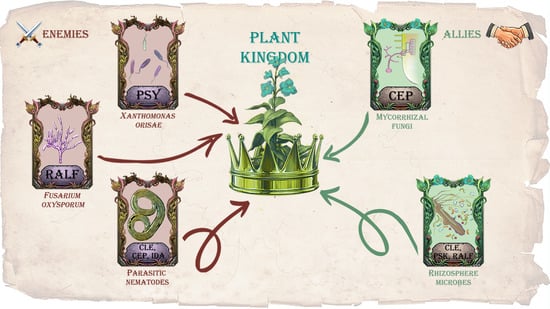
Dialog between Kingdoms: Enemies, Allies and Peptide Phytohormones
Abstract
:1. Introduction
2. Peptide Phytohormones from Plant Pathogens: Divide et impera
2.1. Peptide Phytohormones of Plant Parasitic Nematodes
2.1.1. CLE Peptides of Plant Parasitic Nematodes
2.1.2. CEP Peptides of Plant Parasitic Nematodes
2.1.3. IDA Peptides of Plant Parasitic Nematodes
2.2. Peptide Phytohormones of Plant Pathogenic Fungi
2.2.1. RALF Peptides of Plant Pathogenic Fungi
2.2.2. PSK, IDA and PEP Peptides of Plant Pathogenic Fungi
2.3. Peptide Phytohormones of Plant Pathogenic Bacteria
2.3.1. PSY Peptides of Plant Pathogenic Bacteria
2.3.2. RALF and CEP Peptides of Plant Pathogenic Bacteria
3. Peptide Phytohormones from Plant Symbiotic and Beneficial Microbes: Si vis pacem, para bellum
3.1. Possible Homologues of Peptide Phytohormones in Plant Beneficial and Plant-Associated Non-Pathogenic Bacteria
3.2. The Homologues of Plant Peptide Phytohormones in Arbuscular Mycorrhizal Fungi
4. Discussion: Strategy of Information War and “Spy Games” with Peptide Phytohormones
5. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nobori, T.; Mine, A.; Tsuda, K. Molecular networks in plant–pathogen holobiont. FEBS Lett. 2018, 592, 1937–1953. [Google Scholar] [CrossRef]
- Chialva, M.; Lanfranco, L.; Bonfante, P. The plant microbiota: Composition, functions, and engineering. Curr. Opin. Biotechnol. 2021, 73, 135–142. [Google Scholar] [CrossRef]
- Lo Presti, L.; Lanver, D.; Schweizer, G.; Tanaka, S.; Liang, L.; Tollot, M.; Zuccaro, A.; Reissmann, S.; Kahmann, R. Fungal Effectors and Plant Susceptibility. Annu. Rev. Plant Biol. 2015, 66, 513–545. [Google Scholar] [CrossRef]
- Langin, G.; Gouguet, P.; Üstün, S. Microbial Effector Proteins—A Journey through the Proteolytic Landscape. Trends Microbiol. 2020, 28, 523–535. [Google Scholar] [CrossRef]
- Plett, J.M.; Martin, F. Reconsidering mutualistic plant–fungal interactions through the lens of effector biology. Curr. Opin. Plant Biol. 2015, 26, 45–50. [Google Scholar] [CrossRef] [Green Version]
- Miwa, H.; Okazaki, S. How effectors promote beneficial interactions. Curr. Opin. Plant Biol. 2017, 38, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Pieterse, C.M.J.; Van der Does, D.; Zamioudis, C.; Leon-Reyes, A.; Van Wees, S.C.M. Hormonal Modulation of Plant Immunity. Annu. Rev. Cell Dev. Biol. 2012, 28, 489–521. [Google Scholar] [CrossRef] [Green Version]
- Edenancé, N.; Esánchez-Vallet, A.; Egoffner, D.; Emolina, A. Disease resistance or growth: The role of plant hormones in balancing immune responses and fitness costs. Front. Plant Sci. 2013, 4, 155. [Google Scholar] [CrossRef] [Green Version]
- Beckers, G.J.M.; Spoel, S.H. Fine-Tuning Plant Defence Signalling: Salicylate versus Jasmonate. Plant Biol. 2006, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Vlot, A.C.; Sales, J.H.; Lenk, M.; Bauer, K.; Brambilla, A.; Sommer, A.; Chen, Y.; Wenig, M.; Nayem, S. Systemic propagation of immunity in plants. New Phytol. 2021, 229, 1234–1250. [Google Scholar] [CrossRef]
- Gutjahr, C.; Paszkowski, U. Weights in the Balance: Jasmonic Acid and Salicylic Acid Signaling in Root-Biotroph Interactions. Mol. Plant-Microbe Interact. 2009, 22, 763–772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, S.; Yao, J.; Ma, K.-W.; Zhou, H.; Song, J.; He, S.Y.; Ma, W. Bacterial Effector Activates Jasmonate Signaling by Directly Targeting JAZ Transcriptional Repressors. PLoS Pathog. 2013, 9, e1003715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gimenez-Ibanez, S.; Boter, M.; Fernández-Barbero, G.; Chini, A.; Rathjen, J.P.; Solano, R. The Bacterial Effector HopX1 Targets JAZ Transcriptional Repressors to Activate Jasmonate Signaling and Promote Infection in Arabidopsis. PLoS Biol. 2014, 12, e1001792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geng, X.; Jin, L.; Shimada, M.; Kim, M.G.; Mackey, D. The phytotoxin coronatine is a multifunctional component of the virulence armament of Pseudomonas syringae. Planta 2014, 240, 1149–1165. [Google Scholar] [CrossRef] [Green Version]
- Kloppholz, S.; Kuhn, H.; Requena, N. A Secreted Fungal Effector of Glomus intraradices Promotes Symbiotic Biotrophy. Curr. Biol. 2011, 21, 1204–1209. [Google Scholar] [CrossRef] [Green Version]
- Shaharoona, B.; Arshad, M.; Zahir, Z.A. Effect of plant growth promoting rhizobacteria containing ACC-deaminase on maize (Zea mays L.) growth under axenic conditions and on nodulation in mung bean (Vigna radiata L.). Lett. Appl. Microbiol. 2006, 42, 155–159. [Google Scholar] [CrossRef]
