Comparative Analysis of Root Phenolic Profiles and Antioxidant Activity of Five Native and Invasive Solidago L. Species
Abstract
:1. Introduction
2. Results
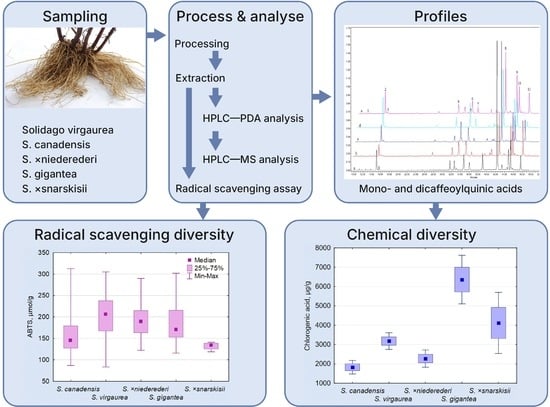
2.1. Phenolic Profiling and Comparative Quantitative of Solidago spp. Roots
2.2. Intraspecific Chemical Differences
2.3. Principal Component Analysis: Relationships between Species
2.4. Antioxidant Activity
3. Discussion
4. Material and Methods
4.1. Plant Material and Sampling
4.2. Chemicals and Reagents
4.3. Extraction
4.4. HPLC—PDA Analysis
4.5. HPLC—MS Analysis
4.6. ABTS Radical-Scavenging Assay
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Site No | (C) | (V) | (H) | (G) | (S) | Locality | Latitude (N) | Longitude (E) | Habitat |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 3 | 3 | 3 | – | – | Barčiai, Trakai distr. | 54.599774 | 24.808421 | Abandoned field |
| 2 | 3 | 2 | 3 | 3 | – | Bajorai, Vilnius | 54.749690 | 25.223965 | Disturbed dry site |
| 3 | 3 | 3 | 3 | – | – | Baltoji Vokė, Vilnius distr. | 54.571394 | 25.197609 | Disturbed dry site |
| 4 | 3 | 3 | 2 | – | – | Aukštieji Paneriai, Vilnius | 54.614087 | 25.070075 | Disturbed dry site |
| 5 | 3 | 2 | 3 | – | – | Velbiškės, Utena distr. | 55.415368 | 25.524135 | Abandoned dry grassland |
| 6 | 3 | 3 | 3 | – | – | Paberžė, Vilnius distr. | 54.958758 | 25.222206 | Abandoned mesic grassland |
| 7 | 3 | 3 | – | 3 | – | Pumaliai, Klaipėda distr. | 55.775298 | 21.156373 | Abandoned mesic grassland |
| 8 | 3 | 3 | 4 | – | – | Miegonys, Rokiškis distr. | 55.997043 | 25.620712 | Disturbed dry site |
| 9 | 3 | 3 | 3 | 3 | – | Užugriovis, Vilnius distr. | 54.827827 | 25.246075 | Abandoned mesic grassland |
| 10 | 3 | 3 | 3 | – | – | Sklėriškės, Vilnius distr. | 54.906741 | 25.559781 | Abandoned dry grassland |
| 11 | 3 | 4 | 3 | – | – | Ažulaukė, Vilnius distr. | 54.869584 | 25.375344 | Abandoned dry grassland |
| 12 | 3 | 3 | 2 | 3 | – | Gulbinai, Vilnius | 54.773923 | 25.285961 | Abandoned dry grassland |
| 13 | 3 | 3 | 3 | – | – | Keblonys, Anykščiai distr. | 55.517329 | 25.127204 | Abandoned dry grassland |
| 14 | 3 | 3 | 2 | 3 | – | Salininkai, Vilnius | 54.618889 | 25.289712 | Disturbed dry site |
| 15 | 3 | 3 | 3 | – | – | Molėtai | 55.217762 | 25.416347 | Abandoned dry grassland |
| 16 | – | – | – | 3 | – | Pakrovai, Prienai distr. | 54.571531 | 24.293226 | Abandoned mesic grassland |
| 17 | – | – | – | 3 | – | Vaišvydava, Kaunas | 54.840549 | 23.991645 | Disturbed dry site |
| 18 | – | – | – | – | 3 | Zabarauskai, Trakai distr. | 54.555114 | 24.512660 | Abandoned mesic grassland |
| 19 | – | – | – | – | 1 | Mažieji Gulbinai, Vilnius | 54.777352 | 25.292256 | Field collection |
References
- Semple, J.C.; Beck, J.B. Revised infrageneric classification of Solidago (Asteraceae: Astereae). Phytoneuron 2021, 10, 1–6. [Google Scholar]
- Cheek, M.D.; Semple, J.C. First official record of naturalised populations of Solidago altissima L. var. pluricephala M.C. Johnst. (Asteraceae: Astereae) in Africa. S. Afr. J. Bot. 2016, 105, 333–336. [Google Scholar] [CrossRef]
