. Bot. 1999, 50, 1653–1661. [Google Scholar] [CrossRef]
- Holscher, C.; Lutterbey, M.C.; Lansing, H.; Meyer, T.; Fischer, K.; von Schaewen, A. Defects in peroxisomal 6-phosphogluconate dehydrogenase isoform PGD2 prevent gametophytic interaction in arabidopsis thaliana. Plant. Physiol. 2016, 171, 192–205. [Google Scholar] [CrossRef] [Green Version]
- Spielbauer, G.; Li, L.; Romisch-Margl, L.; Do, P.T.; Fouquet, R.; Fernie, A.R.; Eisenreich, W.; Gierl, A.; Settles, A.M. Chloroplast-localized 6-phosphogluconate dehydrogenase is critical for maize endosperm starch accumulation. J. Exp. Bot. 2013, 64, 2231–2242. [Google Scholar] [CrossRef] [Green Version]
- Tanksley, S.D.; Kuehn, G.D. Genetics, subcellular localization, and molecular characterization of 6-phosphogluconate dehydrogenase isozymes in tomato. Biochem. Genet. 1985, 23, 441–454. [Google Scholar] [CrossRef]
- Krepinsky, K.; Plaumann, M.; Martin, W.; Schnarrenberger, C. Purification and cloning of chloroplast 6-phosphogluconate dehydrogenase from spinach-cyanobacterial genes for chloroplast and cytosolic isoenzymes encoded in eukaryotic chromosomes. Eur. J. Biochem. 2001, 268, 2678–2686. [Google Scholar] [CrossRef] [PubMed]
- Hou, F.Y.; Huang, J.; Yu, S.L.; Zhang, H.S. The 6-phosphogluconate dehydrogenase genes are responsive to abiotic stresses in rice. J. Integr. Plant. Biol. 2007, 49, 655–663. [Google Scholar] [CrossRef]
- Huang, J.; Zhang, H.S.; Wang, J.F.; Yang, J.S. Molecular cloning and characterization of rice 6-phosphogluconate dehydrogenase gene that is up-regulated by salt stress. Mol. Biol. Rep. 2003, 30, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Sindelar, L.; Sindelarova, M.; Burketova, L. Changes in activity of glucose-6-phosphate and 6-phosphogluconate dehydrogenase isozymes upon potato virus Y infection in tobacco leaf tissues and protoplasts. Plant. Physiol. Biochem. 1999, 37, 195–201. [Google Scholar] [CrossRef]
- Fahrendorf, T.; Ni, W.; Shorrosh, B.S.; Dixon, R.A. Stress responses in alfalfa (Medicago sativa L.) XIX. Transcriptional activation of oxidative pentose phosphate pathway genes at the onset of the isoflavonoid phytoalexin response. Plant. Mol. Biol. 1995, 28, 885–900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danson, J.; Wasano, K.; Nose, A. Infection of rice plants with the sheath blight fungus causes an activation of pentose phosphate and glycolytic pathways. Eur. J. Plant. Pathol. 2000, 106, 555–561. [Google Scholar] [CrossRef]
- Zhou, G.X.; Qi, J.F.; Ren, N.; Cheng, J.A.; Erb, M.; Mao, B.Z.; Lou, Y.G. Silencing OsHI-LOX makes rice more susceptible to chewing herbivores, but enhances resistance to a phloem feeder. Plant. J. 2009, 60, 638–648. [Google Scholar] [CrossRef]
- Lu, J.; Ju, H.P.; Zhou, G.X.; Zhu, C.S.; Erb, M.; Wang, X.P.; Wang, P.; Lou, Y.G. An EAR-motif-containing ERF transcription factor affects herbivore-induced signaling, defense and resistance in rice. Plant. J. 2011, 68, 583–596. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Zhang, J.; Li, J.C.; Zhou, G.X.; Wang, Q.; Bian, W.B.; Erb, M.; Lou, Y.G. Prioritizing plant defence over growth through WRKY regulation facilitates infestation by non-target herbivores. Elife 2015, 4, e04805. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Cao, T.T.; Zhang, J.; Lou, Y.G. Overexpression of OsGID1 enhances the resistance of rice to the brown planthopper Nilaparvata lugens. Int. J. Mol. Sci. 2018, 19, 2744. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.J.; Wang, X.; Lou, Y.G.; Cheng, J.A. Role of ethylene signaling in the production of rice volatiles induced by the rice brown planthopper Nilaparvata lugens. Chin. Sci. Bull. 2006, 51, 2457–2465. [Google Scholar] [CrossRef]
- Guo, J.P.; Xu, C.X.; Wu, D.; Zhao, Y.; Qiu, Y.F.; Wang, X.X.; Ouyang, Y.D.; Cai, B.D.; Liu, X.; Jing, S.L.; et al. Bph6 encodes an exocyst-localized protein and confers broad resistance to planthoppers in rice. Nat. Genet. 2018, 50, 297–306. [Google Scholar] [CrossRef]