- Barry, G.F.; Rogers, S.G.; Fraley, R.T.; Brand, L. Identification of a cloned cytokinin biosynthetic gene. Proc. Natl. Acad. Sci. USA 1984, 81, 4776–4780. [Google Scholar] [CrossRef] [Green Version]
- Kemper, E.; Wafenschmidt, S.; Weiler, E.W.; Rausch, T.; Schroder, J. T-DNA-encoded auxin formation in crown-gall cells. Planta 1985, 163, 257–262. [Google Scholar] [CrossRef]
- Ludwig-Müller, J. Bacteria and fungi controlling plant growth by manipulating auxin: Balance between development and defense. J. Plant Physiol. 2015, 172, 4–12. [Google Scholar] [CrossRef]
- Frébortová, J.; Frébort, I. Biochemical and Structural Aspects of Cytokinin Biosynthesis and Degradation in Bacteria. Microorganisms 2021, 9, 1314. [Google Scholar] [CrossRef]
- Lee, C.; Chronis, D.; Kenning, C.; Peret, B.; Hewezi, T.; Davis, E.L.; Baum, T.J.; Hussey, R.; Bennett, M.; Mitchum, M.G. The Novel Cyst Nematode Effector Protein 19C07 Interacts with the Arabidopsis Auxin Influx Transporter LAX3 to Control Feeding Site Development. Plant Physiol. 2011, 155, 866–880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oh, E.; Seo, P.J.; Kim, J. Signaling Peptides and Receptors Coordinating Plant Root Development. Trends Plant Sci. 2018, 23, 337–351. [Google Scholar] [CrossRef]
- Matsubayashi, Y. Posttranslationally Modified Small-Peptide Signals in Plants. Annu. Rev. Plant Biol. 2014, 65, 385–413. [Google Scholar] [CrossRef] [PubMed]
- Stührwohldt, N.; Schaller, A. Regulation of plant peptide hormones and growth factors by post-translational modification. Plant Biol. 2019, 21 (Suppl. 1), 49–63. [Google Scholar] [CrossRef]
- Bircheneder, S.; Dresselhaus, T. Why cellular communication during plant reproduction is particularly mediated by CRP signalling. J. Exp. Bot. 2016, 67, 4849–4861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gancheva, M.S.; Malovichko, Y.V.; Poliushkevich, L.O.; Dodueva, I.E.; Lutova, L.A. Plant Peptide Hormones. Russ. J. Plant Physiol. 2019, 66, 171–189. [Google Scholar] [CrossRef]
- Chakraborty, S.; Nguyen, B.; Wasti, S.D.; Xu, G. Plant Leucine-Rich Repeat Receptor Kinase (LRR-RK): Structure, Ligand Perception, and Activation Mechanism. Molecules 2019, 24, 3081. [Google Scholar] [CrossRef] [Green Version]
- Poliushkevich, L.O.; Gancheva, M.S.; Dodueva, I.E.; Lutova, L.A. Receptors of CLE Peptides in Plants. Russ. J. Plant Physiol. 2020, 67, 1–16. [Google Scholar] [CrossRef]
- Corrado, G.; Sasso, R.; Pasquariello, M.; Iodice, L.; Carretta, A.; Cascone, P.; Ariati, L.; Digilio, M.C.; Guerrieri, E.; Rao, R. Systemin Regulates Both Systemic and Volatile Signaling in Tomato Plants. J. Chem. Ecol. 2007, 33, 669–681. [Google Scholar] [CrossRef]
- Pearce, G.; Strydom, D.; Johnson, S.; Ryan, C.A. A Polypeptide from Tomato Leaves Induces Wound-Inducible Proteinase Inhibitor Proteins. Science 1991, 253, 895–897. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Huffaker, A.; Bryan, A.C.; Tax, F.E.; Ryan, C.A. PEPR2 Is a Second Receptor for the Pep1 and Pep2 Peptides and Contributes to Defense Responses in Arabidopsis. Plant Cell 2010, 22, 508–522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, S.; Wang, X.; Chen, D.; Yang, X.; Wang, M.; Turrà, D.; Di Pietro, A.; Zhang, W. The Secreted Peptide PIP1 Amplifies Immunity through Receptor-Like Kinase 7. PLoS Pathog. 2014, 10, e1004331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pearce, G. Systemin, hydroxyproline-rich systemin and the induction of protease inhibitors. Curr. Protein Pept. Sci. 2011, 12, 399–408. [Google Scholar] [CrossRef]
- Matsubayashi, Y.; Morita, A.; Matsunaga, E.; Furuya, A.; Hanai, N.; Sakagami, Y. Physiological relationships between auxin, cytokinin, and a peptide growth factor, phytosulfokine-α, in stimulation of asparagus cell proliferation. Planta 1999, 207, 559–565. [Google Scholar] [CrossRef]
- Amano, Y.; Tsubouchi, H.; Shinohara, H.; Ogawa, M.; Matsubayashi, Y. Tyrosine-sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 18333–18338. [Google Scholar] [CrossRef] [Green Version]
- Mosher, S.; Kemmerling, B. PSKR1 and PSY1R-mediated regulation of plant defense responses. Plant Signal. Behav. 2013, 8, e24119. [Google Scholar] [CrossRef] [Green Version]