- Rosef, L.; Ingebrigtsen, H.H.; Heegaard, E. Vegetative propagation of Solidago canadensis—Do fragment size and burial depth matter? Weed Res. 2020, 60, 132–141. [Google Scholar] [CrossRef]
- Weber, E. Biological flora of Central Europe: Solidago altissima L. Flora 2000, 195, 123–134. [Google Scholar] [CrossRef]
- Weber, E.; Jakobs, G. Biological flora of central Europe: Solidago gigantea Aiton. Flora 2005, 200, 109–118. [Google Scholar] [CrossRef]
- Ren, G.Q.; Yang, H.Y.; Li, J.; Prabakaran, K.; Dai, Z.C.; Wang, X.P.; Jiang, K.; Zou, C.B.; Du, D.L. The effect of nitrogen and temperature changes on Solidago canadensis phenotypic plasticity and fitness. Plant Species Biol. 2020, 35, 283–299. [Google Scholar] [CrossRef]
- Ren, G.; Yang, B.; Cui, M.; Dai, Z.; Xiang, Y.; Zhang, H.; Li, G.; Li, J.; Javed, Q.; Du, D. Warming and elevated nitrogen deposition accelerate the invasion process of Solidago canadensis L. Ecol. Process. 2022, 11, 62. [Google Scholar] [CrossRef]
- Callaway, R.M.; Ridenour, W.M. Novel weapons: Invasive success and the evolution of increased competitive ability. Front. Ecol. Environ. 2004, 2, 436–443. [Google Scholar] [CrossRef]
- Pliszko, A.; Łazarski, G.; Kalinowski, P.; Adamowski, W.; Rutkowski, L.; Puchałka, R. An updated distribution of Solidago ×niederederi (Asteraceae) in Poland. Acta Mus. Siles. Sci. Natur. 2017, 66, 253–258. [Google Scholar] [CrossRef]
- Pliszko, A.; Kostrakiewicz-Gierałt, K. Resolving the naturalization strategy of Solidago ×niederederi (Asteraceae) by the production of sexual ramets and seedlings. Plant Ecol. 2017, 218, 1243–1253. [Google Scholar] [CrossRef]
- Pliszko, A.; Kostrakiewicz-Gierałt, K.; Makuch-Pietraś, I. The effect of site conditions and type of ramet clusters on sexual and asexual ramets of Solidago ×niederederi (Asteraceae). Neobiota 2023, 85, 125–143. [Google Scholar] [CrossRef]
- Jaźwa, M.; Jędrzejczak, E.; Klichowska, E.; Pliszko, A. Predicting the potential distribution area of Solidago × niederederi (Asteraceae). Turk. J. Bot. 2018, 42, 51–56. [Google Scholar] [CrossRef]
- Gudžinskas, Z.; Žalneravičius, E. Solidago ×snarskisii nothosp. nov. (Asteraceae) from Lithuania and its position in the infrageneric classification of the genus. Phytotaxa 2016, 253, 147–155. [Google Scholar] [CrossRef]
- Pliszko, A. First record of Solidago ×snarskisii (Asteraceae) in Poland. Botanica 2018, 24, 211–213. [Google Scholar] [CrossRef]
- Vinogradova, Y.K.; Galkina, M.A. Hybridization as a factor of invasive activity of alien species of goldenrods (Solidago). Zhurnal Obs. Biol. 2019, 80, 43–56. [Google Scholar] [CrossRef]
- Pyšek, P.; Richardson, D.M.; Rejmánek, M.; Webster, G.L.; Williamson, M.; Kirschner, J. Alien plants in checklists and floras: Towards better communication between taxonomists and ecologists. Taxon 2004, 53, 131–143. [Google Scholar] [CrossRef]
- EPPO Lists of Invasive Alien Plants. EPPO, Paris. Available online: https://www.eppo.int/ACTIVITIES/invasive_alien_plants/iap_lists (accessed on 22 August 2023).
