Gibberellin Promotes Bolting and Flowering via the Floral Integrators RsFT and RsSOC1-1 under Marginal Vernalization in Radish
Abstract
:1. Introduction
2. Results
2.1. GA Effects on Bolting Time Significantly Differed between the Two Radish Inbred Lines
2.2. GA Regulation Prioritizes the Floral Transition over Vegetative Growth in Radish
2.3. Transcriptome Sequencing Identifies Genes Responding to GA in Radish
2.4. Comparative Analysis of DEGs in the Two Inbred Radish Lines Treated with GA under Marginal Vernalization
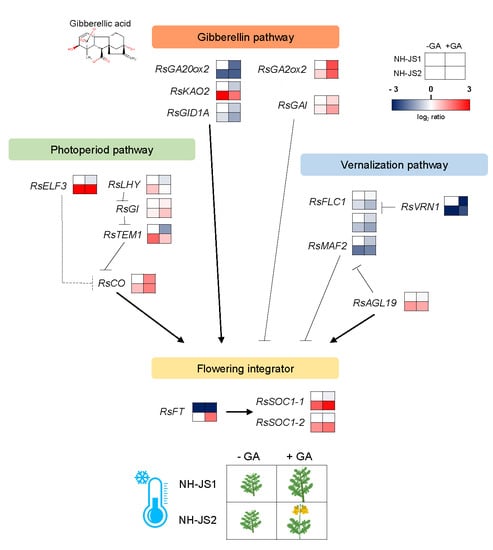
2.5. Comprehensive Analysis of DEGs Related to GA-Responsive Flowering Pathways in Radish
2.6. RsFT and RsSOC1-1 Floral Integrators Were Responsive to GA under Marginal Vernalization in Radish Flowering Pathways
3. Discussion
3.1. The Two Inbred Radish Lines Display Different Bolting Times in Response to GA Treatment
3.2. GA-Responsive Flowering Time DEGs in Radish
3.3. A Gene Regulatory Network Model for GA-Responsive Flowering in Radish
4. Materials and Methods
4.1. Plant Materials, Exogenous GA Treatment, and Bolting Trait Analysis
4.2. Evaluating the GA Effect on Hypocotyl Elongation
4.3. Preparation and Sequencing of the RNA-Seq Library
4.4. Reference-Guided Assembly and Mapping of the Radish Transcriptome
4.5. Functional Annotation Analysis
4.6. Analysis of DEGs
4.7. Identification of Flowering Time- and GA-Related Genes in Radish
4.8. Quantitative PCR (qPCR) Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kasahara, H.; Hanada, A.; Kuzuyama, T.; Takagi, M.; Kamiya, Y.; Yamaguchi, S. Contribution of the mevalonate and methylerythritol phosphate pathways to the biosynthesis of gibberellins in arabidopsis. J. Biol. Chem 2002, 277, 45188–45194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldberg-Moeller, R.; Shalom, L.; Shlizerman, L.; Samuels, S.; Zur, N.; Ophir, R.; Blumwald, E.; Sadka, A. Effects of gibberellin treatment during flowering induction period on global gene expression and the transcription of flowering-control genes in citrus buds. Plant Sci. An. Int. J. Exp. Plant Biol. 2013, 198, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.P.; Gubler, F. Molecular mechanism of gibberellin signaling in plants. Annu. Rev. Plant Biol. 2004, 55, 197–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daviere, J.M.; Achard, P. A pivotal role of dellas in regulating multiple hormone signals. Mol. Plant 2016, 9, 10–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Q.; Krishnan, S.; Merewitz, E.; Xu, J.; Huang, B. Gibberellin-regulation and genetic variations in leaf elongation for tall fescue in association with differential gene expression controlling cell expansion. Sci. Rep. 2016, 6, 30258. [Google Scholar] [CrossRef] [Green Version]
- Achard, P.; Gusti, A.; Cheminant, S.; Alioua, M.; Dhondt, S.; Coppens, F.; Beemster, G.T.; Genschik, P. Gibberellin signaling controls cell proliferation rate in arabidopsis. Curr. Biol. 2009, 19, 1188–1193. [Google Scholar] [CrossRef]
- Achard, P.; Cheng, H.; De Grauwe, L.; Decat, J.; Schoutteten, H.; Moritz, T.; Van Der Straeten, D.; Peng, J.; Harberd, N.P. Integration of plant responses to environmentally activated phytohormonal signals. Science (N. Y.) 2006, 311, 91–94. [Google Scholar] [CrossRef]
- Achard, P.; Gong, F.; Cheminant, S.; Alioua, M.; Hedden, P.; Genschik, P. The cold-inducible cbf1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing della proteins via its effect on gibberellin metabolism. Plant. Cell 2008, 20, 2117–2129. [Google Scholar] [CrossRef] [Green Version]