- Aalen, R.B.; Wildhagen, M.; Stø, I.M.; Butenko, M.A. IDA: A peptide ligand regulating cell separation processes in Arabidopsis. J. Exp. Bot. 2013, 64, 5253–5261. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Hou, S.; Wu, Q.; Lin, M.; Acharya, B.R.; Wu, D.; Zhang, W. IDL6-HAE/HSL2 impacts pectin degradation and resistance to Pseudomonas syringae pv tomato DC3000 in Arabidopsis leaves. Plant J. 2017, 89, 250–263. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.; Nolan, T.M.; Song, G.; Liu, S.; Xie, Z.; Chen, J.; Schnable, P.S.; Walley, J.W.; Yin, Y. FERONIA Receptor Kinase Contributes to Plant Immunity by Suppressing Jasmonic Acid Signaling in Arabidopsis thaliana. Curr. Biol. 2018, 28, 3316–3324.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronald, P.; Joe, A. Molecular mimicry modulates plant host responses to pathogens. Ann. Bot. 2018, 121, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Yuan, N.; Furumizu, C.; Zhang, B.; Sawa, S. Database mining of plant peptide homologues. Plant Biotechnol. 2021, 38, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Le Marquer, M.; Bécard, G.; Frey, N.F.D. Arbuscular mycorrhizal fungi possess a CLAVATA3/embryo surrounding region-related gene that positively regulates symbiosis. New Phytol. 2019, 222, 1030–1042. [Google Scholar] [CrossRef] [PubMed]
- Gheysen, G.; Mitchum, M.G. Phytoparasitic Nematode Control of Plant Hormone Pathways. Plant Physiol. 2019, 179, 1212–1226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutiérrez-Alanís, D.; Yong-Villalobos, L.; Jimenez-Sandoval, P.; Alatorre-Cobos, F.; Oropeza-Aburto, A.; Mora-Macías, J.; Sánchez-Rodríguez, F.; Cruz-Ramírez, L.A.; Herrera-Estrella, L. Phosphate Starvation-Dependent Iron Mobilization Induces CLE14 Expression to Trigger Root Meristem Differentiation through CLV2/PEPR2 Signaling. Dev. Cell 2017, 41, 555–570.e3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, N.; Mohd-Radzman, N.A.; Corcilius, L.; Crossett, B.; Connolly, A.; Cordwell, S.J.; Ivanovici, A.; Taylor, K.; Wlilliams, J.; Binos, S.; et al. Diverse Peptide Hormones Affecting Root Growth Identified in the Medicago truncatula Secreted Peptidome. Mol. Cell. Proteom. 2018, 17, 160–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Allen, R.; Ding, X.; Goellner, M.; Maier, T.; de Boer, J.M.; Baum, T.J.; Hussey, R.S.; Davis, E.L. Signal Peptide-Selection of cDNA Cloned Directly from the Esophageal Gland Cells of the Soybean Cyst Nematode Heterodera glycines. Mol. Plant-Microbe Interact. 2001, 14, 536–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, B.; Allen, R.; Maier, T.; Davis, E.L.; Baum, T.J.; Hussey, R.S. The Parasitome of the PhytonematodeHeterodera glycines. Mol. Plant-Microbe Interact. 2003, 16, 720–726. [Google Scholar] [CrossRef] [Green Version]
- Guo, X.; Wang, J.; Gardner, M.; Fukuda, H.; Kondo, Y.; Etchells, J.P.; Wang, X.; Mitchum, M.G. Identification of cyst nematode B-type CLE peptides and modulation of the vascular stem cell pathway for feeding cell formation. PLoS Pathog. 2017, 13, e1006142. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Lee, C.; Replogle, A.; Joshi, S.; Korkin, D.; Hussey, R.; Baum, T.J.; Davis, E.L.; Wang, X.; Mitchum, M.G. Dual roles for the variable domain in protein trafficking and host-specific recognition of Heterodera glycines CLE effector proteins. New Phytol. 2010, 187, 1003–1017. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Joshi, S.; Korkin, D.; Mitchum, M.G. Variable domain I of nematode CLEs directs post-translational targeting of CLE peptides to the extracellular space. Plant Signal. Behav. 2010, 5, 1633–1635. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Replogle, A.; Hussey, R.; Baum, T.; Wang, X.; Davis, E.L.; Mitchum, M.G. Identification of potential host plant mimics of CLAVATA3/ESR (CLE)-like peptides from the plant-parasitic nematode Heterodera schachtii. Mol. Plant Pathol. 2011, 12, 177–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Replogle, A.; Wang, J.; Bleckmann, A.; Hussey, R.S.; Baum, T.J.; Sawa, S.; Davis, E.L.; Wang, X.; Simon, R.; Mitchum, M.G. Nematode CLE signaling in Arabidopsis requires CLAVATA2 and CORYNE. Plant J. 2011, 65, 430–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Replogle, A.; Wang, J.; Paolillo, V.; Smeda, J.; Kinoshita, A.; Durbak, A.; Tax, F.E.; Wang, X.; Sawa, S.; Mitchum, M.G. Synergistic Interaction of CLAVATA1, CLAVATA2, and RECEPTOR-LIKE PROTEIN KINASE 2 in Cyst Nematode Parasitism of Arabidopsis. Mol. Plant-Microbe Interact. 2013, 26, 87–96. [Google Scholar] [CrossRef] [Green Version]
- Guo, X.; Chronis, D.; De La Torre, C.M.; Smeda, J.; Wang, X.; Mitchum, M.G. Enhanced resistance to soybean cyst nematodeHeterodera glycinesin transgenic soybean by silencing putative CLE receptors. Plant Biotechnol. J. 2015, 13, 801–810. [Google Scholar] [CrossRef]