- Domaradzki, K.; Badowski, M. Możliwość chemicznego ograniczania występowania Solidago gigantea Aiton na terenach odłogowanych. Zesz. Nauk. UP Wroc. Rol. 2012, 584, 17. (In Polish) [Google Scholar]
- Apáti, P.; Szentmihályi, K.; Kristó, S.T.; Papp, I.; Vinkler, P.; Szoke, E.; Kéry, A. Herbal remedies of Solidago-correlation of phytochemical characteristics and antioxidative properties. J. Pharm. Biomed. Anal. 2003, 32, 1045–1053. [Google Scholar] [CrossRef] [PubMed]
- ESCOP Monographs. Solidaginis Virgaureae Herba (European Goldenrod). 2018. Available online: https://escop.com/downloads/solidaginis-virgaureae-herba-european-goldenrod (accessed on 17 October 2023).
- Fursenco, C.; Calalb, T.; Uncu, L.; Dinu, M.; Ancuceanu, R. Solidago virgaurea L.: A review of its ethnomedicinal uses, phytochemistry, and pharmacological activities. Biomolecules 2020, 10, 1619. [Google Scholar] [CrossRef]
- Kołodziej, B.; Kowalski, R.; Kedzia, B. Antibacterial and antimutagenic activity of extracts aboveground parts of three Solidago species: Solidago virgaurea L., Solidago canadensis L. and Solidago gigantea Ait. J. Med. Plants Res. 2011, 5, 6770–6779. [Google Scholar] [CrossRef]
- Liu, S.; Shao, X.; Wei, Y.; Li, Y.; Xu, F.; Wang, H. Solidago canadensis L. essential oil vapor effectively inhibits Botrytis cinerea growth and preserves postharvest quality of strawberry as a food model system. Front. Microbiol. 2016, 7, 1179. [Google Scholar] [CrossRef] [PubMed]
- Benelli, G.; Pavela, R.; Cianfaglione, K.; Nagy, D.U.; Canale, A.; Maggi, F. Evaluation of two invasive plant invaders in Europe (Solidago canadensis and Solidago gigantea) as possible sources of botanical insecticides. J. Pest. Sci. 2019, 92, 805–821. [Google Scholar] [CrossRef]
- Werrie, P.-Y.; Durenne, B.; Delaplace, P.; Fauconnier, M.-L. Phytotoxicity of EOs: Opportunities and constraints for the development of biopesticides. A Review. Foods 2020, 9, 1291. [Google Scholar] [CrossRef]
- Krüzselyi, D.; Bakonyi, J.; Ott, P.G.; Darcsi, A.; Csontos, P.; Morlock, G.E.; Moricz, A.M. Goldenrod root compounds active against crop pathogenic fungi. J. Agric. Food Chem. 2021, 69, 12686–12694. [Google Scholar] [CrossRef] [PubMed]
- Móricz, Á.M.; Krüzselyi, D.; Ott, P.G.; Garádi, Z.; Béni, S.; Morlock, G.E.; Bakonyi, J. Bioactive clerodane diterpenes of giant goldenrod (Solidago gigantea Ait.) root extract. J. Chromatogr. A 2021, 1635, 461727. [Google Scholar] [CrossRef]
- Baglyas, M.; Ott, P.G.; Garádi, Z.; Glavnik, V.; Béni, S.; Vovk, I.; Móricz, Á.M. High-performance thin-layer chromatography—Antibacterial assay first reveals bioactive clerodane diterpenes in Giant goldenrod (Solidago gigantea Ait.). J. Chromatogr. A 2022, 1677, 463308. [Google Scholar] [CrossRef]
- Baglyas, M.; Ott, P.G.; Schwarczinger, I.; Nagy, J.K.; Darcsi, A.; Bakonyi, J.; Móricz, Á.M. Antimicrobial diterpenes from Rough goldenrod (Solidago rugosa Mill.). Molecules 2023, 28, 3790. [Google Scholar] [CrossRef]
- Radušienė, J.; Karpavičienė, B.; Marksa, M.; Ivanauskas, L.; Raudonė, L. Distribution patterns of essential oil terpenes in native and invasive Solidago species and their comparative assessment. Plants 2022, 11, 1159. [Google Scholar] [CrossRef]