- Magome, H.; Yamaguchi, S.; Hanada, A.; Kamiya, Y.; Oda, K. The ddf1 transcriptional activator upregulates expression of a gibberellin-deactivating gene, ga2ox7, under high-salinity stress in arabidopsis. Plant J. 2008, 56, 613–626. [Google Scholar] [CrossRef]
- Hedden, P.; Thomas, S.G. Gibberellin biosynthesis and its regulation. Biochem. J. 2012, 444, 11–25. [Google Scholar] [CrossRef] [Green Version]
- Thomas, S.G.; Rieu, I.; Steber, C.M. Gibberellin metabolism and signaling. Vitam. Horm. 2005, 72, 289–338. [Google Scholar] [PubMed]
- Murase, K.; Hirano, Y.; Sun, T.P.; Hakoshima, T. Gibberellin-induced della recognition by the gibberellin receptor gid1. Nature 2008, 456, 459–463. [Google Scholar] [CrossRef]
- Shimada, A.; Ueguchi-Tanaka, M.; Nakatsu, T.; Nakajima, M.; Naoe, Y.; Ohmiya, H.; Kato, H.; Matsuoka, M. Structural basis for gibberellin recognition by its receptor gid1. Nature 2008, 456, 520–523. [Google Scholar] [CrossRef] [PubMed]
- Itoh, H.; Matsuoka, M.; Steber, C.M. A role for the ubiquitin-26s-proteasome pathway in gibberellin signaling. Trends Plant. Sci. 2003, 8, 492–497. [Google Scholar] [CrossRef]
- Du, R.; Niu, S.; Liu, Y.; Sun, X.; Porth, I.; El-Kassaby, Y.A.; Li, W. The gibberellin gid1-della signalling module exists in evolutionarily ancient conifers. Sci. Rep. 2017, 7, 16637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hedden, P. The genes of the green revolution. Trends Genet. 2003, 19, 5–9. [Google Scholar] [CrossRef]
- Peng, J.; Richards, D.E.; Hartley, N.M.; Murphy, G.P.; Devos, K.M.; Flintham, J.E.; Beales, J.; Fish, L.J.; Worland, A.J.; Pelica, F.; et al. ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 1999, 400, 256–261. [Google Scholar] [CrossRef]
- Elias, A.A.; Busov, V.B.; Kosola, K.R.; Ma, C.; Etherington, E.; Shevchenko, O.; Gandhi, H.; Pearce, D.W.; Rood, S.B.; Strauss, S.H. Green revolution trees: Semidwarfism transgenes modify gibberellins, promote root growth, enhance morphological diversity, and reduce competitiveness in hybrid poplar. Plant Physiol. 2012, 160, 1130–1144. [Google Scholar] [CrossRef] [Green Version]
- Zhao, M.; Liu, B.; Wu, K.; Ye, Y.; Huang, S.; Wang, S.; Wang, Y.; Han, R.; Liu, Q.; Fu, X.; et al. Regulation of osmir156h through alternative polyadenylation improves grain yield in rice. PLoS ONE 2015, 10, e0126154. [Google Scholar] [CrossRef]
- Komeda, Y. Genetic regulation of time to flower in arabidopsis thaliana. Annu. Rev. Plant. Biol. 2004, 55, 521–535. [Google Scholar] [CrossRef] [Green Version]
- Davis, S.J. Integrating hormones into the floral-transition pathway of arabidopsis thaliana. Plant. Cell Environ. 2009, 32, 1201–1210. [Google Scholar] [CrossRef] [PubMed]
- Simpson, G.G.; Dean, C. Arabidopsis, the rosetta stone of flowering time? Science (N. Y.) 2002, 296, 285–289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mutasa-Gottgens, E.; Hedden, P. Gibberellin as a factor in floral regulatory networks. J. Exp. Bot. 2009, 60, 1979–1989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilkie, J.D.; Sedgley, M.; Olesen, T. Regulation of floral initiation in horticultural trees. J. Exp. Bot. 2008, 59, 3215–3228. [Google Scholar] [CrossRef] [Green Version]
- Bangerth, K.F. Floral induction in mature, perennial angiosperm fruit trees: Similarities and discrepancies with annual/biennial plants and the involvement of plant hormones. Sci. Hort. 2009, 122, 153–163. [Google Scholar] [CrossRef]