- Lu, S.-W.; Chen, S.; Wang, J.; Yu, H.; Chronis, D.; Mitchum, M.G.; Wang, X. Structural and Functional Diversity of CLAVATA3/ESR (CLE)-Like Genes from the Potato Cyst Nematode Globodera rostochiensis. Mol. Plant-Microbe Interact. 2009, 22, 1128–1142. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Lang, P.; Chronis, D.; Zhang, S.; De Jong, W.S.; Mitchum, M.G.; Wang, X. In Planta Processing and Glycosylation of a Nematode CLAVATA3/ENDOSPERM SURROUNDING REGION-Like Effector and Its Interaction with a Host CLAVATA2-Like Receptor to Promote Parasitism. Plant Physiol. 2015, 167, 262–272. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Ni, J.; Denver, R.; Wang, X.; Clark, S.E. Mechanisms of Molecular Mimicry of Plant CLE Peptide Ligands by the Parasitic Nematode Globodera rostochiensis. Plant Physiol. 2011, 157, 476–484. [Google Scholar] [CrossRef] [Green Version]
- Wubben, M.J.; Gavilano, L.; Baum, T.J.; Davis, E.L. Sequence and Spatiotemporal Expression Analysis of CLE-Motif Containing Genes from the Reniform Nematode (Rotylenchulus reniformis Linford & Oliveira). J. Nematol. 2015, 47, 159–165. [Google Scholar]
- Dinh, P.T.Y.; Zhang, L.; Mojtahedi, H.; Brown, C.R.; Elling, A.A. Broad Meloidogyne Resistance in Potato Based on RNA Interference of Effector Gene 16D10. J. Nematol. 2015, 47, 71–78. [Google Scholar]
- Huang, G.; Dong, R.; Allen, R.; Davis, E.L.; Baum, T.J.; Hussey, R.S. A Root-Knot Nematode Secretory Peptide Functions as a Ligand for a Plant Transcription Factor. Mol. Plant-Microbe Interact. 2006, 19, 463–470. [Google Scholar] [CrossRef] [Green Version]
- Petitot, A.-S.; Dereeper, A.; Da Silva, C.; Guy, J.; Fernandez, D. Analyses of the Root-Knot Nematode (Meloidogyne graminicola) Transcriptome during Host Infection Highlight Specific Gene Expression Profiling in Resistant Rice Plants. Pathogens 2020, 9, 644. [Google Scholar] [CrossRef] [PubMed]
- Rutter, W.B.; Hewezi, T.; Maier, T.R.; Mitchum, M.G.; Davis, E.L.; Hussey, R.S.; Baum, T.J. Members of the Meloidogyne Avirulence Protein Family Contain Multiple Plant Ligand-Like Motifs. Phytopathology 2014, 104, 879–885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Semblat, J.-P.; Rosso, M.-N.; Hussey, R.S.; Abad, P.; Castagnone-Sereno, P. Molecular Cloning of a cDNA Encoding an Amphid-Secreted Putative Avirulence Protein from the Root-Knot Nematode Meloidogyne incognita. Mol. Plant-Microbe Interact. 2001, 14, 72–79. [Google Scholar] [CrossRef] [Green Version]
- Eves-Van Den Akker, S.; Lilley, C.J.; Yusup, H.B.; Jones, J.T.; Urwin, P.E. Functional C-TERMINALLY ENCODED PEPTIDE (CEP) plant hormone domains evolved de novo in the plant parasite Rotylenchulus reniformis. Mol. Plant Pathol. 2016, 17, 1265–1275. [Google Scholar] [CrossRef] [Green Version]
- Bobay, B.G.; Digennaro, P.; Scholl, E.; Imin, N.; Djordjevic, M.A.; Mck Bird, D. Solution NMR studies of the plant peptide hormone CEP inform function. FEBS Lett. 2013, 587, 3979–3985. [Google Scholar] [CrossRef] [Green Version]
- Pruitt, R.N.; Joe, A.; Zhang, W.; Feng, W.; Stewart, V.; Schwessinger, B.; Dinneny, J.R.; Ronald, P.C. A microbially derived tyrosine-sulfated peptide mimics a plant peptide hormone. New Phytol. 2017, 215, 725–736. [Google Scholar] [CrossRef] [Green Version]
- Pruitt, R.N.; Schwessinger, B.; Joe, A.; Thomas, N.; Liu, F.; Albert, M.; Robinson, M.R.; Chan, L.J.G.; Luu, D.D.; Chen, H.; et al. The rice immune receptor XA21 recognizes a tyrosine-sulfated protein from a Gram-negative bacterium. Sci. Adv. 2015, 1, e1500245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, F.; McDonald, M.; Schwessinger, B.; Joe, A.; Pruitt, R.; Erickson, T.; Zhao, X.; Stewart, V.; Ronald, P.C. Variation and inheritance of the Xanthomonas raxX-raxSTAB gene cluster required for activation of XA21-mediated immunity. Mol. Plant Pathol. 2019, 20, 656–672. [Google Scholar] [CrossRef] [Green Version]
- Luu, D.D.; Joe, A.; Chen, Y.; Parys, K.; Bahar, O.; Pruitt, R.; Chan, L.J.G.; Petzold, C.J.; Long, K.; Adamchak, C.; et al. Biosynthesis and secretion of the microbial sulfated peptide RaxX and binding to the rice XA21 immune receptor. Proc. Natl. Acad. Sci. USA 2019, 116, 8525–8534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tucker, M.L.; Yang, R. A gene encoding a peptide with similarity to the plant IDA signaling peptide (AtIDA) is expressed most abundantly in the root-knot nematode (Meloidogyne incognita) soon after root infection. Exp. Parasitol. 2013, 134, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Yang, R.; Chang, C.; Park, Y.; Tucker, M.L. The root-knot nematode Meloidogyne incognita produces a functional mimic of the Arabidopsis INFLORESCENCE DEFICIENT IN ABSCISSION signaling peptide. J. Exp. Bot. 2018, 69, 3009–3021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masachis, S.; Segorbe, D.; Turrá, D.; Leon-Ruiz, M.; Fürst, U.; El Ghalid, M.; Leonard, G.; López-Berges, M.S.; Richards, T.A.; Felix, G.; et al. A fungal pathogen secretes plant alkalinizing peptides to increase infection. Nat. Microbiol. 2016, 1, 16043. [Google Scholar] [CrossRef] [PubMed]