- Hamed, Y.S.; Abdin, M.; Chen, G.; Akhtar, H.M.S.; Zeng, X. Effects of impregnate temperature on extraction of caffeoylquinic acid derivatives from Moringa oleifera leaves and evaluation of inhibitory activity on digestive enzyme, antioxidant, anti-proliferative and antibacterial activities of the extract. Int. J. Food Sci. Technol. 2020, 55, 3082–3090. [Google Scholar] [CrossRef]
- Nzekoue, F.K.; Angeloni, C.; Vitali, L.A.; Sagratini, G.; Vittori, S.; Angeloni, S.; Freschi, M.; Hrelia, S.; Vitali, L.A.; Sagratini, G.; et al. Coffee silverskin extracts: Quantification of 30 bioactive compounds by a new HPLC-MS/MS method and evaluation of their antioxidant and antibacterial activities. Food Res. Int. 2020, 133, 109128. [Google Scholar] [CrossRef]
- Trendafilova, A.; Ivanova, V.; Rangelov, M.; Todorova, M.; Ozek, G.; Yur, S.; Ozek, T.; Aneva, I.; Veleva, R.; Moskova-Doumanova, V.; et al. Caffeoylquinic acids, cytotoxic, antioxidant, acetylcholinesterase and tyrosinase enzyme inhibitory activities of six inula species from Bulgaria. Chem. Biodivers. 2020, 17, e2000051. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Li, J.; Zhang, X.; Zu, Y.; Yang, Y.; Liu, W.; Xu, Z.; Gao, H.; Sun, X.; Jiang, X.; et al. Current advances in naturally occurring caffeoylquinic acids: Structure, bioactivity, and synthesis. J. Agric. Food Chem. 2020, 68, 10489–10516. [Google Scholar] [CrossRef] [PubMed]
- Matthews, D.G.; Caruso, M.; Alcazar Magana, A.; Wright, K.M.; Maier, C.S.; Stevens, J.F.; Gray, N.E.; Quinn, J.F.; Soumyanath, A. Caffeoylquinic acids in Centella asiatica reverse cognitive deficits in male 5XFAD Alzheimer’s disease model mice. Nutrients 2020, 12, 3488. [Google Scholar] [CrossRef] [PubMed]
- Gray, N.E.; Morrè, J.; Kelley, J.; Maier, C.S.; Stevens, J.F.; Quinn, J.F.; Soumyanath, A. Caffeoylquinic acids in Centella asiatica protect against amyloid-β toxicity. J. Alzheimers Dis. 2014, 40, 359–373. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, K.; Han, J.; Shimozono, H.; Villareal, M.O.; Isoda, H. Caffeoylquinic acid-rich purple sweet potato extract, with or without anthocyanin, imparts neuroprotection and contributes to the improvement of spatial learning and memory of SAMP8 mouse. J. Agric. Food Chem. 2013, 61, 5037–5045. [Google Scholar] [CrossRef] [PubMed]
- Metwally, D.M.; Alajmi, R.A.; El-Khadragy, M.F.; Yehia, H.M.; Al-Megrin, W.A.; Akabawy, A.M.A.; Amin, H.K.; Abdel Moneim, A.E. Chlorogenic acid confers robust neuroprotection against arsenite toxicity in mice by reversing oxidative stress, inflammation, and apoptosis. J. Funct. Foods 2020, 75, 104202. [Google Scholar] [CrossRef]
- Mishra, D.; Joshi, S.; Bisht, G.; Pilkhwal, S. Chemical composition and antimicrobial activity of Solidago canadensis Linn. root essential oil. J. Basic. Clin. Pharm. 2010, 1, 187–190. [Google Scholar]
- Lu, T.S.; Menelaou, M.A.; Vargas, D.; Fronczek, F.R.; Fischer, N.H. Polyacetylenes and diterpenes from Solidago canadensis. Phytochemistry 1993, 32, 1483–1488. [Google Scholar]
- Móricz, Á.M.; Jamshidi-Aidji, M.; Krüzselyi, D.; Darcsi, A.; Böszörményi, A.; Csontos, P.; Béni, S.; Ott, P.G.; Morlock, G.E. Distinction and valorization of 30 root extracts of five goldenrod (Solidago) species. J. Chromatogr. A 2020, 1611, 460602. [Google Scholar] [CrossRef]
