Myrtucommulones and Related Acylphloroglucinols from Myrtaceae as a Promising Source of Multitarget SARS-CoV-2 Cycle Inhibitors
Abstract
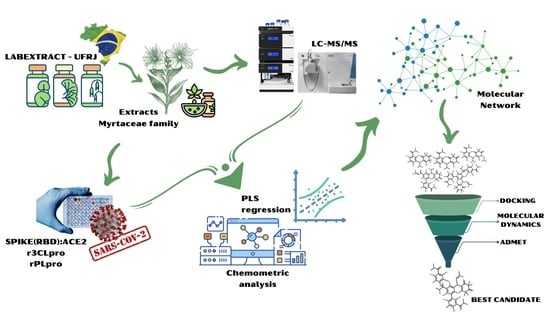
:1. Introduction
2. Results and Discussion
2.1. Myrtaceae Extracts Inhibit RBD:ACE2 Complex Formation and SARS-CoV-2 Proteases Activity In Vitro
2.2. Myrtaceae Extracts Inhibit SARS-CoV-2 Spike-Pseudotyped VSV Entry into VERO ACE2 Cells
2.3. Chemometric Analyses Using Partial Least Squares Regression Prediction Model
2.4. Annotated Compounds for the Predicted Ions in the Multitarget Model Using Molecular Networking Analyses
2.5. Myrtaceae Extracts Inhibit SARS-CoV-2 Replication in Infected Lung Cells
2.6. Docking Analysis and Ligand Interactions of Compounds 1–5 with Spike Protein and ACE2
2.6.1. Molecular Docking of ACE2 and Spike Protein
2.6.2. Interactions of the Ligand Compound 5 with ACE2
2.6.3. Interactions of the Ligand Compound 5 with the Spike Protein
2.7. PLpro and 3CLpro Interaction Analysis and Molecular Dynamics
3. Materials and Methods
3.1. Chemicals, Materials and Plant Extracts
3.2. Human Lung Cells Culture
3.3. SARS-CoV-2 Virus
3.4. SARS-CoV-2 RBD:ACE2 Interaction
3.5. SARS-CoV-2 Spike-Pseudotyped VSV Neutralization Assay
3.6. In Vitro Inhibition of SARS-CoV-2 3CLpro and PLpro
3.7. Cytotoxicity Assays
3.8. LC-MS/MS Analysis
3.9. Data Processing, Statistical Analysis and Molecular Networking
3.10. Anti-SARS-CoV-2 Assay in Calu-3 Cells
3.11. Virus Titration
3.12. Statistical Analysis
3.13. Assessment of Pharmacokinetic Characteristics
3.14. Molecular Docking Analysis
3.15. Molecular Dynamics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wagner, M.D.A.; Bogoni, J.A.; Fiaschi, P. Myrtaceae Richness and Distribution across the Atlantic Forest Domain Are Constrained by Geoclimatic Variables. Plant Ecol. 2022, 223, 1079–1092. [Google Scholar] [CrossRef]
- Celaj, O.; Durán, A.G.; Cennamo, P.; Scognamiglio, M.; Fiorentino, A.; Esposito, A.; D’Abrosca, B. Phloroglucinols from Myrtaceae: Attractive Targets for Structural Characterization, Biological Properties and Synthetic Procedures. Phytochem. Rev. 2021, 20, 259–299. [Google Scholar] [CrossRef]
- Leal, C.M.; Leitão, S.G.; Sausset, R.; Mendonça, S.C.; Nascimento, P.H.A.; De Araujo, R.; Cheohen, C.F.; Esteves, M.E.A.; Leal Da Silva, M.; Gondim, T.S.; et al. Flavonoids from Siparuna cristata as Potential Inhibitors of SARS-CoV-2 Replication. Rev. Bras. Farmacogn. 2021, 31, 658–666. [Google Scholar] [CrossRef]
- Cheohen, C.F.D.A.R.; Esteves, M.E.A.; Da Fonseca, T.S.; Leal, C.M.; Assis, F.D.L.F.; Campos, M.F.; Rebelo, R.S.; Allonso, D.; Leitão, G.G.; Da Silva, M.L.; et al. In Silico Screening of Phenylethanoid Glycosides, a Class of Pharmacologically Active Compounds as Natural Inhibitors of SARS-CoV-2 Proteases. Comput. Struct. Biotechnol. J. 2023, 21, 1461–1472. [Google Scholar] [CrossRef]