- Achard, P.; Herr, A.; Baulcombe, D.C.; Harberd, N.P. Modulation of floral development by a gibberellin-regulated microrna. Development 2004, 131, 3357–3365. [Google Scholar] [CrossRef] [Green Version]
- Moon, J.; Suh, S.S.; Lee, H.; Choi, K.R.; Hong, C.B.; Paek, N.C.; Kim, S.G.; Lee, I. The soc1 mads-box gene integrates vernalization and gibberellin signals for flowering in arabidopsis. Plant. J. 2003, 35, 613–623. [Google Scholar] [CrossRef]
- Macmillan, C.P.; Blundell, C.A.; King, R.W. Flowering of the grass lolium perenne: Effects of vernalization and long days on gibberellin biosynthesis and signaling. Plant. Physiol. 2005, 138, 1794–1806. [Google Scholar] [CrossRef] [Green Version]
- Kitashiba, H.; Li, F.; Hirakawa, H.; Kawanabe, T.; Zou, Z.; Hasegawa, Y.; Tonosaki, K.; Shirasawa, S.; Fukushima, A.; Yokoi, S.; et al. Draft sequences of the radish (raphanus sativus l.) genome. DNA Res. 2014, 21, 481–490. [Google Scholar] [CrossRef] [Green Version]
- Mun, J.H.; Chung, H.; Chung, W.H.; Oh, M.; Jeong, Y.M.; Kim, N.; Ahn, B.O.; Park, B.S.; Park, S.; Lim, K.B.; et al. Construction of a reference genetic map of raphanus sativus based on genotyping by whole-genome resequencing. Theor. Appl. Genet. 2015, 128, 259–272. [Google Scholar] [CrossRef]
- Nie, S.; Li, C.; Xu, L.; Wang, Y.; Huang, D.; Muleke, E.M.; Sun, X.; Xie, Y.; Liu, L. De novo transcriptome analysis in radish (raphanus sativus l.) and identification of critical genes involved in bolting and flowering. BMC Genom. 2016, 17, 389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, W.Y.; Park, H.J.; Lee, A.; Lee, S.S.; Kim, Y.S.; Cho, H.S. Identification of flowering-related genes responsible for differences in bolting time between two radish inbred lines. Front. Plant Sci. 2016, 7, 1844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yi, G.; Park, H.; Kim, J.-S.; Chae, W.; Park, S.; Huh, J. Identification of three flowering locus c genes responsible for vernalization response in radish (raphanus sativus l.). Hortic. Environ. Biotechnol. 2014, 55, 548–556. [Google Scholar] [CrossRef]
- Erwin, J.E.; Warner, R.M.; Smith, A.G. Vernalization, photoperiod and ga3 interact to affect flowering of japanese radish (raphanus sativus chinese radish jumbo scarlet). Physiol. Plant. 2002, 115, 298–302. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Wan, H.; Cheng, T.; Wang, J.; Yang, W.; Pan, H.; Zhang, Q. Comparative rna-seq analysis of transcriptome dynamics during petal development in rosa chinensis. Sci. Rep. 2017, 7, 43382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qu, J.; Kang, S.G.; Hah, C.; Jang, J.C. Molecular and cellular characterization of ga-stimulated transcripts gasa4 and gasa6 in arabidopsis thaliana. Plant Sci. An. Int. J. Exp. Plant Biol. 2016, 246, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Toh, S.; Imamura, A.; Watanabe, A.; Nakabayashi, K.; Okamoto, M.; Jikumaru, Y.; Hanada, A.; Aso, Y.; Ishiyama, K.; Tamura, N.; et al. High temperature-induced abscisic acid biosynthesis and its role in the inhibition of gibberellin action in arabidopsis seeds. Plant. Physiol. 2008, 146, 1368–1385. [Google Scholar] [CrossRef] [Green Version]
- Peng, J.; Harberd, N.P. Gibberellin deficiency and response mutations suppress the stem elongation phenotype of phytochrome-deficient mutants of arabidopsis. Plant. Physiol. 1997, 113, 1051–1058. [Google Scholar] [CrossRef] [Green Version]