- Schmeda-Hirschmann, G.; Rodriguez, J.; Astudillo, L. Gastroprotective activity of the diterpene solidagenone and its derivatives on experimentally induced gastric lesions in mice. J. Ethnopharmacol. 2002, 81, 111–115. [Google Scholar] [CrossRef]
- Marksa, M.; Zymone, K.; Ivanauskas, L.; Radušienė, J.; Pukalskas, A.; Raudonė, L. Antioxidant profiles of leaves and inflorescences of native, invasive and hybrid Solidago species. Ind. Crop. Prod. 2020, 145, 112123. [Google Scholar] [CrossRef]
- Raudone, L.; Vilkickyte, G.; Pitkauskaite, L.; Raudonis, R.; Vainoriene, R.; Motiekaityte, V. Antioxidant activities of Vaccinium vitis-idaea L. leaves within cultivars and their phenolic compounds. Molecules 2019, 24, 844. [Google Scholar] [CrossRef] [PubMed]
- Rice-Evans, C.A.; Miller, N.J.; Paganga, G. Structure-antioxidant activity relationship of flavonoids and phenolic acids. Free Radic. Biol. Med. 1996, 20, 933–956. [Google Scholar] [CrossRef] [PubMed]
- Kundu, A.; Vadassery, J. Chlorogenic acid-mediated chemical defence of plants against insect herbivores. Plant Biol. 2019, 21, 185–189. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Shahrajabian, M.H. Therapeutic potential of phenolic compounds in medicinal plants—Natural health products for human health. Molecules 2023, 28, 1845. [Google Scholar] [CrossRef] [PubMed]
- Radušienė, J.; Marksa, M.; Karpavičienė, B. Assessment of Solidago ×niederederi origin based on the accumulation of phenolic compounds in plant raw materials. Weed Sci. 2018, 66, 324–330. [Google Scholar] [CrossRef]
- Hull-Sanders, H.M.; Johnson, R.H.; Owen, H.A.; Meyer, G.A. Effects of polyploidy on secondary chemistry, physiology, and performance of native and invasive genotypes of Solidago gigantea (Asteraceae). Am. J. Bot. 2009, 96, 762–770. [Google Scholar] [CrossRef]
- Madani, H.; Escrich, A.; Hosseini, B.; Sanchez-Muñoz, R.; Khojasteh, A.; Palazon, J. Effect of polyploidy induction on natural metabolite production in medicinal plants. Biomolecules 2021, 11, 899. [Google Scholar] [CrossRef]
- Stefanowicz, A.M.; Kapusta, P.; Stanek, M.; Frąc, M.; Oszust, K.; Woch, M.W.; Zubek, S. Invasive plant Reynoutria japonica produces large amounts of phenolic compounds and reduces the biomass but not activity of soil microbial communities. Sci. Total Environ. 2021, 767, 145439. [Google Scholar] [CrossRef]
- Kiełtyk, P.; Mirek, Z. Taxonomy of the Solidago virgaurea group (Asteraceae) in Poland, with special reference to variability along an altitudinal gradient. Folia Geobot. 2014, 49, 259–282. [Google Scholar] [CrossRef]
- Szymura, M.; Szymura, T.H.; Rajsz, A.; Świerszcz, S. Is phenotypic plasticity an explanation for the invasiveness of goldenrods (Solidago and Euthamia) in Europe? Plant Species Biol. 2019, 34, 73–84. [Google Scholar] [CrossRef]
- Szymura, M.; Szymura, T.H. Interactions between alien goldenrods (Solidago and Euthamia species) and comparison with native species in Central Europe. Flora: Morphol. Distrib. Funct. Ecol. 2016, 218, 51–61. [Google Scholar] [CrossRef]
- Judžentienė, A. Allelopathic activity of canadian goldenrod (Solidago canadensis L.) extracts on seed germination and growth of lettuce (Lactuca sativa L.) and garden pepper cress (Lepidium sativum L.). Plants 2023, 12, 1421. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Wang, B.; Zhang, S.; Tang, J.; Tu, C.; Hu, S.; Yong, J.W.H.; Chen, X. Enhanced allelopathy and competitive ability of invasive plant Solidago canadensis in its introduced range. J. Plant Ecol. 2013, 6, 253–263. [Google Scholar] [CrossRef]