- Zhou, S.X.; Chen, M.T.; Zhang, Y.B.; Gao, Q.; Noman, A.; Wang, Q.; Li, H.; Chen, L.; Zhou, P.Y.; Lu, J.; et al. OsMKK3, a stress-responsive protein kinase, positively regulates rice resistance to Nilaparvata lugens via phytohormone dynamics. Int. J. Mol. Sci. 2019, 20, 3023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baileyserres, J.; Nguyen, M.T. Purification and characterization of cytosolic 6-phosphogluconate dehydrogenase isozymes from maize. Plant. Physiol. 1992, 100, 1580–1583. [Google Scholar] [CrossRef]
- Ribeiro, C. Engineering 6-phosphogluconate Dehydrogenase to Improve Heat Sability of Starch Accumulation in Maize Seed Development. Ph.D. Thesis, University of Florida, Gainesville, FL, USA, 2017. [Google Scholar]
- Wallstrom, S.V.; Florez-Sarasa, I.; Araujo, W.L.; Aidemark, M.; Fernandez-Fernandez, M.; Fernie, A.R.; Ribas-Carbo, M.; Rasmusson, A.G. Suppression of the external mitochondrial NADPH dehydrogenase, NDB1, in Arabidopsis thaliana affects central metabolism and vegetative growth. Mol. Plant. 2014, 7, 356–368. [Google Scholar] [CrossRef] [Green Version]
- Züst, T.; Agrawal, A.A. Trade-offs between plant growth and defense against insect herbivory: An emerging mechanistic synthesis. Annu. Rev. Plant. Biol. 2017, 68, 513–534. [Google Scholar] [CrossRef] [Green Version]
- Guo, Q.; Major, I.T.; Howe, G.A. Resolution of growth–defense conflict: Mechanistic insights from jasmonate signaling. Curr. Opin. Plant. Biol. 2018, 44, 72–81. [Google Scholar] [CrossRef]
- Corpas, F.J.; Barroso, J.B. Peroxisomal plant metabolism-an update on nitric oxide, Ca2+ and the NADPH recycling network. J. Cell Sci. 2018, 131. [Google Scholar] [CrossRef] [Green Version]
- Pugin, A.; Frachisse, J.M.; Tavernier, E.; Bligny, R.; Gout, E.; Douce, R.; Guern, J. Early events induced by the elicitor cryptogein in tobacco cells: Involvement of a plasma membrane NADPH oxidase and activation of glycolysis and the pentose phosphate pathway. Plant. Cell 1997, 9, 2077–2091. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.W.; Tian, H.; Yu, X.; Zheng, L.P. Glucose-6-phosphate dehydrogenase plays critical role in artemisinin production of Artemisia annua under salt stress. Biol. Plant. 2017, 61, 529–539. [Google Scholar] [CrossRef]
- Schaller, F.; Biesgen, C.; Mussig, C.; Altmann, T.; Weiler, E.W. 12-oxophytodienoate reductase 3 (OPR3) is the isoenzyme involved in jasmonate biosynthesis. Planta 2000, 210, 979–984. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Ye, M.; Li, R.; Lou, Y. OsWRKY53, a versatile switch in regulating herbivore-induced defense responses in rice. Plant. Signal. Behav. 2016, 11, e1169357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, J.; Li, J.C.; Ju, H.P.; Liu, X.L.; Erb, M.; Wang, X.; Lou, Y.G. Contrasting effects of ethylene biosynthesis on induced plant resistance against a chewing and a piercing-sucking herbivore in rice. Mol. Plant. 2014, 7, 1670–1682. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, S.; Forno, D.A.; Cock, J.H. Laboratory Manual for Physiological Studies of Rice, 3rd ed.; International Rice Research Institute: Los Baños, Philippines, 1976. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. Mega X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Hiei, Y.; Ohta, S.; Komari, T.; Kumashiro, T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence-analysis of the boundaries of the T-DNA. Plant. J. 1994, 6, 271–282. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Luo, T.; Wang, W.W.; Cao, T.T.; Li, R.; Lou, Y.G. Silencing OsSLR1 enhances the resistance of rice to the brown planthopper Nilaparvata lugens. Plant. Cell Environ. 2017, 40, 2147–2159. [Google Scholar] [CrossRef] [PubMed]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, L.; Kuai, P.; Ye, M.; Zhou, S.; Lu, J.; Lou, Y. Overexpression of a Cytosolic 6-Phosphogluconate Dehydrogenase Gene Enhances the Resistance of Rice to Nilaparvata lugens. Plants 2020, 9, 1529. https://doi.org/10.3390/plants9111529
Chen L, Kuai P, Ye M, Zhou S, Lu J, Lou Y. Overexpression of a Cytosolic 6-Phosphogluconate Dehydrogenase Gene Enhances the Resistance of Rice to Nilaparvata lugens. Plants. 2020; 9(11):1529. https://doi.org/10.3390/plants9111529
Chicago/Turabian StyleChen, Lin, Peng Kuai, Miaofen Ye, Shuxing Zhou, Jing Lu, and Yonggen Lou. 2020. "Overexpression of a Cytosolic 6-Phosphogluconate Dehydrogenase Gene Enhances the Resistance of Rice to Nilaparvata lugens" Plants 9, no. 11: 1529. https://doi.org/10.3390/plants9111529