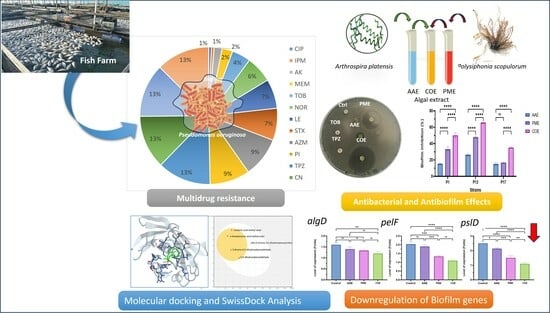
Unveiling the Potential of Algal Extracts as Promising Antibacterial and Antibiofilm Agents against Multidrug-Resistant Pseudomonas aeruginosa: In Vitro and In Silico Studies including Molecular Docking
Abstract
:1. Introduction
2. Results
2.1. Isolation, Quantification, and Characterization of Pseudomonas aeruginosa
2.2. Antibiotic Susceptibility Profile of P. aeruginosa Isolates (Disc Diffusion Method)
2.3. Biofilm-Producing Potential of P. aeruginosa Isolates
2.4. The Antibacterial Efficacy of Crude Extracts
2.5. Determination of MIC of Crude Extracts
2.6. Determination of the Antibiofilm Activity of Algal Extracts
2.7. Molecular Identification of MDR P. aeruginosa Strain P13
2.8. Effect of the Algal Extract on the Biofilm Architecture of P. aeruginosa Using SEM
2.9. Determination of Gene Expression with Quantitative RT-PCR
2.10. Chemical Analysis of the Algal Extract Used in This Study Using GC–MS Chromatograms
2.11. Physicochemical Properties of the Algal Extracts
2.12. Water Solubility, Pharmacokinetics, Drug Likeness, and Toxicity
2.13. Docking Studies
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Algal Collection
4.3. Preparation of Organic Algal Extracts
4.4. Bacterial Sample Collection
4.5. Quantification and Identification of Pseudomonas aeruginosa
4.6. Antibiotic Susceptibility Test (Disc Diffusion Method)
4.7. Quantitative Assessment of Biofilm Formation
4.8. Detection of Specific Biofilm Genes in Selected Bacterial Isolates
4.9. Antibacterial Efficacy of Algal Extracts: Individual and Combination Assessments
4.10. Determination of the Minimum Inhibitory Concentration (MIC) Using Broth Microdilution Technique
4.11. Determination of Antibiofilm Activity and Biofilm Inhibitory Percentage for the Algal Extracts
4.12. Molecular Identification of the Selected MDR P. aeruginosa Strain
4.13. Scanning Electron Microscopy (SEM)
4.14. Determination of Gene Expression with Quantitative Real-Time PCR
4.15. GC–MS Characterization
4.16. Evaluation of Pharmacokinetics and Toxicity
4.17. Molecular Docking Studies
4.18. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Paul, D.; Gopal, J.; Kumar, M.; Manikandan, M. Nature to the natural rescue: Silencing microbial chats. Chem. Biol. Interact. 2018, 280, 86–98. [Google Scholar] [CrossRef] [PubMed]
- Woolhouse, M.; Farrar, J. Policy: An intergovernmental panel on antimicrobial resistance. Nature 2014, 509, 555–557. [Google Scholar] [CrossRef] [PubMed]
- Algammal, A.M.; Mabrok, M.; Sivaramasamy, E.; Youssef, F.M.; Atwa, M.H.; El-kholy, A.W.; Hetta, H.F.; Hozzein, W.N. Emerging MDR-Pseudomonas aeruginosa in fish commonly harbor oprL and toxA virulence genes and blaTEM, blaCTX-M, and tetA antibiotic-resistance genes. Sci. Rep. 2020, 10, 15961. [Google Scholar] [CrossRef]
- Tümmler, B. What Makes Pseudomonas aeruginosa a Pathogen? In Pseudomonas aeruginosa. Advances in Experimental Medicine and Biology; Filloux, A., Ramos, J.L., Eds.; Springer: Berlin/Heidelberg, Germany, 2022; pp. 283–301. Available online: https://link.springer.com/chapter/10.1007/978-3-031-08491-1_10 (accessed on 15 July 2023).