- Campos, M.F.; Mendonça, S.C.; Peñaloza, E.M.C.; De Oliveira, B.A.C.; Rosa, A.S.; Leitão, G.G.; Tucci, A.R.; Ferreira, V.N.S.; Oliveira, T.K.F.; Miranda, M.D.; et al. Anti-SARS-CoV-2 Activity of Ampelozizyphus amazonicus (Saracura-Mirá): Focus on the Modulation of the Spike-ACE2 Interaction by Chemically Characterized Bark Extracts by LC-DAD-APCI-MS/MS. Molecules 2023, 28, 3159. [Google Scholar] [CrossRef]
- Kaul, R.; Paul, P.; Kumar, S.; Büsselberg, D.; Dwivedi, V.D.; Chaari, A. Promising Antiviral Activities of Natural Flavonoids against SARS-CoV-2 Targets: Systematic Review. Int. J. Mol. Sci. 2021, 22, 11069. [Google Scholar] [CrossRef]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 Entry into Cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Scialo, F.; Daniele, A.; Amato, F.; Pastore, L.; Matera, M.G.; Cazzola, M.; Castaldo, G.; Bianco, A. ACE2: The Major Cell Entry Receptor for SARS-CoV-2. Lung 2020, 198, 867–877. [Google Scholar] [CrossRef]
- Salian, V.S.; Wright, J.A.; Vedell, P.T.; Nair, S.; Li, C.; Kandimalla, M.; Tang, X.; Carmona Porquera, E.M.; Kalari, K.R.; Kandimalla, K.K. COVID-19 Transmission, Current Treatment, and Future Therapeutic Strategies. Mol. Pharm. 2021, 18, 754–771. [Google Scholar] [CrossRef]
- Zhang, L.; Lin, D.; Sun, X.; Curth, U.; Drosten, C.; Sauerhering, L.; Becker, S.; Rox, K.; Hilgenfeld, R. Crystal Structure of SARS-CoV-2 Main Protease Provides a Basis for Design of Improved α-Ketoamide Inhibitors. Science 2020, 368, 409–412. [Google Scholar] [CrossRef]
- Khan, R.J.; Jha, R.K.; Amera, G.M.; Jain, M.; Singh, E.; Pathak, A.; Singh, R.P.; Muthukumaran, J.; Singh, A.K. Targeting SARS-CoV-2: A Systematic Drug Repurposing Approach to Identify Promising Inhibitors against 3C-like Proteinase and 2′-O-Ribose Methyltransferase. J. Biomol. Struct. Dyn. 2021, 39, 2679–2692. [Google Scholar] [CrossRef] [PubMed]
- Bosken, Y.K.; Cholko, T.; Lou, Y.-C.; Wu, K.-P.; Chang, C.A. Insights Into Dynamics of Inhibitor and Ubiquitin-Like Protein Binding in SARS-CoV-2 Papain-Like Protease. Front. Mol. Biosci. 2020, 7, 174. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Qin, B.; Chen, P.; Zhu, K.; Hou, P.; Wojdyla, J.A.; Wang, M.; Cui, S. Crystal Structure of SARS-CoV-2 Papain-like Protease. Acta Pharm. Sin. B 2021, 11, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Osipiuk, J.; Azizi, S.-A.; Dvorkin, S.; Endres, M.; Jedrzejczak, R.; Jones, K.A.; Kang, S.; Kathayat, R.S.; Kim, Y.; Lisnyak, V.G.; et al. Structure of Papain-like Protease from SARS-CoV-2 and Its Complexes with Non-Covalent Inhibitors. Nat. Commun. 2021, 12, 743. [Google Scholar] [CrossRef] [PubMed]
- Santos, L.S.; Alves Filho, E.G.; Ribeiro, P.R.V.; Zocolo, G.J.; Silva, S.M.; De Lucena, E.M.P.; Alves, R.E.; De Brito, E.S. Chemotaxonomic Evaluation of Different Species from the Myrtaceae Family by UPLC-qToF/MS-MS Coupled to Supervised Classification Based on Genus. Biochem. Syst. Ecol. 2020, 90, 104028. [Google Scholar] [CrossRef]
- Alipour, G.; Dashti, S.; Hosseinzadeh, H. Review of Pharmacological Effects of Myrtus communis L. and Its Active Constituents: Myrtus communis L. and Its Active Constituents. Phytother. Res. 2014, 28, 1125–1136. [Google Scholar] [CrossRef] [PubMed]