- Du, B.; Peng, F.; Cheng, C.; Chen, Y.; Wu, J.; Zhu, F.; Yang, Y. Phenolic profiles and antioxidant activities of exocarp, endocarp, and hypanthium of three pear cultivars grown in China. J. Food Bioact. 2021, 14, 75–80. [Google Scholar] [CrossRef]
- Yuan, Y.; Jin, H.; Li, J. Effects of latitude and soil microbes on the resistance of invasive Solidago canadensis to its co-evolved insect herbivore Corythucha marmorata. J. Plant Ecol. 2022, 15, 549–560. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, Y.; Zou, J.; Siemann, E. Decomposition of Phragmites australis litter retarded by invasive Solidago canadensis in mixtures: An antagonistic non-additive effect. Sci. Rep. 2014, 4, 5488. [Google Scholar] [CrossRef]
- Zhang, S.; Zhu, W.; Wang, B.X.; Tang, J.; Chen, X. Secondary metabolites from the invasive Solidago canadensis L. accumulation in soil and contribution to inhibition of soil pathogen Pythium ultimum. Appl. Soil. Ecol. 2011, 48, 280–286. [Google Scholar] [CrossRef]
- Yang, R.Y.; Mei, L.X.; Tang, J.J.; Chen, X. Allelopathic effects of invasive Solidago canadensis L. on germination and growth of native Chinese plant species. Allelopath. J. 2007, 19, 241–248. [Google Scholar]
- Hamilton, J.A.; Miller, J.M. Adaptive introgression as a resource for management and genetic conservation in a changing climate. Conserv. Biol. 2016, 30, 33–41. [Google Scholar] [CrossRef]
- Bautista, C.; Marsit, S.; Landry, C.R. Interspecific hybrids show a reduced adaptive potential under DNA damaging conditions. Evol. Appl. 2021, 14, 758–769. [Google Scholar] [CrossRef] [PubMed]
- Karpavičienė, B.; Radušienė, J. Morphological and anatomical characterization of Solidago ×niederederi and other sympatric Solidago species. Weed Sci. 2016, 64, 61–70. [Google Scholar] [CrossRef]
- Musiał, K.; Pagitz, K.; Gudžinskas, Z.; Łazarski, G.; Pliszko, A. Chromosome numbers in hybrids between invasive and native Solidago (Asteraceae) species in Europe. Phytotaxa 2020, 471, 267–275. [Google Scholar] [CrossRef]
- Orians, C.M. The effects of hybridization in plants on secondary chemistry: Implications for the ecology and evolution of plant-herbivore interactions. Am. J. Bot. 2000, 87, 1749–1756. [Google Scholar] [CrossRef] [PubMed]
- Cheng, D.; Vrieling, K.; Klinkhamer, P.G.L. The effect of hybridization on secondary metabolites and herbivore resistance: Implications for the evolution of chemical diversity in plants. Phytochem. Rev. 2011, 10, 107–117. [Google Scholar] [CrossRef]
- Vilkickyte, G.; Raudone, L.; Petrikaite, V. Phenolic fractions from Vaccinium vitis-idaea L. and their antioxidant and anticancer activities assessment. Antioxidants 2020, 9, 1261. [Google Scholar] [CrossRef]
- Raudone, L.; Radušiene, J.; Seyis, F.; Yayla, F.; Vilkickyte, G.; Marksa, M.; Ivanauskas, L.; Cırak, C. Distribution of phenolic compounds and antioxidant activity in plant parts and populations of seven underutilized wild Achillea species. Plants 2022, 11, 447. [Google Scholar] [CrossRef]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, K. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Rad. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef]