- Thynne, E.; Saur, I.M.L.; Simbaqueba, J.; Ogilvie, H.A.; Gonzalez-Cendales, Y.; Mead, O.; Taranto, A.; Catanzariti, A.-M.; McDonald, M.C.; Schwessinger, B.; et al. Fungal phytopathogens encode functional homologues of plant rapid alkalinization factor (RALF) peptides. Mol. Plant Pathol. 2017, 18, 811–824. [Google Scholar] [CrossRef] [PubMed]
- Wood, A.K.M.; Walker, C.; Lee, W.-S.; Urban, M.; Hammond-Kosack, K.E. Functional evaluation of a homologue of plant rapid alkalinisation factor (RALF) peptides in Fusarium graminearum. Fungal Biol. 2020, 124, 753–765. [Google Scholar] [CrossRef]
- Chisholm, S.T.; Coaker, G.; Day, B.; Staskawicz, B.J. Host-Microbe Interactions: Shaping the Evolution of the Plant Immune Response. Cell 2006, 124, 803–814. [Google Scholar] [CrossRef] [Green Version]
- van Megen, H.; van den Elsen, S.; Holterman, M.; Karssen, G.; Mooyman, P.; Bongers, T.; Holovachov, O.; Bakker, J.; Helder, J. A phylogenetic tree of nematodes based on about 1200 full-length small subunit ribosomal DNA sequences. Nematology 2009, 11, 927–950. [Google Scholar] [CrossRef]
- Ali, M.A.; Azeem, F.; Li, H.; Bohlmann, H. Smart Parasitic Nematodes Use Multifaceted Strategies to Parasitize Plants. Front. Plant Sci. 2017, 8, 1699. [Google Scholar] [CrossRef] [Green Version]
- Haegeman, A.; Mantelin, S.; Jones, J.T.; Gheysen, G. Functional roles of effectors of plant-parasitic nematodes. Gene 2012, 492, 19–31. [Google Scholar] [CrossRef]
- de Almeida Engler, J.; Kyndt, T.; Vieira, P.; Van Cappelle, E.; Boudolf, V.; Sanchez, V.; Escobar, C.; De Veylder, L.; Engler, G.; Abad, P.; et al. CCS52andDEL1genes are key components of the endocycle in nematode-induced feeding sites. Plant J. 2012, 72, 185–198. [Google Scholar] [CrossRef]
- Redding, N.W.; Agudelo, P.; Wells, C.E. Multiple Nodulation Genes Are Up-Regulated During Establishment of Reniform Nematode Feeding Sites in Soybean. Phytopathology 2018, 108, 275–291. [Google Scholar] [CrossRef] [Green Version]
- De Meutter, J.; Tytgat, T.; Witters, E.; Gheysen, G.; Van Onckelen, H.; Gheysen, G. Identification of cytokinins produced by the plant parasitic nematodes Heterodera schachtii and Meloidogyne incognita. Mol. Plant Pathol. 2003, 4, 271–277. [Google Scholar] [CrossRef]
- Patel, N.; Hamamouch, N.; Li, C.; Hussey, R.; Mitchum, M.; Baum, T.; Wang, X.; Davis, E.L. Similarity and Functional Analyses of Expressed Parasitism Genes in Heterodera Schachtii and Heterodera Glycines. J. Nematol. 2008, 40, 299–310. [Google Scholar]
- Pokhare, S.S.; Thorpe, P.; Hedley, P.; Morris, J.; Habash, S.S.; Elashry, A.; Eves-van den Akker, S.; Grundler, F.M.W.; Jones, J.T. Signatures of adaptation to a monocot host in the plant-parasitic cyst nematode Heterodera sacchari. Plant J. 2020, 103, 1263–1274. [Google Scholar] [CrossRef]
- Ito, Y.; Nakanomyo, I.; Motose, H.; Iwamoto, K.; Sawa, S.; Dohmae, N.; Fukuda, H. Dodeca-CLE Peptides as Suppressors of Plant Stem Cell Differentiation. Science 2006, 313, 842–845. [Google Scholar] [CrossRef]
- Brand, U.; Fletcher, J.C.; Hobe, M.; Meyerowitz, E.M.; Simon, R. Dependence of Stem Cell Fate in Arabidopsis on a Feedback Loop Regulated by CLV3 Activity. Science 2000, 289, 617–619. [Google Scholar] [CrossRef]
- Stahl, Y.; Wink, R.H.; Ingram, G.C.; Simon, R. A Signaling Module Controlling the Stem Cell Niche in Arabidopsis Root Meristems. Curr. Biol. 2009, 19, 909–914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirakawa, Y.; Kondo, Y.; Fukuda, H. TDIF Peptide Signaling Regulates Vascular Stem Cell Proliferation via the WOX4 Homeobox Gene in Arabidopsis. Plant Cell 2010, 22, 2618–2629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whitford, R.; Fernandez, A.; De Groodt, R.; Ortega, E.; Hilson, P. Plant CLE peptides from two distinct functional classes synergistically induce division of vascular cells. Proc. Natl. Acad. Sci. USA 2008, 105, 18625–18630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bird, D.M.; Jones, J.T.; Opperman, C.H.; Kikuchi, T.; Danchin, E.G.J. Signatures of adaptation to plant parasitism in nematode genomes. Parasitology 2015, 142 (Suppl. 1), S71–S84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamaguchi, Y.L.; Suzuki, R.; Cabrera, J.; Nakagami, S.; Sagara, T.; Ejima, C.; Sano, R.; Aoki, Y.; Olmo, R.; Kurata, T.; et al. Root-Knot and Cyst Nematodes Activate Procambium-Associated Genes in Arabidopsis Roots. Front. Plant Sci. 2017, 8, 1195. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Liu, X.; Engstrom, E.M.; Nimchuk, Z.L.; Pruneda-Paz, J.L.; Tarr, P.T.; Yan, A.; Kay, S.A.; Meyerowitz, E.M. Control of plant stem cell function by conserved interacting transcriptional regulators. Nature 2015, 517, 377–380. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.W.; Lim, J. Control of Asymmetric Cell Divisions during Root Ground Tissue Maturation. Mol. Cells 2016, 39, 524–529. [Google Scholar] [CrossRef] [Green Version]