- Nicoletti, R.; Salvatore, M.; Ferranti, P.; Andolfi, A. Structures and Bioactive Properties of Myrtucommulones and Related Acylphloroglucinols from Myrtaceae. Molecules 2018, 23, 3370. [Google Scholar] [CrossRef] [PubMed]
- Knauthe, A.; Mittag, S.; Bloch, L.; Albring, K.F.; Schmidt, M.; Werz, O.; Huber, O. Hyperforin and Myrtucommulone Derivatives Act as Natural Modulators of Wnt/β-Catenin Signaling in HCT116 Colon Cancer Cells. Int. J. Mol. Sci. 2022, 23, 2984. [Google Scholar] [CrossRef]
- Moradi, M.T.; Karimi, A.; Rafieian, M.; Kheiri, S.; Saedi, M. The Inhibitory Effects of Myrtle (Myrtus communis) Extract on Herpes Simplex Virus-1 Replication in Baby Hamster Kidney Cells. J. Shahrekord Univ. Med. Sci. 2011, 12, 54–61. [Google Scholar]
- Chen, M.; Chen, L.-F.; Li, M.-M.; Li, N.-P.; Cao, J.-Q.; Wang, Y.; Li, Y.-L.; Wang, L.; Ye, W.-C. Myrtucomvalones A–C, Three Unusual Triketone–Sesquiterpene Adducts from the Leaves of Myrtus communis ‘Variegata’. RSC Adv. 2017, 7, 22735–22740. [Google Scholar] [CrossRef]
- Liu, J.; Song, J.-G.; Su, J.-C.; Huang, X.-J.; Ye, W.-C.; Wang, Y. Tomentodione E, a New Sec-Pentyl Syncarpic Acid-Based Meroterpenoid from the Leaves of Rhodomyrtus tomentosa. J. Asian Nat. Prod. Res. 2018, 20, 67–74. [Google Scholar] [CrossRef]
- Peron, G.; López, A.M.; Cabada-Aquirre, P.; Garay Buenrosto, K.D.; Ostos Mendoza, K.C.; Mahady, G.B.; Seidel, V.; Sytar, O.; Koirala, N.; Gurung, R.; et al. Antiviral and Antibacterial Properties of Phloroglucinols: A Review on Naturally Occurring and (Semi)Synthetic Derivatives with Potential Therapeutic Interest. Crit. Rev. Biotechnol. 2023, 44, 319–336. [Google Scholar] [CrossRef]
- Alves, J.; Engel, L.; De Vasconcelos Cabral, R.; Rodrigues, E.L.; De Jesus Ribeiro, L.; Higa, L.M.; Da Costa Ferreira Júnior, O.; Castiñeiras, T.M.P.P.; De Carvalho Leitão, I.; Tanuri, A.; et al. A Bioluminescent and Homogeneous SARS-CoV-2 Spike RBD and hACE2 Interaction Assay for Antiviral Screening and Monitoring Patient Neutralizing Antibody Levels. Sci. Rep. 2021, 11, 18428. [Google Scholar] [CrossRef]
- Hariono, M.; Rollando, R.; Yoga, I.; Harjono, A.; Suryodanindro, A.; Yanuar, M.; Gonzaga, T.; Parabang, Z.; Hariyono, P.; Febriansah, R.; et al. Bioguided Fractionation of Local Plants against Matrix Metalloproteinase9 and Its Cytotoxicity against Breast Cancer Cell Models: In Silico and In Vitro Study (Part II). Molecules 2021, 26, 1464. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and Community Curation of Mass Spectrometry Data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef]
- Afoullouss, S.; Balsam, A.; Allcock, A.L.; Thomas, O.P. Optimization of LC-MS2 Data Acquisition Parameters for Molecular Networking Applied to Marine Natural Products. Metabolites 2022, 12, 245. [Google Scholar] [CrossRef]