- Qin, S.; Xiao, W.; Zhou, C.; Pu, Q.; Deng, X.; Lan, L.; Liang, H.; Song, X.; Wu, M. Pseudomonas aeruginosa: Pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics. Signal Transduct. Target. Ther. 2022, 7, 199. [Google Scholar] [CrossRef]
- WHO. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Develipment of New Antibiotics; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Breidenstein, E.B.M.; de la Fuente-Núñez, C.; Hancock, R.E.W. Pseudomonas aeruginosa: All roads lead to resistance. Trends Microbiol. 2011, 19, 419–426. [Google Scholar] [CrossRef]
- Moradali, M.F.; Ghods, S.; Rehm, B.H.A. Pseudomonas aeruginosa lifestyle: A paradigm for adaptation, survival, and persistence. Front. Cell. Infect. Microbiol. 2017, 7, 39. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.S.; Mukherjee, P.K.; Ghannoum, M.A. Biofilm antimicrobial resistance. In Microbial Biofilms; Wiley: Hoboken, NJ, USA, 2004; pp. 250–268. [Google Scholar]
- Ma, L.; Conover, M.; Lu, H.; Parsek, M.R.; Bayles, K.; Wozniak, D.J. Assembly and Development of the Pseudomonas aeruginosa Biofilm Matrix. PLoS Pathog. 2009, 5, e1000354. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef]
- Campanac, C.; Pineau, L.; Payard, A.; Baziard-Mouysset, G.; Roques, C. Interactions between biocide cationic agents and bacterial biofilms. Antimicrob. Agents Chemother. 2002, 46, 1469–1474. [Google Scholar] [CrossRef]
- Rima, M.; Trognon, J.; Latapie, L.; Chbani, A.; Roques, C.; Garah, F. El Seaweed Extracts: A Promising Source of Antibiofilm Agents with Distinct Mechanisms of Action against Pseudomonas aeruginosa. Mar. Drugs 2022, 20, 92. [Google Scholar] [CrossRef]
- Mishra, R.; Panda, A.K.; De Mandal, S.; Shakeel, M.; Bisht, S.S.; Khan, J. Natural anti-biofilm agents: Strategies to control biofilm-forming pathogens. Front. Microbiol. 2020, 11, 566325. [Google Scholar] [CrossRef] [PubMed]
- Dahms, H.; Dobretsov, S. Antifouling Compounds from Marine Macroalgae. Mar. Drugs 2017, 15, 265. [Google Scholar] [CrossRef]
- Bamunuarachchi, N.I.; Khan, F.; Kim, Y.-M. Bactericidal Activity of Sargassum aquifolium (Turner) C. Agardh against Gram-positive and Gram-negative Biofilm-forming Pathogenic Bacteria. Curr. Pharm. Biotechnol. 2021, 22, 1628–1640. [Google Scholar] [CrossRef]
- Leandro, A.; Pereira, L.; Gonçalves, A.M.M. Diverse Applications of Marine Macroalgae. Mar. Drugs 2019, 18, 17. [Google Scholar] [CrossRef] [PubMed]
- Elshobary, M.E.; El-Shenody, R.A.; Ashour, M.; Zabed, H.M.; Qi, X. Antimicrobial and antioxidant characterization of bioactive components from Chlorococcum minutum. Food Biosci. 2020, 35, 100567. [Google Scholar] [CrossRef]
- Ashour, M.; Al-Souti, A.S.; Hassan, S.M.; Ammar, G.A.G.; Goda, A.M.A.; El-Shenody, R.; Abomohra, A.E.-F.; El-Haroun, E.; Elshobary, M.E. Commercial Seaweed Liquid Extract as Strawberry Biostimulants and Bioethanol Production. Life 2022, 13, 85. [Google Scholar] [CrossRef] [PubMed]
- Montalvo, G.E.B.; de Souza Vandenberghe, L.P.; Soccol, V.T.; de Carvalho, J.C.; Soccol, C.R. The Antihypertensive, Antimicrobial and Anticancer Peptides from Arthrospira with Therapeutic Potential: A Mini Review. Curr. Mol. Med. 2020, 20, 593–606. [Google Scholar] [CrossRef] [PubMed]
- Nag, M.; Lahiri, D.; Dey, A.; Sarkar, T.; Joshi, S.; Ray, R.R. Evaluation of algal active compounds as potent antibiofilm agent. J. Basic Microbiol. 2022, 62, 1098–1109. [Google Scholar] [CrossRef] [PubMed]
- Laverty, G.; Gorman, S.; Gilmore, B. Biomolecular Mechanisms of Pseudomonas aeruginosa and Escherichia coli Biofilm Formation. Pathogens 2014, 3, 596–632. [Google Scholar] [CrossRef] [PubMed]
- Campisano, A.; Schroeder, C.; Schemionek, M.; Overhage, J.; Rehm, B.H.A. PslD Is a Secreted Protein Required for Biofilm Formation by Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2006, 72, 3066–3068. [Google Scholar] [CrossRef] [PubMed]