- Tomalová, I.; Iachia, C.; Mulet, K.; Castagnone-Sereno, P. The map-1 Gene Family in Root-Knot Nematodes, Meloidogyne spp.: A Set of Taxonomically Restricted Genes Specific to Clonal Species. PLoS ONE 2012, 7, e38656. [Google Scholar] [CrossRef]
- Doehlemann, G.; Ökmen, B.; Zhu, W.; Sharon, A. Plant Pathogenic Fungi. Microbiol. Spectr. 2017, 5, 703–726. [Google Scholar] [CrossRef]
- Chanclud, E.; Morel, J.-B. Plant hormones: A fungal point of view. Mol. Plant Pathol. 2016, 17, 1289–1297. [Google Scholar] [CrossRef] [PubMed]
- Pearce, G.; Yamaguchi, Y.; Munske, G.; Ryan, C.A. Structure–activity studies of RALF, Rapid Alkalinization Factor, reveal an essential-YISY-motif. Peptides 2010, 31, 1973–1977. [Google Scholar] [CrossRef] [PubMed]
- Haruta, M.; Sabat, G.; Stecker, K.; Minkoff, B.B.; Sussman, M.R. A Peptide Hormone and Its Receptor Protein Kinase Regulate Plant Cell Expansion. Science 2014, 343, 408–411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, Y.; Yuan, Q.; Tang, J.; Huang, J.; Hsiang, T.; Wei, Y.; Zheng, L. Colletotrichum higginsianum as a Model for Understanding Host–Pathogen Interactions: A Review. Int. J. Mol. Sci. 2018, 19, 2142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, H.D.T.; Sultana, T.; Kesanakurti, P.; Hambleton, S. Genome sequencing and comparison of five Tilletia species to identify candidate genes for the detection of regulated species infecting wheat. IMA Fungus 2019, 10, 1–17. [Google Scholar] [CrossRef]
- Zheng, W.; Peng, Z.; Peng, S.; Yu, Z.; Cao, Z. Multinuclei Occurred Under Cryopreservation and Enhanced the Pathogenicity of Melampsora larici-populina. Front. Microbiol. 2021, 12, 650902. [Google Scholar] [CrossRef] [PubMed]
- EFSA Panel on Plant Health (PLH); Bragard, C.; Dehnen-Schmutz, K.; Di Serio, F.; Gonthier, P.; Jacques, M.-A.; Jaques Miret, J.A.; Justesen, A.F.; MacLeod, A.; Magnusson, C.S.; et al. Pest categorisation of Colletotrichum fructicola. EFSA J. 2021, 19, e06803. [Google Scholar] [CrossRef] [PubMed]
- Bull, C.T.; de Boer, S.H.; Denny, T.P.; Firrao, G.; Fischer-Le Saux, M.; Saddler, G.S.; Scortichini, M.; Stead, D.E.; Takikawa, Y. Comprehensive List of Names of Plant Pathogenic Bacteria, 1980–2007. J. Plant Pathol. 2010, 92, 551–592. [Google Scholar]
- Mansfield, J.; Genin, S.; Magori, S.; Citovsky, V.; Sriariyanum, M.; Ronald, P.; Dow, M.; Verdier, V.; Beer, S.V.; Machado, M.A.; et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 614–629. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.-L.; Song, W.Y.; Ruan, D.L.; Sideris, S.; Ronald, P.C. The Cloned Gene, Xa21, Confers Resistance to Multiple Xanthomonas oryzae pv. oryzae Isolates in Transgenic Plants. Mol. Plant-Microbe Interact. 1996, 9, 850–855. [Google Scholar] [CrossRef] [Green Version]
- Safni, I.; Subandiyah, S.; Fegan, M. Ecology, Epidemiology and Disease Management of Ralstonia syzygii in Indonesia. Front. Microbiol. 2018, 9, 419. [Google Scholar] [CrossRef] [Green Version]
- Van Wees, S.C.M.; Van der Ent, S.; Pieterse, C.M.J. Plant immune responses triggered by beneficial microbes. Curr. Opin. Plant Biol. 2008, 11, 443–448. [Google Scholar] [CrossRef] [Green Version]
- Mabood, F.; Souleimanov, A.; Khan, W.; Smith, D.L. Jasmonates induce Nod factor production by Bradyrhizobium japonicum. Plant Physiol. Biochem. 2006, 44, 759–765. [Google Scholar] [CrossRef]
- Gourion, B.; Berrabah, F.; Ratet, P.; Stacey, G. Rhizobium–legume symbioses: The crucial role of plant immunity. Trends Plant Sci. 2015, 20, 186–194. [Google Scholar] [CrossRef] [Green Version]
- Kinkema, M.; Gresshoff, P.M. Investigation of Downstream Signals of the Soybean Autoregulation of Nodulation Receptor Kinase GmNARK. Mol. Plant-Microbe Interact. 2008, 21, 1337–1348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oldroyd, G.E.D.; Murray, J.D.; Poole, P.S.; Downie, J.A. The Rules of Engagement in the Legume-Rhizobial Symbiosis. Annu. Rev. Genet. 2011, 45, 119–144. [Google Scholar] [CrossRef] [PubMed]
- Frugier, F.; Kosuta, S.; Murray, J.D.; Crespi, M.; Szczyglowski, K. Cytokinin: Secret agent of symbiosis. Trends Plant Sci. 2008, 13, 115–120. [Google Scholar] [CrossRef] [PubMed]