| Peak no. | Rt (min) | Tentative Identity | UV Max (nm) | [M-H]− (m/z) | Other Ions (m/z) |
|---|---|---|---|---|---|
| 8 | 42.3 | * Derivative of dicaffeoylquinic acid | 217, 245, 292 (sh), 325 | 629 | 585, 543, 113 |
| 9 | 44.9 | Derivative of phenolic acid I (Leiocarposide isomer) | 218, 292 (sh), 314 (sh), 325 | 613 | 569 |
| 10 | 45.3 | Derivative of phenolic acid II (Leiocarposide isomer) | 218, 292 (sh), 314 (sh), 325 | 613 | 569 |
| 11 | 47.6 | Derivative of phenolic acid III (Leiocarposide isomer) | 217, 240, 292 (sh), 316 (sh), 325 | 613 | 569 |
| 12 | 61.9 | Derivative of phenolic acid IV (Methyl 3,5-dicaffeoylquinate) | 209, 245, 311, 325 (sh) | 361 | 113 |
| Compounds | S. virgaurea (42) 1 | S. candensis (44) | S. ×niederederi (39) | S. gigantea (21) | S. ×snarskisii (4) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| M (±SD) | Range | M (±SD) | Range | M (±SD) | Range | M (±SD) | Range | M (±SD) | Range | |
| Neochlorogenic acid | 312.3 b (244.7) | 94.3–1108.9 | 88.6 a 2 (38.4) | 61.3–301.2 * | 106.0 ac (33.8) | 64.0–212.1 ** | 141.7 c (93.2) | 80.2–448.8 | 112.7 abc (19.5) | 83.9–125.5 * |
| Chlorogenic acid | 3181.3 b (1428.0) | 731.8–6229.2 * | 1827.7 a (1190.5) | 696.2–8509.0 | 2273.0 ac (1421.0) | 696.5–6526.4 * | 6360.9 d (2935.1) | 2566.8–13,252.0 | 4118.3 bcd (1611.5) | 2748.7–6447.0 |
| Cryptochlorogenic acid | 452.4 b (282.0) | 100.7–1185.5 | 120.7 a (46.5) | 44.0–240.5 | 180.0 a (95.1) | 81.7–515.7 | 433.5 b (302.8) | 171.0–1449.1 | 249.0 b (52.8) | 214.8–327.3 |
| 4,5-dicaffeoylquinic acid | 1752.4 ab (1212.7) | 201.6–4826.7 | 1371.0 a (1104.2) | 180.7–5955.6 | 1683.8 ab (1018.1) | 464.3–4716.7 | 2354.6 b (1401.2) | 495.1–5688.3 | 1723.7 ab (313.3) | 1483.2–2182.0 |
| 1,5-dicaffeoylquinic acid | 69.7 b (29.2) | 19.7–145.9 * | 43.6 a (17.9) | 22.0–95.4 | 49.4 a (28.7) | 26.8–170.8 | 72.1 b (24.7) | 46.8–135.6 | 86.5 b (14.1) | 68.6–103.2 |
| 3,5-dicaffeoylquinic acid | 1622.3 ab (942.0) | 351.0–3988.0 | 1229.6 a (649.0) | 412.7–3584.6 | 2064.4 bc (1469.1) | 312.5–7472.1 | 2836.7 c (1310.0) | 1094.5–5604.4 | 2160.6 abc (1078.5) | 1033.0–3623.5 |
| 3,4-dicaffeoylquinic acid | 950.4 a (630.5) | 116.4–2953.8 | 588.0 a (377.5) | 93.7–1557.6 | 854.9 a (658.5) | 259.8–3134.1 | 2197.3 b (1635.1) | 672.6–7039.6 | 1210.0 ab (587.3) | 743.5–2068.5 |
| Dicaffeoylquinic acid derivative | 13,666.3 b (5106.7) | 5796.8–25,411.0 * | 5209.6 a (1652.8) | 1829.5–8635.3 * | 9143.7 c (2813.5) | 4444.9–17,185.1 | 8714.1 c (3020.2) | 2429.9–13,267.3 * | 5974.8 ac (655.3) | 5134.3–6717.9 |
| Phenolic acid I derivative | 3411.3 ab (1488.4) | 1050.0–9318.8 | 2971.1 a (1054.3) | 867.5–5157.4 | 4052.3 b (1664.2) | 1777.9–9153.9 | 3272.0 ab (830.5) | 1850.4–4924.6 | 3875.0 ab (1067.2) | 2948.5–5414.6 * |
| Phenolic acid II derivative | 3539.3 b (1216.8) | 1263.7–5728.5 | 2346.5 a (576.2) | 1075.8–3514.6 | 3673.4 b (1346.6) | 2095.4–7532.2 | 3201.1 b (1319.3) | 1049.3–5706.5 * | 3695.5 b (409.5) | 3331.8–4265.6 |
| Phenolic acid III derivative | 2170.2 a (741.8) | 593.2–3915.3 * | 2765.6 ab (1019.6) | 942.6–4848.4 | 3620.2 b (1930.7) | 971.5–9347.3 | 1267.3 c (528.3) | 659.1–2305.8 | 1895.0 abc (1152.2) | 860.2–3020.1 |