- Jennings, L.K.; Storek, K.M.; Ledvina, H.E.; Coulon, C.; Marmont, L.S.; Sadovskaya, I.; Secor, P.R.; Tseng, B.S.; Scian, M.; Filloux, A.; et al. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc. Natl. Acad. Sci. USA 2015, 112, 11353–11358. [Google Scholar] [CrossRef]
- Friedman, L.; Kolter, R. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 2003, 51, 675–690. [Google Scholar] [CrossRef] [PubMed]
- Ghafoor, A.; Jordens, Z.; Rehm, B.H.A. Role of PelF in Pel Polysaccharide Biosynthesis in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2013, 79, 2968–2978. [Google Scholar] [CrossRef] [PubMed]
- Gil-Perotin, S.; Ramirez, P.; Marti, V.; Sahuquillo, J.M.; Gonzalez, E.; Calleja, I.; Menendez, R.; Bonastre, J. Implications of endotracheal tube biofilm in ventilator-associated pneumonia response: A state of concept. Crit. Care 2012, 16, R93. [Google Scholar] [CrossRef] [PubMed]
- Talebi Bezmin Abadi, A.; Rizvanov, A.A.; Haertlé, T.; Blatt, N.L. World Health Organization report: Current crisis of antibiotic resistance. Bionanoscience 2019, 9, 778–788. [Google Scholar] [CrossRef]
- de Macedo, J.L.S.; Santos, J.B. Bacterial and fungal colonization of burn wounds. Mem. Inst. Oswaldo Cruz 2005, 100, 535–539. [Google Scholar] [CrossRef]
- Selvadoss, P.P.; Nellore, J.; Ravindrran, M.B.; Sekar, U.; Tippabathani, J. Enhancement of antimicrobial activity by liposomal oleic acid-loaded antibiotics for the treatment of multidrug-resistant Pseudomonas aeruginosa. Artif. CellsNanomed. Biotechnol. 2018, 46, 268–273. [Google Scholar] [CrossRef]
- Vital-Lopez, F.G.; Reifman, J.; Wallqvist, A. Biofilm Formation Mechanisms of Pseudomonas aeruginosa Predicted via Genome-Scale Kinetic Models of Bacterial Metabolism. PLOS Comput. Biol. 2015, 11, e1004452. [Google Scholar] [CrossRef]
- Thi, M.T.T.; Wibowo, D.; Rehm, B.H.A. Pseudomonas aeruginosa Biofilms. Int. J. Mol. Sci. 2020, 21, 8671. [Google Scholar] [CrossRef]
- Kamali, E.; Jamali, A.; Ardebili, A.; Ezadi, F.; Mohebbi, A. Evaluation of antimicrobial resistance, biofilm forming potential, and the presence of biofilm-related genes among clinical isolates of Pseudomonas aeruginosa. BMC Res. Notes 2020, 13, 27. [Google Scholar] [CrossRef]
- Ciofu, O.; Moser, C.; Jensen, P.Ø.; Høiby, N. Tolerance and resistance of microbial biofilms. Nat. Rev. Microbiol. 2022, 20, 621–635. [Google Scholar] [CrossRef]
- Rather, M.A.; Gupta, K.; Mandal, M. Microbial biofilm: Formation, architecture, antibiotic resistance, and control strategies. Brazilian J. Microbiol. 2021, 52, 1701–1718. [Google Scholar] [CrossRef] [PubMed]
- Barakat, K.M.; Ismail, M.M.; Abou El Hassayeb, H.E.; El Sersy, N.A.; Elshobary, M.E. Chemical characterization and biological activities of ulvan extracted from Ulva fasciata (Chlorophyta). Rend. Lincei. Sci. Fis. Nat. 2022, 33, 829–841. [Google Scholar] [CrossRef]
- Afzal, S.; Yadav, A.K.; Poonia, A.K.; Choure, K.; Yadav, A.N.; Pandey, A. Antimicrobial therapeutics isolated from algal source: Retrospect and prospect. Biologia 2023, 78, 291–305. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, H.S.; Elshobary, M.E.; Barakat, K.M.; Khairy, H.M.; El-Sheikh, M.A.; Czaja, R.; Allam, B.; Senousy, H.H. Ocean acidification induced changes in Ulva fasciata biochemistry may improve Dicentrarchus labrax aquaculture via enhanced antimicrobial activity. Aquaculture 2022, 560, 738474. [Google Scholar] [CrossRef]
- Osman, M.E.H.; Abo-Shady, A.M.; Elshobary, M.E.; Abd El-Ghafar, M.O.; Abomohra, A.E.-F.F. Screening of seaweeds for sustainable biofuel recovery through sequential biodiesel and bioethanol production. Environ. Sci. Pollut. Res. 2020, 27, 32481–32493. [Google Scholar] [CrossRef] [PubMed]
- Ismail, M.M.; Ismail, G.A.; Elshobary, M.E. Morpho-anatomical, and chemical characterization of some calcareous Mediterranean red algae species. Bot. Stud. 2023, 64, 10. [Google Scholar] [CrossRef] [PubMed]
- Ponnanikajamideen, M.; Malini, M.; Malarkodi, C.; Rajeshkumar, S. Bioactivity and phytochemical constituents of marine brown seaweed (Padina tetrastromatica) extract from various organic solvents. Int. J. Pharm. Ther. 2014, 5, 108–112. [Google Scholar]