- Peng, J.; Harberd, N.P. Derivative alleles of the arabidopsis gibberellin-insensitive (gai) mutation confer a wild-type phenotype. The Plant. Cell 1993, 5, 351–360. [Google Scholar] [CrossRef]
- Silverstone, A.L.; Tseng, T.S.; Swain, S.M.; Dill, A.; Jeong, S.Y.; Olszewski, N.E.; Sun, T.P. Functional analysis of spindly in gibberellin signaling in arabidopsis. Plant. Physiol. 2007, 143, 987–1000. [Google Scholar] [CrossRef] [Green Version]
- Sun, T.P. Gibberellin-gid1-della: A pivotal regulatory module for plant growth and development. Plant. Physiol. 2010, 154, 567–570. [Google Scholar] [CrossRef] [PubMed]
- Takata, N.; Saito, S.; Saito, C.T.; Nanjo, T.; Shinohara, K.; Uemura, M. Molecular phylogeny and expression of poplar circadian clock genes, lhy1 and lhy2. New Phytol. 2009, 181, 808–819. [Google Scholar] [CrossRef] [PubMed]
- Mouradov, A.; Cremer, F.; Coupland, G. Control of flowering time: Interacting pathways as a basis for diversity. Plant. Cell 2002, 14 (Suppl. 1), S111–S130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osnato, M.; Castillejo, C.; Matias-Hernandez, L.; Pelaz, S. Tempranillo genes link photoperiod and gibberellin pathways to control flowering in arabidopsis. Nat. Commun. 2012, 3, 808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castillejo, C.; Pelaz, S. The balance between constans and tempranillo activities determines ft expression to trigger flowering. Curr. Biol. CB 2008, 18, 1338–1343. [Google Scholar] [CrossRef] [Green Version]
- King, R.W.; Moritz, T.; Evans, L.T.; Junttila, O.; Herlt, A.J. Long-day induction of flowering in lolium temulentum involves sequential increases in specific gibberellins at the shoot apex. Plant. Physiol. 2001, 127, 624–632. [Google Scholar] [CrossRef]
- Eriksson, S.; Bohlenius, H.; Moritz, T.; Nilsson, O. Ga4 is the active gibberellin in the regulation of leafy transcription and arabidopsis floral initiation. Plant. Cell 2006, 18, 2172–2181. [Google Scholar] [CrossRef] [Green Version]
- Guo, D.-P.; Ali Shah, G.; Zeng, G.-W.; Zheng, S.-J. The interaction of plant growth regulators and vernalization on the growth and flowering of cauliflower (brassica oleracea var. Botrytis). Plant. Growth Regul. 2004, 43, 163–171. [Google Scholar] [CrossRef]
- Xie, J.; Tian, J.; Du, Q.; Chen, J.; Li, Y.; Yang, X.; Li, B.; Zhang, D. Association genetics and transcriptome analysis reveal a gibberellin-responsive pathway involved in regulating photosynthesis. J. Exp. Bot. 2016, 67, 3325–3338. [Google Scholar] [CrossRef] [Green Version]
- Hui, W.K.; Wang, Y.; Chen, X.Y.; Zayed, M.Z.; Wu, G.J. Analysis of transcriptional responses of the inflorescence meristems in jatropha curcas following gibberellin treatment. Int. J. Mol. Sci. 2018, 19, 432. [Google Scholar] [CrossRef] [Green Version]
- Singh, V.K.; Jain, M. Transcriptome profiling for discovery of genes involved in shoot apical meristem and flower development. Genom Data 2014, 2, 135–138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Lin, C.; Fu, F.; Zhong, X.; Peng, B.; Yan, H.; Zhang, J.; Zhang, W.; Wang, P.; Ding, X.; et al. Comparative transcriptome analysis of flower heterosis in two soybean f1 hybrids by rna-seq. PLoS ONE 2017, 12, e0181061. [Google Scholar] [CrossRef] [Green Version]