- Goodger, J.Q.D.; Seneratne, S.L.; Nicolle, D.; Woodrow, I.E. Foliar Essential Oil Glands of Eucalyptus Subgenus Eucalyptus (Myrtaceae) Are a Rich Source of Flavonoids and Related Non-Volatile Constituents. PLoS ONE 2016, 11, e0151432. [Google Scholar] [CrossRef]
- Appendino, G.; Bianchi, F.; Minassi, A.; Sterner, O.; Ballero, M.; Gibbons, S. Oligomeric Acylphloroglucinols from Myrtle (Myrtus communis). J. Nat. Prod. 2002, 65, 334–338. [Google Scholar] [CrossRef]
- Hans, M.; Charpentier, M.; Huch, V.; Jauch, J.; Bruhn, T.; Bringmann, G.; Quandt, D. Stereoisomeric Composition of Natural Myrtucommulone A. J. Nat. Prod. 2015, 78, 2381–2389. [Google Scholar] [CrossRef] [PubMed]
- Zaki, M.A.; Nanayakkara, N.P.D.; Hetta, M.H.; Jacob, M.R.; Khan, S.I.; Mohammed, R.; Ibrahim, M.A.; Samoylenko, V.; Coleman, C.; Fronczek, F.R.; et al. Bioactive Formylated Flavonoids from Eugenia rigida: Isolation, Synthesis, and X-Ray Crystallography. J. Nat. Prod. 2016, 79, 2341–2349. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, F.M.G.; Romão, W.; Kuster, R.M. Identification of Phenolic Compounds in Eugenia uniflora Leaves by FTICR MS in Association with Different Ionization Sources. Anal. Methods 2018, 10, 1647–1655. [Google Scholar] [CrossRef]
- Yoshida, T.; Amakura, Y.; Yoshimura, M. Structural Features and Biological Properties of Ellagitannins in Some Plant Families of the Order Myrtales. Int. J. Mol. Sci. 2010, 11, 79–106. [Google Scholar] [CrossRef] [PubMed]
- Ogando, N.S.; Dalebout, T.J.; Zevenhoven-Dobbe, J.C.; Limpens, R.W.A.L.; Van Der Meer, Y.; Caly, L.; Druce, J.; De Vries, J.J.C.; Kikkert, M.; Bárcena, M.; et al. SARS-Coronavirus-2 Replication in Vero E6 Cells: Replication Kinetics, Rapid Adaptation and Cytopathology. J. Gen. Virol. 2020, 101, 925–940. [Google Scholar] [CrossRef]
- Mellott, D.M.; Tseng, C.-T.; Drelich, A.; Fajtová, P.; Chenna, B.C.; Kostomiris, D.H.; Hsu, J.; Zhu, J.; Taylor, Z.W.; Kocurek, K.I.; et al. A Clinical-Stage Cysteine Protease Inhibitor Blocks SARS-CoV-2 Infection of Human and Monkey Cells. ACS Chem. Biol. 2021, 16, 642–650. [Google Scholar] [CrossRef]
- Mautner, L.; Hoyos, M.; Dangel, A.; Berger, C.; Ehrhardt, A.; Baiker, A. Replication Kinetics and Infectivity of SARS-CoV-2 Variants of Concern in Common Cell Culture Models. Virol. J. 2022, 19, 76. [Google Scholar] [CrossRef]
- White, J.M.; Schiffer, J.T.; Bender Ignacio, R.A.; Xu, S.; Kainov, D.; Ianevski, A.; Aittokallio, T.; Frieman, M.; Olinger, G.G.; Polyak, S.J. Drug Combinations as a First Line of Defense against Coronaviruses and Other Emerging Viruses. mBio 2021, 12, e03347-21. [Google Scholar] [CrossRef]
- Kumar, S.; Sarma, P.; Kaur, H.; Prajapat, M.; Bhattacharyya, A.; Avti, P.; Sehkhar, N.; Kaur, H.; Bansal, S.; Mahendiratta, S.; et al. Clinically Relevant Cell Culture Models and Their Significance in Isolation, Pathogenesis, Vaccine Development, Repurposing and Screening of New Drugs for SARS-CoV-2: A Systematic Review. Tissue Cell 2021, 70, 101497. [Google Scholar] [CrossRef] [PubMed]