- Zammuto, V.; Rizzo, M.G.; Spanò, A.; Spagnuolo, D.; Di Martino, A.; Morabito, M.; Manghisi, A.; Genovese, G.; Guglielmino, S.; Calabrese, G.; et al. Effects of crude polysaccharides from marine macroalgae on the adhesion and biofilm formation of Pseudomonas aeruginosa and Staphylococcus aureus. Algal Res. 2022, 63, 102646. [Google Scholar] [CrossRef]
- Ghafoor, A.; Hay, I.D.; Rehm, B.H.A. Role of Exopolysaccharides in Pseudomonas aeruginosa Biofilm Formation and Architecture. Appl. Environ. Microbiol. 2011, 77, 5238–5246. [Google Scholar] [CrossRef]
- El Zawawy, N.A.; El-Shenody, R.A.; Ali, S.S.; El-Shetehy, M. A novel study on the inhibitory effect of marine macroalgal extracts on hyphal growth and biofilm formation of candidemia isolates. Sci. Rep. 2020, 10, 9339. [Google Scholar] [CrossRef] [PubMed]
- Sonbol, H.; Ameen, F.; AlYahya, S.; Almansob, A.; Alwakeel, S. Padina boryana mediated green synthesis of crystalline palladium nanoparticles as potential nanodrug against multidrug resistant bacteria and cancer cells. Sci. Rep. 2021, 11, 5444. [Google Scholar] [CrossRef] [PubMed]
- Elshobary, M.E.; El-Shenody, R.A.; Abomohra, A.E.; El-Shenody, R.; Abomohra, A.E. Sequential biofuel production from seaweeds enhances the energy recovery: A case study for biodiesel and bioethanol production. Int. J. Energy Res. 2021, 45, 6457–6467. [Google Scholar] [CrossRef]
- El-Khodary, G.M.; El-Sayed, H.S.; Khairy, H.M.; El-Sheikh, M.A.; Qi, X.; Elshobary, M.E. Comparative study on growth, survival and pigmentation of Solea aegyptiaca larvae by using four different microalgal species with emphasize on water quality and nutritional value. Aquac. Nutr. 2021, 27, 615–629. [Google Scholar] [CrossRef]
- Elshobary, M.E.; Zabed, H.M.; Qi, X.; El-Shenody, R.A. Enhancing biomass and lipid productivity of a green microalga Parachlorella kessleri for biodiesel production using rapid mutation of atmospheric and room temperature plasma. Biotechnol. Biofuels Bioprod. 2022, 15, 122. [Google Scholar] [CrossRef] [PubMed]
- Zaki, M.A.; Ashour, M.; Heneash, A.M.M.; Mabrouk, M.M.; Alprol, A.E.; Khairy, H.M.; Nour, A.M.; Mansour, A.T.; Hassanien, H.A.; Gaber, A.; et al. Potential applications of native cyanobacterium isolate (Arthrospira platensis NIOF17/003) for biodiesel production and utilization of its byproduct in marine rotifer (Brachionus plicatilis) production. Sustainability 2021, 13, 1769. [Google Scholar] [CrossRef]
- Casillas-Vargas, G.; Ocasio-Malavé, C.; Medina, S.; Morales-Guzmán, C.; Del Valle, R.G.; Carballeira, N.M.; Sanabria-Ríos, D.J. Antibacterial fatty acids: An update of possible mechanisms of action and implications in the development of the next-generation of antibacterial agents. Prog. Lipid Res. 2021, 82, 101093. [Google Scholar] [CrossRef]
- Faris, N.S.; Al-Zeidaneen, S.; Abu-Huriarh, H.A.; Hadieh, M.M.; Hadieh, O.M. The sensitivity and specificity of available Helicobacter pylori diagnosis techniques considering the emerging multidrug resistance. Drug Invent. Today 2019, 12, 2736–2740. [Google Scholar]
- Kim, H.S.; Cha, E.; Ham, S.Y.; Park, J.H.; Nam, S.J.; Kwon, H.; Byun, Y.; Park, H.D. Linoleic acid inhibits Pseudomonas aeruginosa biofilm formation by activating diffusible signal factor-mediated quorum sensing. Biotechnol. Bioeng. 2021, 118, 82–93. [Google Scholar] [CrossRef]
- Kumar, P.; Lee, J.J.-H.; Beyenal, H.; Lee, J.J.-H. Fatty Acids as Antibiofilm and Antivirulence Agents. Trends Microbiol. 2020, 28, 753–768. [Google Scholar] [CrossRef]
- Rehman Nengroo, Z.; Rauf, A.; Danish, M.; Sharief Dar, M. Evaluation of Fatty Acid Composition and Antimicrobial Activity of Eight Medicinal Plants from Kashmir. Orient. J. Chem. 2020, 36, 44–53. [Google Scholar] [CrossRef]
- Al-Juhaimi, F.Y.; Özcan, M.M.; Mohamed Ahmed, I.A.; Alsawmahia, O.N.; Özcan, M.M.; Ghafoor, K.; Babiker, E.E. Bioactive compounds, antioxidant activity, fatty acid composition, and antimicrobial activity of propolis from different locations in Turkey. J. Apic. Res. 2022, 61, 246–254. [Google Scholar] [CrossRef]