- Liu, K.; Feng, S.; Pan, Y.; Zhong, J.; Chen, Y.; Yuan, C.; Li, H. Transcriptome analysis and identification of genes associated with floral transition and flower development in sugar apple (annona squamosa l.). Front. Plant Sci. 2016, 7, 1695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olszewski, N.; Sun, T.P.; Gubler, F. Gibberellin signaling: Biosynthesis, catabolism, and response pathways. Plant. Cell 2002, 14 (Suppl. 1), S61–S80. [Google Scholar] [CrossRef] [Green Version]
- Alabadi, D.; Oyama, T.; Yanovsky, M.J.; Harmon, F.G.; Mas, P.; Kay, S.A. Reciprocal regulation between toc1 and lhy/cca1 within the arabidopsis circadian clock. Science (N. Y.) 2001, 293, 880–883. [Google Scholar] [CrossRef]
- Sawa, M.; Nusinow, D.A.; Kay, S.A.; Imaizumi, T. Fkf1 and gigantea complex formation is required for day-length measurement in arabidopsis. Science 2007, 318, 261–265. [Google Scholar] [CrossRef] [Green Version]
- Cha, J.Y.; Kim, J.; Kim, T.S.; Zeng, Q.; Wang, L.; Lee, S.Y.; Kim, W.Y.; Somers, D.E. Gigantea is a co-chaperone which facilitates maturation of zeitlupe in the arabidopsis circadian clock. Nat. Commun. 2017, 8, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levy, Y.Y.; Mesnage, S.; Mylne, J.S.; Gendall, A.R.; Dean, C. Multiple roles of arabidopsis vrn1 in vernalization and flowering time control. Science 2002, 297, 243–246. [Google Scholar] [CrossRef] [PubMed]
- Cox, M.P.; Peterson, D.A.; Biggs, P.J. Solexaqa: At-a-glance quality assessment of illumina second-generation sequencing data. BMC Bioinform. 2010, 11, 485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped blast and psi-blast: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using david bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, M. Kaas: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007, 35, W182–W185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [Green Version]
- Nilsson, O.; Lee, I.; Blazquez, M.A.; Weigel, D. Flowering-time genes modulate the response to leafy activity. Genetics 1998, 150, 403–410. [Google Scholar]
- Amasino, R.M.; Michaels, S.D. The timing of flowering. Plant. Physiol. 2010, 154, 516–520. [Google Scholar] [CrossRef]
- Yamauchi, Y.; Ogawa, M.; Kuwahara, A.; Hanada, A.; Kamiya, Y.; Yamaguchi, S. Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of arabidopsis thaliana seeds. Plant. Cell 2004, 16, 367–378. [Google Scholar] [CrossRef] [Green Version]
- Colebrook, E.H.; Thomas, S.G.; Phillips, A.L.; Hedden, P. The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Biol. 2014, 217, 67–75. [Google Scholar] [CrossRef] [Green Version]






© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jung, H.; Jo, S.H.; Jung, W.Y.; Park, H.J.; Lee, A.; Moon, J.S.; Seong, S.Y.; Kim, J.-K.; Kim, Y.-S.; Cho, H.S. Gibberellin Promotes Bolting and Flowering via the Floral Integrators RsFT and RsSOC1-1 under Marginal Vernalization in Radish. Plants 2020, 9, 594. https://doi.org/10.3390/plants9050594
Jung H, Jo SH, Jung WY, Park HJ, Lee A, Moon JS, Seong SY, Kim J-K, Kim Y-S, Cho HS. Gibberellin Promotes Bolting and Flowering via the Floral Integrators RsFT and RsSOC1-1 under Marginal Vernalization in Radish. Plants. 2020; 9(5):594. https://doi.org/10.3390/plants9050594
Chicago/Turabian StyleJung, Haemyeong, Seung Hee Jo, Won Yong Jung, Hyun Ji Park, Areum Lee, Jae Sun Moon, So Yoon Seong, Ju-Kon Kim, Youn-Sung Kim, and Hye Sun Cho. 2020. "Gibberellin Promotes Bolting and Flowering via the Floral Integrators RsFT and RsSOC1-1 under Marginal Vernalization in Radish" Plants 9, no. 5: 594. https://doi.org/10.3390/plants9050594