- van Zeijl, A.; den Camp, R.H.M.; Deinum, E.E.; Charnikhova, T.; Franssen, H.; den Camp, H.J.M.; Bouwmeester, H.; Kohlen, W.; Bisseling, T.; Geurts, R.; et al. Rhizobium Lipo-chitooligosaccharide Signaling Triggers Accumulation of Cytokinins in Medicago truncatula Roots. Mol. Plant 2015, 8, 1213–1226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boiero, L.; Perrig, D.; Masciarelli, O.; Penna, C.; Cassán, F.; Luna, V. Phytohormone production by three strains of Bradyrhizobium japonicum and possible physiological and technological implications. Appl. Microbiol. Biotechnol. 2007, 74, 874–880. [Google Scholar] [CrossRef]
- Kisiala, A.; Laffont, C.; Emery, R.J.N.; Frugier, F. Bioactive Cytokinins Are Selectively Secreted by Sinorhizobium meliloti Nodulating and Nonnodulating Strains. Mol. Plant-Microbe Interact. 2013, 26, 1225–1231. [Google Scholar] [CrossRef] [Green Version]
- Theunis, M.; Kobayashi, H.; Broughton, W.J.; Prinsen, E. Flavonoids, NodD1, NodD2, and Nod-Box NB15 Modulate Expression of the y4wEFG Locus That Is Required for Indole-3-Acetic Acid Synthesis in Rhizobium sp. strain NGR234. Mol. Plant-Microbe Interact. 2004, 17, 1153–1161. [Google Scholar] [CrossRef] [Green Version]
- Jaiswal, S.K.; Mohammed, M.; Ibny, F.Y.I.; Dakora, F.D. Rhizobia as a Source of Plant Growth-Promoting Molecules: Potential Applications and Possible Operational Mechanisms. Front. Sustain. Food Syst. 2021, 4, 311. [Google Scholar] [CrossRef]
- Sathya, A.; Vijayabharathi, R.; Gopalakrishnan, S. Plant growth-promoting actinobacteria: A new strategy for enhancing sustainable production and protection of grain legumes. 3 Biotech 2017, 7, 102. [Google Scholar] [CrossRef] [Green Version]
- Hastwell, A.H.; Gresshoff, P.M.; Ferguson, B.J. The structure and activity of nodulation-suppressing CLE peptide hormones of legumes. Funct. Plant Biol. 2014, 42, 229–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brundrett, M.C.; Tedersoo, L. Evolutionary history of mycorrhizal symbioses and global host plant diversity. New Phytol. 2018, 220, 1108–1115. [Google Scholar] [CrossRef] [PubMed]
- Pons, S.; Fournier, S.; Chervin, C.; Bécard, G.; Rochange, S.; Frey, N.F.D.; Pagès, V.P. Phytohormone production by the arbuscular mycorrhizal fungus Rhizophagus irregularis. PLoS ONE 2020, 15, e0240886. [Google Scholar] [CrossRef]
- Reid, D.E.; Ferguson, B.J.; Hayashi, S.; Lin, Y.-H.; Gresshoff, P.M. Molecular mechanisms controlling legume autoregulation of nodulation. Ann. Bot. 2011, 108, 789–795. [Google Scholar] [CrossRef] [Green Version]
- Müller, L.M.; Floková, K.; Schnabel, E.; Sun, X.; Fei, Z.; Frugoli, J.; Bouwmeester, H.J.; Harrison, M.J. A CLE–SUNN module regulates strigolactone content and fungal colonization in arbuscular mycorrhiza. Nat. Plants 2019, 5, 933–939. [Google Scholar] [CrossRef] [PubMed]
- Samorodova, A.P.; Tvorogova, V.E.; Tkachenko, A.A.; Potsenkovskaya, E.A.; Lebedeva, M.A.; Tikhonovich, I.A.; Lutova, L. Agrobacterial tumors interfere with nodulation and demonstrate the expression of nodulation-induced CLE genes in pea. J. Plant Physiol. 2018, 221, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, Y.; Shinohara, H.; Kondo, Y.; Inoue, A.; Nakanomyo, I.; Ogawa, M.; Sawa, S.; Ohashi-Ito, K.; Matsubayashi, Y.; Fukuda, H. Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc. Natl. Acad. Sci. USA 2008, 105, 15208–15213. [Google Scholar] [CrossRef] [Green Version]
- Lohar, D.P.; Bird, D.M. Lotus japonicus: A New Model to Study Root-Parasitic Nematodes. Plant Cell Physiol. 2003, 44, 1176–1184. [Google Scholar] [CrossRef] [Green Version]
- Soucy, S.M.; Huang, J.; Gogarten, J.P. Horizontal gene transfer: Building the web of life. Nat. Rev. Genet. 2015, 16, 472–482. [Google Scholar] [CrossRef] [PubMed]
- Husnik, F.; McCutcheon, J.P. Functional horizontal gene transfer from bacteria to eukaryotes. Nat. Rev. Microbiol. 2018, 16, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Spíchal, L. Cytokinins—recent news and views of evolutionally old molecules. Funct. Plant Biol. 2012, 39, 267–284. [Google Scholar] [CrossRef] [Green Version]
- Gao, C.; Ren, X.; Mason, A.S.; Liu, H.; Xiao, M.; Li, J.; Fu, D. Horizontal gene transfer in plants. Funct. Integr. Genom. 2014, 14, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Matveeva, T.V.; Otten, L. Widespread occurrence of natural genetic transformation of plants by Agrobacterium. Plant Mol. Biol. 2019, 101, 415–437. [Google Scholar] [CrossRef]