- Mahoney, M.; Damalanka, V.C.; Tartell, M.A.; Chung, D.H.; Lourenco, A.L.; Pwee, D.; Mayer Bridwell, A.E.; Hoffmann, M.; Voss, J.; Karmakar, P.; et al. A Novel Class of TMPRSS2 Inhibitors Potently Block SARS-CoV-2 and MERS-CoV Viral Entry and Protect Human Epithelial Lung Cells. Proc. Natl. Acad. Sci. USA 2021. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, P.K.; Efferth, T.; Das, B.; Kar, A.; Ghosh, S.; Singha, S.; Debnath, P.; Sharma, N.; Bhardwaj, P.K.; Haldar, P.K. Role of Medicinal Plants in Inhibiting SARS-CoV-2 and in the Management of Post-COVID-19 Complications. Phytomedicine 2022, 98, 153930. [Google Scholar] [CrossRef]
- Jamal, Q.M.S. Antiviral Potential of Plants against COVID-19 during Outbreaks—An Update. Int. J. Mol. Sci. 2022, 23, 13564. [Google Scholar] [CrossRef]
- Gupta, S.; Singh, V.; Varadwaj, P.K.; Chakravartty, N.; Katta, A.V.S.K.M.; Lekkala, S.P.; Thomas, G.; Narasimhan, S.; Reddy, A.R.; Reddy Lachagari, V.B. Secondary Metabolites from Spice and Herbs as Potential Multitarget Inhibitors of SARS-CoV-2 Proteins. J. Biomol. Struct. Dyn. 2022, 40, 2264–2283. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.-T.; Chen, Y.; Chang, W.-C.; Chen, H.-F.; Lai, H.-C.; Lin, Y.-C.; Wang, W.-J.; Wang, Y.-C.; Yang, C.-S.; Wang, S.-C.; et al. Scutellaria barbata D. Don Inhibits the Main Proteases (Mpro and TMPRSS2) of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infection. Viruses 2021, 13, 826. [Google Scholar] [CrossRef]
- Tallei, T.E.; Tumilaar, S.G.; Niode, N.J.; Fatimawali; Kepel, B.J.; Idroes, R.; Effendi, Y.; Sakib, S.A.; Emran, T.B. Potential of Plant Bioactive Compounds as SARS-CoV-2 Main Protease (Mpro) and Spike (S) Glycoprotein Inhibitors: A Molecular Docking Study. Scientifica 2020, 2020, 6307457. [Google Scholar] [CrossRef]
- Llivisaca-Contreras, S.A.; Naranjo-Morán, J.; Pino-Acosta, A.; Pieters, L.; Vanden Berghe, W.; Manzano, P.; Vargas-Pérez, J.; León-Tamariz, F.; Cevallos-Cevallos, J.M. Plants and Natural Products with Activity against Various Types of Coronaviruses: A Review with Focus on SARS-CoV-2. Molecules 2021, 26, 4099. [Google Scholar] [CrossRef]
- Fornari Laurindo, L.; Taynara Marton, L.; Minniti, G.; Dogani Rodrigues, V.; Buzinaro Suzuki, R.; Maria Cavallari Strozze Catharin, V.; Joshi, R.K.; Barbalho, S.M. Exploring the Impact of Herbal Therapies on COVID-19 and Influenza: Investigating Novel Delivery Mechanisms for Emerging Interventions. Biologics 2023, 3, 158–186. [Google Scholar] [CrossRef]
- Kumar, Y.; Singh, H.; Patel, C.N. In Silico Prediction of Potential Inhibitors for the Main Protease of SARS-CoV-2 Using Molecular Docking and Dynamics Simulation Based Drug-Repurposing. J. Infect. Public Health 2020, 13, 1210–1223. [Google Scholar] [CrossRef]
- Ullrich, S.; Nitsche, C. The SARS-CoV-2 Main Protease as Drug Target. Bioorg. Med. Chem. Lett. 2020, 30, 127377. [Google Scholar] [CrossRef]
- WHO—World Health Organization Coronavirus Dashboard. Available online: https://covid19.who.int (accessed on 5 March 2024).