| Phenolic acid IV derivative | 6606.5 b (4188.0) | 1842.9–21,480.2 * | 868.0 ac (770.0) | 98.8–4683.0 | 1834.0 a (2323.0) | 216.0–14,780.1 | 658.8 c (671.0) | 66.2–2585.2 | 196.0 c (69.0) | 131.1–278.8 |
| Total | 37,734.4 b (11,977.2) | 16,200.3–61,935.5 | 19,429.9 a (4912.8) | 7794.5–32,284.8 * | 29,535.2 b (8382.3) | 16,467.4–51,243.8 | 31,510.2 b (9895.2) | 13,256.2–50,049.7 * | 25,297.1 ab (2201.0) | 23,301.7–28,300.0 |
| ABTS | 205.0 b (52.5) | 83.3–305.0 | 154.4 a (42.2) | 86.7–312.2 | 193.1 b (39.8) | 122.2–289.4 | 187.4 ab (50.1) | 115.6–301.7 * | 131.9 a (9.5) | 118.9–141.1 |
| Variables | PCA1 | PCA2 | ||
|---|---|---|---|---|
| PC 1 | PC 2 | PC 1 | PC 2 | |
| Neochlorogenic acid | 0.72 | −0.48 | 0.60 | −0.48 |
| Chlorogenic acid | 0.79 | −0.05 | 0.65 | 0.66 |
| Cryptochlorogenic acid | 0.82 | −0.41 | 0.83 | −0.11 |
| 4,5-dicaffeoylquinic acid | 0.73 | 0.22 | 0.82 | 0.26 |
| 1,5-dicaffeoylquinic acid | 0.48 | −0.23 | 0.27 | 0.02 |
| 3,5-dicaffeoylquinic acid | 0.70 | 0.41 | 0.77 | 0.50 |
| 3,4-dicaffeoylquinic acid | 0.84 | 0.24 | 0.78 | 0.50 |
| Dicaffeoylquinic acid derivative | 0.83 | −0.26 | 0.59 | −0.64 |
| Phenolic acid I derivative | 0.47 | 0.58 | 0.55 | −0.47 |
| Phenolic acid II derivative | 0.75 | 0.35 | 0.73 | −0.34 |
| Phenolic acid III derivative | 0.08 | 0.75 | 0.21 | −0.76 |
| Phenolic acid IV derivative | 0.44 | −0.45 | −0.01 | −0.75 |
| Variables | S. virgaurea | S. canadensis | S. ×niederederi | S. gigantea | S. ×snarskisii | Total |
|---|---|---|---|---|---|---|
| Neochlorogenic acid | 0.52 ** | −0.19 | 0.27 | 0.31 | −0.69 | 0.43 ** |
| Chlorogenic acid | 0.49 ** | 0.04 | 0.41 * | 0.46 * | −0.24 | 0.33 ** |
| Cryptochlorogenic acid | 0.68 ** | −0.01 | 0.37 * | 0.37 | −0.17 | 0.51 ** |
| 4,5-dicaffeoylquinic acid | 0.64 ** | 0.22 | 0.38 * | 0.15 | −0.23 | 0.40 ** |
| 1,5--dicaffeoylquinic acid | 0.13 | −0.10 | −0.07 | −0.29 | −0.42 | 0.09 |
| 3,5-dicaffeoylquinic acid | 0.51 ** | 0.27 | 0.41 * | 0.31 | −0.36 | 0.38 ** |
| 3,4-dicaffeoylquinic acid | 0.70 ** | 0.29 | 0.50 ** | 0.30 | −0.21 | 0.38 ** |
| Dicaffeoylquinic acid derivative | 0.74 ** | 0.37 * | 0.42 * | 0.28 | 0.25 | 0.63 ** |
| Phenolic acid I derivative | 0.67 ** | 0.45 * | 0.46 * | 0.07 | 0.55 | 0.49 ** |
| Phenolic acid II derivative | 0.70 ** | 0.44 * | 0.61 ** | 0.07 | 0.10 | 0.57 ** |
| Phenolic acid III derivative | 0.53 ** | 0.40 * | 0.36 * | −0.15 | 0.84 | 0.24 * |
| Phenolic acid IV derivative | 0.2 | −0.05 | 0.03 | 0.09 | 0.09 | 0.30 ** |
| Total | 0.8 ** | 0.46 * | 0.65 ** | 0.36 | 0.35 | 0.70 ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Radušienė, J.; Karpavičienė, B.; Vilkickytė, G.; Marksa, M.; Raudonė, L. Comparative Analysis of Root Phenolic Profiles and Antioxidant Activity of Five Native and Invasive Solidago L. Species. Plants 2024, 13, 132. https://doi.org/10.3390/plants13010132
Radušienė J, Karpavičienė B, Vilkickytė G, Marksa M, Raudonė L. Comparative Analysis of Root Phenolic Profiles and Antioxidant Activity of Five Native and Invasive Solidago L. Species. Plants. 2024; 13(1):132. https://doi.org/10.3390/plants13010132
Chicago/Turabian StyleRadušienė, Jolita, Birutė Karpavičienė, Gabrielė Vilkickytė, Mindaugas Marksa, and Lina Raudonė. 2024. "Comparative Analysis of Root Phenolic Profiles and Antioxidant Activity of Five Native and Invasive Solidago L. Species" Plants 13, no. 1: 132. https://doi.org/10.3390/plants13010132