- Chanda, W.; Joseph, T.; Padhiar, A.; Guo, X.; Min, L.; Wang, W.; Lolokote, S.; Ning, A.; Cao, J.; Huang, M.; et al. Combined effect of linolenic acid and tobramycin on Pseudomonas aeruginosa biofilm formation and quorum sensing. Exp. Ther. Med. 2017, 14, 4328–4338. [Google Scholar] [CrossRef] [PubMed]
- Pompilio, A.; Scribano, D.; Sarshar, M.; Di Bonaventura, G.; Palamara, A.T.; Ambrosi, C. Gram-Negative Bacteria Holding Together in a Biofilm: The Acinetobacter baumannii Way. Microorganisms 2021, 9, 1353. [Google Scholar] [CrossRef] [PubMed]
- Cheung Lam, A.H.; Sandoval, N.; Wadhwa, R.; Gilkes, J.; Do, T.Q.; Ernst, W.; Chiang, S.-M.; Kosina, S.; Howard Xu, H.; Fujii, G. Assessment of free fatty acids and cholesteryl esters delivered in liposomes as novel class of antibiotic. BMC Res. Notes 2016, 9, 337. [Google Scholar] [CrossRef]
- Huang, C.B.; Alimova, Y.; Myers, T.M.; Ebersole, J.L. Short-and medium-chain fatty acids exhibit antimicrobial activity for oral microorganisms. Arch. Oral Biol. 2011, 56, 650–654. [Google Scholar] [CrossRef] [PubMed]
- Soliman, H.M.; Abdel-Wahhab, M.A. Synthesis of Antibacterial Bioactive Compounds Using Linoleic Acid Extracted from Melon Seeds Oil and Evaluation of Its Waste Meal Ash for Fried Oil Regeneration. Waste Biomass Valorization 2023. [Google Scholar] [CrossRef]
- Hemmati, F.; Rezaee, M.A.; Ebrahimzadeh, S.; Yousefi, L.; Nouri, R.; Kafil, H.S.; Gholizadeh, P. Novel Strategies to Combat Bacterial Biofilms. Mol. Biotechnol. 2021, 63, 569–586. [Google Scholar] [CrossRef]
- Adeosun, I.J.; Baloyi, I.T.; Cosa, S. Anti-Biofilm and Associated Anti-Virulence Activities of Selected Phytochemical Compounds against Klebsiella pneumoniae. Plants 2022, 11, 1429. [Google Scholar] [CrossRef]
- Nasab, S.B.; Homaei, A.; Pletschke, B.I.; Salinas-Salazar, C.; Castillo-Zacarias, C.; Parra-Saldívar, R. Marine resources effective in controlling and treating diabetes and its associated complications. Process Biochem. 2020, 92, 313–342. [Google Scholar] [CrossRef]
- Qin, S.-G.; Tian, H.-Y.; Wei, J.; Han, Z.-H.; Zhang, M.-J.; Hao, G.-H.; Liu, X.; Pan, L.-F. 3-Bromo-4,5-Dihydroxybenzaldehyde Protects Against Myocardial Ischemia and Reperfusion Injury Through the Akt-PGC1α-Sirt3 Pathway. Front. Pharmacol. 2018, 9, 722. [Google Scholar] [CrossRef]
- Choi, Y.K.; Ye, B.-R.; Kim, E.-A.; Kim, J.; Kim, M.; Lee, W.W.; Ahn, G.-N.; Kang, N.; Jung, W.-K.; Heo, S.-J. Bis (3-bromo-4,5-dihydroxybenzyl) ether, a novel bromophenol from the marine red alga Polysiphonia morrowii that suppresses LPS-induced inflammatory response by inhibiting ROS-mediated ERK signaling pathway in RAW 264.7 macrophages. Biomed. Pharmacother. 2018, 103, 1170–1177. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-Y.; Kim, S.R.; Oh, M.-J.; Jung, S.-J.; Kang, S.Y. In vitro antiviral activity of red alga, Polysiphonia morrowii extract and its bromophenols against fish pathogenic infectious hematopoietic necrosis virus and infectious pancreatic necrosis virus. J. Microbiol. 2011, 49, 102–106. [Google Scholar] [CrossRef]
- Zheng, Y.; Zhou, J.; Sayre, D.A.; Sintim, H.O. Identification of bromophenol thiohydantoin as an inhibitor of DisA, a c-di-AMP synthase, from a 1000 compound library, using the coralyne assay. Chem. Commun. 2014, 50, 11234–11237. [Google Scholar] [CrossRef]
- Gándara, C.; de Lucena, D.K.C.; Torres, R.; Serrano, E.; Altenburger, S.; Graumann, P.L.; Alonso, J.C. Activity and in vivo dynamics of Bacillus subtilis DisA are affected by RadA/Sms and by Holliday junction-processing proteins. DNA Repair 2017, 55, 17–30. [Google Scholar] [CrossRef] [PubMed]
- Prasath, K.G.; Tharani, H.; Kumar, M.S.; Pandian, S.K. Palmitic Acid Inhibits the Virulence Factors of Candida tropicalis: Biofilms, Cell Surface Hydrophobicity, Ergosterol Biosynthesis, and Enzymatic Activity. Front. Microbiol. 2020, 11, 864. [Google Scholar] [CrossRef]