- Danchin, E.G.J. What Nematode genomes tell us about the importance of horizontal gene transfers in the evolutionary history of animals. Mob. Genet. Elements 2011, 1, 269–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danchin, E.G.J.; Rosso, M.-N.; Vieira, P.; de Almeida-Engler, J.; Coutinho, P.M.; Henrissat, B.; Abad, P. Multiple lateral gene transfers and duplications have promoted plant parasitism ability in nematodes. Proc. Natl. Acad. Sci. USA 2010, 107, 17651–17656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paganini, J.; Campan-Fournier, A.; DA Rocha, M.; Gouret, P.; Pontarotti, P.; Wajnberg, E.; Abad, P.; Danchin, E.G.J. Contribution of Lateral Gene Transfers to the Genome Composition and Parasitic Ability of Root-Knot Nematodes. PLoS ONE 2012, 7, e50875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dieterich, C.; Clifton, S.W.; Schuster, L.N.; Chinwalla, A.; Delehaunty, K.; Dinkelacker, I.; Fulton, L.; Fulton, R.; Godfrey, J.; Minx, P.; et al. The Pristionchus pacificus genome provides a unique perspective on nematode lifestyle and parasitism. Nat. Genet. 2008, 40, 1193–1198. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Family of Peptide Phytohormones | Species Name | Examples of Functional Domain Sequences | Domain Structure of Precursor Protein * | Putative Plant Receptor | References |
|---|---|---|---|---|---|
| CLE | Bacterial CLEs | ||||
| Actinobacteria sp. | RLSPGGPDPARH RTIPTGSNPLRN |  | Unknown | [41] | |
| Thiotrichales sp. Acidimicrobiaceae sp. | RGIPTGPDPTHN | ||||
| Gemmatimonadetes sp. Actinobacteria sp. | RVVPGGPRPVHH RVVPGGPRPIYH |  | |||
| Fungal CLEs | |||||
| Rhizophagus irregularis | RTVPSGPNPLHN RIVPSGPNPLHN |  | CLV2 | [42,44,45] | |
| Rhizophagus diaphanus | RLVPSGPNPLHN | ||||
| Rhizophagus cerebriforme | RIVPSGPNPLHN | ||||
| Rhizophagus clarus | RTVPTGSNPLHN | ||||
| Nematode CLEs | |||||
| Cyst nematodes: | |||||
| Heterodera spp. | RLSPSGPDPHHH RVSPSGPDPQHH RLSPSGPDPRHH HEVPSGPNPTQN |  | CLV1, CLV2/CRN, RPK1, BAM1, BAM2, TDR | [46,47,48,49,50,51,52,53,54] | |
| Globodera spp. | RVTPGGPDPLHN RVTPGVPDRQHR RVAGAGPDPIHN RAVPAGPDPKHN RGAPAGPDPIHN RVVVGGPDPQHH |  from 1 to 5 CLE domains | CLV2 | [55,56,57] | |
| Reniform nematodes: | |||||
| Rotylenchulus spp. | RESPGGPDPKHH |  | Unknown | [58] | |
| Root-knot nematodes: | |||||
| Meloidogyne spp. 16D10 | GKKPSGPNPGGN |  | Unknown | [59,60,61] | |
| Meloidogyne spp. MAP | NVYPSGPEPSTP NLYPSGPEPTPK NPYQSGPEPSTQ NPYPSGPEPTPK NVYPSGPESSTP GKYPSGPPSHNQ |  from 1 to 9 CLE domains | Unknown | [62,63] | |
| CEP | Bacterial CEPs | ||||
| Ralstonia syzygii | DSQPTTPGHSPGVGH |  | Unknown | [41] | |
| Nematode CEPs | |||||
| Reniform nematodes: | |||||
| Rotylenchulus spp. | AFKPTTPGHSPGAGH AFKPTTPGHSPGDGH GFRPTTPGHSPGVGN GFRPTTPGHAPGFGN AFRPTTPVHSPGAGQ PFKPTAPGHSPGVGH |  from 1 to 9 CLE domains | Unknown | [64] | |
| Root-knot nematodes: | |||||
| Meloidogyne spp. | DPRPTNPGHSPGIGH AFRPTAPGHSPGVGH GYQPTNPGHSPGIGH PFKTVPGQSSPGVGH VIKPACIGNSPGVGH AFRPTNPGPSPAIGN |  | Unknown | [65] | |
| PSY | Bacterial PSY | ||||
| Xanthomonas oryzae pv. oryzae other Xanthomonas species | DYPPPGANPKHDP |  | Xa21 | [66,67,68,69] | |
| PSK | Fungal PSKs | ||||
| Tilletia sp. | YIYTQ |  | Unknown | [41] | |
| Colletotrichum sp. Lasiodiplodia sp. Diplodia sp. Macrophomina phaseolina Cercospora sp. Ramularia collo-cygni Pseudocercospora sp. Zymoseptoria sp. | YIYTQ |  from 2 to 4 PSK domains | |||
| Bacterial PSKs | |||||
| Proteobacteria sp. | YIYTQ |  | Unknown | [41] | |
| IDA | Fungal IDAs | ||||
| Melampsora larici-populina | PIPPSTTSKRHA |  | Unknown | [41] | |
| Colletotrichum fructicola | KIPPSGPSKKHN |  | |||
| Nematode IDAs | |||||
| Meloidogyne spp. | KNIPVPASGPSKRHN RGEPIPGSGPSVRN KGVPPNSGPSRRGN KGVPHNSAPSRRGN KGNKKSSGPSHRGN KGKKESSGPSLGGN |  | HAE, HSL2 | [70,71] | |
| RALF | Fungal RALFs | ||||
| Fusarium oxysporum other 26 fungi species | ..ISY…-[PCS…….NCR]- -[RGC…CRG]- |  ISY—‘ISY’ motif, C—cysteine residues predicted to form disulfide bonds | FER | [72,73,74] | |
| Bacterial RALFs | |||||
| Streptomyces acidiscabies other 8 species of Actinobacteria | ..ISY…-[PCS…….NCR]- -[RGC…CRG]- |  ISY—‘ISY’ motif, C—cysteine residues predicted to form disulfide bonds | Unknown | [73] | |
| PEP | Fungal PEPs | ||||
| Metschnikowia sp. JCM 33374 | SSGIGGADN |  | [41] | ||
| Bacterial PEPs | |||||
| Mycolicibacterium conceptionense | SSGTSGGSN |  | Unknown | [41] | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dodueva, I.; Lebedeva, M.; Lutova, L. Dialog between Kingdoms: Enemies, Allies and Peptide Phytohormones. Plants 2021, 10, 2243. https://doi.org/10.3390/plants10112243
Dodueva I, Lebedeva M, Lutova L. Dialog between Kingdoms: Enemies, Allies and Peptide Phytohormones. Plants. 2021; 10(11):2243. https://doi.org/10.3390/plants10112243
Chicago/Turabian StyleDodueva, Irina, Maria Lebedeva, and Lyudmila Lutova. 2021. "Dialog between Kingdoms: Enemies, Allies and Peptide Phytohormones" Plants 10, no. 11: 2243. https://doi.org/10.3390/plants10112243