- Alreshidi, M.; Badraoui, R.; Adnan, M.; Patel, M.; Alotaibi, A.; Saeed, M.; Ghandourah, M.; Al-Motair, K.A.; Arif, I.A.; Albulaihed, Y.; et al. Phytochemical profiling, antibacterial, and antibiofilm activities of Sargassum sp. (brown algae) from the Red Sea: ADMET prediction and molecular docking analysis. Algal Res. 2023, 69, 102912. [Google Scholar] [CrossRef]
- Geethalakshmi, R.; Sundaramurthi, J.C.; Sarada, D.V.L. Antibacterial activity of flavonoid isolated from Trianthema decandra against Pseudomonas aeruginosa and molecular docking study of FabZ. Microb. Pathog. 2018, 121, 87–92. [Google Scholar] [CrossRef]
- Leesong, M.; Henderson, B.S.; Gillig, J.R.; Schwab, J.M.; Smith, J.L. Structure of a dehydratase–isomerase from the bacterial pathway for biosynthesis of unsaturated fatty acids: Two catalytic activities in one active site. Structure 1996, 4, 253–264. [Google Scholar] [CrossRef]
- Heath, R.J.; Rock, C.O. Roles of the FabA and FabZ β-Hydroxyacyl-Acyl Carrier Protein Dehydratases in Escherichia coli Fatty Acid Biosynthesis*. J. Biol. Chem. 1996, 271, 27795–27801. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, W.; Hu, T.; Du, L.; Luo, C.; Chen, K.; Shen, X.; Jiang, H. Structural Basis for Catalytic and Inhibitory Mechanisms of β-Hydroxyacyl-acyl Carrier Protein Dehydratase (FabZ). J. Biol. Chem. 2008, 283, 5370–5379. [Google Scholar] [CrossRef] [PubMed]
- Aiba, S.; Ogawa, T. Assessment of Growth Yield of a Blue--green Alga, Spirulina platensis, in Axenic and Continuous Culture. J. Gen. Microbiol. 1977, 102, 179–182. [Google Scholar] [CrossRef]
- Holt, J.G.; Krieg, N.R.; Sneath, P.H.A.; Staley, J.T.; Williams, S.T. Bergey’s Manual of Determinate Bacteriology; 1994; Lippincott Williams and Wilkins: Baltimore, MD, USA. [Google Scholar]
- Karatuna, O.; Yagci, A. Analysis of quorum sensing-dependent virulence factor production and its relationship with antimicrobial susceptibility in Pseudomonas aeruginosa respiratory isolates. Clin. Microbiol. Infect. 2010, 16, 1770–1775. [Google Scholar] [CrossRef] [PubMed]
- EUCAST European Committee on antimicrobial susceptibility testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 2.0. Växjö: EUCAST 2012. Available online: http://www.congrex-switzerland.com/uploads/tx_mnmcxpwprotecteddl/EW03_Handout_EUCAST_01.pdf (accessed on 15 July 2023).
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Ali, S.S.; Moawad, M.S.; Hussein, M.A.; Azab, M.; Abdelkarim, E.A.; Badr, A.; Sun, J.; Khalil, M. Efficacy of metal oxide nanoparticles as novel antimicrobial agents against multi-drug and multi-virulent Staphylococcus aureus isolates from retail raw chicken meat and giblets. Int. J. Food Microbiol. 2021, 344, 109116. [Google Scholar] [CrossRef] [PubMed]
- Stepanović, S.; Vuković, D.; Hola, V.; Bonaventura, G.D.; Djukić, S.; Ćirković, I.; Ruzicka, F. Quantification of biofilm in microtiter plates: Overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. Apmis 2007, 115, 891–899. [Google Scholar] [CrossRef]
- Elshikh, M.; Ahmed, S.; Funston, S.; Dunlop, P.; McGaw, M.; Marchant, R.; Banat, I.M. Resazurin-based 96-well plate microdilution method for the determination of minimum inhibitory concentration of biosurfactants. Biotechnol. Lett. 2016, 38, 1015–1019. [Google Scholar] [CrossRef]
- Gurunathan, S.; Han, J.W.; Kwon, D.-N.; Kim, J.-H. Enhanced antibacterial and anti-biofilm activities of silver nanoparticles against Gram-negative and Gram-positive bacteria. Nanoscale Res. Lett. 2014, 9, 373. [Google Scholar] [CrossRef] [PubMed]
- Barapatre, A.; Aadil, K.R.; Jha, H. Synergistic antibacterial and antibiofilm activity of silver nanoparticles biosynthesized by lignin-degrading fungus. Bioresour. Bioprocess. 2016, 3, 8. [Google Scholar] [CrossRef]
- Barakat, K.M.; El-Sayed, H.S.; Khairy, H.M.; El-Sheikh, M.A.; Al-Rashed, S.A.; Arif, I.A.; Elshobary, M.E. Effects of ocean acidification on the growth and biochemical composition of a green alga (Ulva fasciata) and its associated microbiota. Saudi. J. Biol. Sci. 2021, 28, 5106–5114. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Elshobary, M.E.; Becker, M.G.; Kalichuk, J.L.; Chan, A.C.; Belmonte, M.F.; Piercey-normore, M.D. Tissue-specific localization of polyketide synthase and other associated genes in the lichen, Cladonia rangiferina, using laser microdissection. Phytochemistry 2018, 156, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2023 update. Nucleic Acids Res. 2023, 51, D1373–D1380. [Google Scholar] [CrossRef] [PubMed]
- Saeed, M.; Saeed, A.; Alam, M.J.; Alreshidi, M. Receptor-Based Pharmacophore Modeling in the Search for Natural Products for COVID-19 Mpro. Molecules 2021, 26, 1549. [Google Scholar] [CrossRef]
- Hchicha, K.; Korb, M.; Badraoui, R.; Naïli, H. A novel sulfate-bridged binuclear copper (II) complex: Structure, optical, ADMET and in vivo approach in a murine model of bone metastasis. New J. Chem. 2021, 45, 13775–13784. [Google Scholar] [CrossRef]
- Snoussi, M.; Noumi, E.; Hajlaoui, H.; Bouslama, L.; Hamdi, A.; Saeed, M.; Alreshidi, M.; Adnan, M.; Al-Rashidi, A.; Aouadi, K.; et al. Phytochemical Profiling of Allium subhirsutum L. Aqueous Extract with Antioxidant, Antimicrobial, Antibiofilm, and Anti-Quorum Sensing Properties: In Vitro and In Silico Studies. Plants 2022, 11, 495. [Google Scholar] [CrossRef]
- Ghannay, S.; Kadri, A.; Aouadi, K. Synthesis, in vitro antimicrobial assessment, and computational investigation of pharmacokinetic and bioactivity properties of novel trifluoromethylated compounds using in silico ADME and toxicity prediction tools. Monatshefte Für Chem.-Chem. Mon. 2020, 151, 267–280. [Google Scholar] [CrossRef]









| Antibiotic Class | Antimicrobial Agent | Disc Potency (µg/Disc) | Resistant Strains |
Susceptible Strains | ||
|---|---|---|---|---|---|---|
| No. | % | No. | % | |||
| Cephalosporin | Cephalexin (CN) | 3 | 20 | 86.9% | 3 | 13.1% |
| Cefoxitin (Fox) | 30 | 20 | 86.9% | 3 | 13.1% | |
| Cefsulodin (CFS) | 30 | 20 | 86.9% | 3 | 13.1% | |
| Aminoglycosides | Tobramycin (TOB) | 10 | 7 | 30.4% | 16 | 69.5% |
| Amikacin (AK) | 30 | 3 | 13.1% | 20 | 86.9% | |
| Fluoroquinolones | Ciprofloxacin (CIP) | 5 | 1 | 4.34% | 22 | 95.6% |
| Norfloxacine (NOR) | 10 | 10 | 43.4% | 13 | 56.5% | |
| Levofloxacin (LE) | 5 | 11 | 47.8% | 12 | 52.1% | |
| Carbapenem | Meropenem (MEM) | 10 | 4 | 17.3% | 19 | 82.6% |
| Imipenem (IPM) | 10 | 1 | 4.34% | 22 | 95.6% | |
| Macrolide | Azithromycin (AZM) | 30 | 15 | 65.2% | 8 | 34.7% |
| Penicillin | Piperacillin–tazobactam (TPZ) | 75/10 | 20 | 86.9% | 3 | 13.1% |
| Sulfonamides–trimethoprim | Sulfonamides–trimethoprim (STX) | 25 | 12 | 52.1% | 11 | 47.8% |
| Quinolones | Pipemidic acid (PI) | 20 | 15 | 65.2% | 11 | 34.7% |
| Pattern Code | Resistance Pattern Profile | No. of MDR Isolates (23) | Isolate Code Exhibited by Pattern | |
|---|---|---|---|---|
| I | a | TPZ, CN, FOX, CFS, and AZM | 3 | 3, 11, 16 |
| b | TPZ, CN, FOX, CFS, and PI | 3 | 10, 21, 22 | |
| c | TPZ, CN, FOX, CFS, and LE | 2 | 4, 5 | |
| II | a | TPZ, CN, FOX, CFS, LE, AZM, STX, and PI | 2 | 6, 9 |
| b | TPZ, CN, FOX, CFS, NOR, AZM, STX, and PI | 3 | 15, 20, 23 | |
| III | TPZ, CN, FOX, CFS, LE, NOR, AZM, STX, PI, and TOB | 4 | 12,14, 18, 19 | |
| IV | TPZ, CN, FOX, CFS, LE, NOR, AZM, STX, PI, TOB, MEM, and AK | 2 | 1, 13 | |
| V | TPZ, CN, FOX, CFS LE, NOR, AZM, STX, PI, TOB, MEM, AK, IPM, and CIP | 1 | 17 | |
| Bacterial Strain Code | Biofilm Producing Ability (BPA) | Classification according to Antibiotic Resistance | |
|---|---|---|---|
| OD ± SD | Strength | ||
| P1 | 0.58 ± 0.02 a | Strong | MDR |
| P2 | 0.22 ± 0.04 h | Weak | Sensitive |
| P3 | 0.14 ± 0.06 k | Weak | MDR |
| P4 | 0.22 ± 0.07 h | Weak | MDR |
| P5 | 0.15 ± 0.04 k | Weak | MDR |
| P6 | 0.32 ± 0.02 f | Moderate | MDR |
| P7 | 0.22 ± 0.03 h | Weak | Sensitive |
| P8 | 0.22 ± 0.05 h | Weak | Sensitive |
| P9 | 0.26 ± 0.04 g | Moderate | MDR |
| P10 | 0.19 ± 0.05 i | Weak | MDR |
| P11 | 0.21 ± 0.05 h | Weak | MDR |
| P12 | 0.37 ± 0.05 d | Moderate | MDR |
| P13 | 0.54 ± 0.02 b | Strong | MDR |
| P14 | 0.32 ± 0.05 f | Moderate | MDR |
| P15 | 0.32 ± 0.01 f | Moderate | MDR |
| P16 | 0.15 ± 0.06 k | Weak | MDR |
| P17 | 0.60 ± 0.04 a | Strong | PDR |
| P18 | 0.37 ± 0.04 d | Moderate | MDR |
| P19 | 0.25 ± 0.04 g | Moderate | MDR |
| P20 | 0.34 ± 0.05 e | Moderate | MDR |
| P21 | 0.18± 0.05 j | Weak | MDR |
| P22 | 0.21 ± 0.05 h | Weak | MDR |
| P23 | 0.43 ± 0.05 c | Moderate | MDR |
| F-value | 3114.33 | p-value | <0.001 |
| Microbial Strain | Mean of Diameter of Inhibition Zones (mm) | ||||||
|---|---|---|---|---|---|---|---|
| A. platensis | P. scopulorum | AAE + PME (COE) | TPZ | TOB | |||
| Methanol | Acetone | Methanol | Acetone | ||||
| P1 | 6.1± 0.02 a | 11.66 ± 0.94 a | 16 ± 0.81 a | 6.8 ± 0.21 c | 27.66 ± 0.94 b | 0.0 ± 0.0 | 0.0 ± 0.0 |
| P13 | 6.8 ± 0.13 b | 10.66 ± 0.47 c | 25.66 ± 0.94 b | 7.3 ± 0.11 b | 36 ± 0.81 a | 0.0 ± 0.0 | 0.0 ± 0.0 |
| P17 | 6.5 ± 0.05 c | 11 ± 0.47 b | 15.33 ± 0.47 a | 7.5±0.23 a | 18.33 ± 0.47 c | 0.0 ± 0.0 | 0.0 ± 0.0 |
| F-value | 9676.42 | 811.71 | 55.84 | 5207.11 | 245,428.04 | ||
| P-value | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | ||
| No. | Compound Name | Peak Area % | Molecular Formula |
|---|---|---|---|
| 1 | 3,7,11,15-Tetramethyl-2-hexadecen-1-ol (phytol) | 36.6 | C20H40O |
| 2 | n-Hexadecanoic acid (palmitic acid) | 21.01 | C16H32O2 |
| 3 | 11,14-Eicosadienoic acid, methyl ester | 12.68 | C21H38O2 |
| 4 | Trans-13-octadecenoic acid (elaidic acid) | 7.67 | C18H34O2 |
| 5 | Linoleic acid ethyl ester | 7.42 | C18H32O2 |
| 6 | 9,12,15-Octadecatrienoic acid | 3.44 | C18H30O2 |
| 7 | 8,11,14-Eicosatrienoic acid, (Z,Z,Z)- | 3.11 | C20H34O2 |
| 8 | Cholesta-8,24-dien-3-ol, 4-methyl-, (3á,4à)- | 2.89 | C28H46O |
| 9 | Hexadecenoic acid, Z-11- | 1.72 | C16H30O2 |
| 10 | Octadecanoic acid | 1.08 | C18H36O2 |
| 11 | Decane, 2,9-dimethyl | 1.03 | C12H26 |
| 12 | Tridecane | 0.88 | C13H28 |
| 13 | Hexadecane | 0.47 | C16H34 |
| No. | Compound Name | Peak Area % | Molecular Formula |
|---|---|---|---|
| 1 | n-Hexadecanoic acid methyl ester | 22.75 | C16H32O2 |
| 2 | Bis (3-bromo-4,5-dihydroxybenzyl) ether | 13.25 | C14H12Br2O5 |
| 3 | 3-Bromo-4,5-dihydroxybenzaldehyde | 10.76 | C7H5BrO3 |
| 4 | 3,4-Dihydroxybenzaldehyde | 9.71 | C7H6O3 |
| 5 | Linolenic acid methyl ester | 8.7 | C18H30O4 |
| 6 | Eicosane | 3.95 | C20H42 |
| 7 | Myristic acid methyl ester | 3.63 | C14H28O2 |
| 8 | Linoleic acid | 3.52 | C18H32O2 |
| 9 | Octadecanoic acid | 3.24 | C18H36O2 |
| 10 | Hexadecane | 3.11 | C16H34 |
| 11 | 24-Methyl-cholesta-5,22Z-dien-3ß-ol | 3.08 | C31H52O |
| 12 | Octadecane | 1.89 | C18H38 |
| 13 | Nonadecane | 1.83 | C19H40 |
| 14 | Phytol | 1.77 | C20H40O |
| 15 | Dodecane, 2,6,10-trimethyl | 1.71 | C15H32 |
| 16 | Cis-13-octadecenoic acid | 1.19 | C18H34O2 |
| 17 | Phthalic acid, | 1.11 | C8H6O4 |
| 18 | Fucosterol | 1.09 | C29H48O |
| 19 | 24-Ethyl-cholest-5-en-3ß-ol | 0.71 | C29H50O |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El-Sapagh, S.; El-Shenody, R.; Pereira, L.; Elshobary, M. Unveiling the Potential of Algal Extracts as Promising Antibacterial and Antibiofilm Agents against Multidrug-Resistant Pseudomonas aeruginosa: In Vitro and In Silico Studies including Molecular Docking. Plants 2023, 12, 3324. https://doi.org/10.3390/plants12183324
El-Sapagh S, El-Shenody R, Pereira L, Elshobary M. Unveiling the Potential of Algal Extracts as Promising Antibacterial and Antibiofilm Agents against Multidrug-Resistant Pseudomonas aeruginosa: In Vitro and In Silico Studies including Molecular Docking. Plants. 2023; 12(18):3324. https://doi.org/10.3390/plants12183324
Chicago/Turabian StyleEl-Sapagh, Shimaa, Rania El-Shenody, Leonel Pereira, and Mostafa Elshobary. 2023. "Unveiling the Potential of Algal Extracts as Promising Antibacterial and Antibiofilm Agents against Multidrug-Resistant Pseudomonas aeruginosa: In Vitro and In Silico Studies including Molecular Docking" Plants 12, no. 18: 3324. https://doi.org/10.3390/plants12183324