- Tucci, A.R.; Da Rosa, R.M.; Rosa, A.S.; Augusto Chaves, O.; Ferreira, V.N.S.; Oliveira, T.K.F.; Coutinho Souza, D.D.; Borba, N.R.R.; Dornelles, L.; Rocha, N.S.; et al. Antiviral Effect of 5′-Arylchalcogeno-3-Aminothymidine Derivatives in SARS-CoV-2 Infection. Molecules 2023, 28, 6696. [Google Scholar] [CrossRef]
- Caleffi, G.S.; Rosa, A.S.; De Souza, L.G.; Avelar, J.L.S.; Nascimento, S.M.R.; De Almeida, V.M.; Tucci, A.R.; Ferreira, V.N.; Da Silva, A.J.M.; Santos-Filho, O.A.; et al. Aurones: A Promising Scaffold to Inhibit SARS-CoV-2 Replication. J. Nat. Prod. 2023, 86, 1536–1549. [Google Scholar] [CrossRef] [PubMed]
- Xiong, G.; Wu, Z.; Yi, J.; Fu, L.; Yang, Z.; Hsieh, C.; Yin, M.; Zeng, X.; Wu, C.; Lu, A.; et al. ADMETlab 2.0: An Integrated Online Platform for Accurate and Comprehensive Predictions of ADMET Properties. Nucleic Acids Res. 2021, 49, W5–W14. [Google Scholar] [CrossRef] [PubMed]
- da-Costa-Rodrigues, B.; Cheohen, C.; Sciammarella, F.; Pierre-Bonetti-Pozzobon, A.; Ribeiro, L.; Nepomuceno-Silva, J.L.; Medeiros, M.; Mury, F.; Monteiro-de-Barros, C.; Lazoski, C.; et al. SARS-CoV-2 Spatiotemporal Genomic and Molecular Analysis of the First Wave of the COVID-19 Pandemic in Macaé, the Brazilian Capital of Oil. Int. J. Mol. Sci. 2022, 23, 11497. [Google Scholar] [CrossRef]
- Case, D.A.; Aktulga, H.M.; Belfon, K.; Cerutti, D.S.; Cisneros, G.A.; Cruzeiro, V.W.D.; Forouzesh, N.; Giese, T.J.; Götz, A.W.; Gohlke, H.; et al. AmberTools. J. Chem. Inf. Model. 2023, 63, 6183–6191. [Google Scholar] [CrossRef]
- Anandakrishnan, R.; Aguilar, B.; Onufriev, A.V. H++ 3.0: Automating pK Prediction and the Preparation of Biomolecular Structures for Atomistic Molecular Modeling and Simulations. Nucleic Acids Res. 2012, 40, W537–W541. [Google Scholar] [CrossRef]
- Gordon, J.C.; Myers, J.B.; Folta, T.; Shoja, V.; Heath, L.S.; Onufriev, A. H++: A Server for Estimating pKas and Adding Missing Hydrogens to Macromolecules. Nucleic Acids Res. 2005, 33, W368–W371. [Google Scholar] [CrossRef]
- Myers, J.; Grothaus, G.; Narayanan, S.; Onufriev, A. A Simple Clustering Algorithm Can Be Accurate Enough for Use in Calculations of pKs in Macromolecules. Proteins Struct. Funct. Bioinform. 2006, 63, 928–938. [Google Scholar] [CrossRef]
- Li, P.; Merz, K.M. MCPB.Py: A Python Based Metal Center Parameter Builder. J. Chem. Inf. Model. 2016, 56, 599–604. [Google Scholar] [CrossRef]
- Barca, G.M.J.; Bertoni, C.; Carrington, L.; Datta, D.; De Silva, N.; Deustua, J.E.; Fedorov, D.G.; Gour, J.R.; Gunina, A.O.; Guidez, E.; et al. Recent Developments in the General Atomic and Molecular Electronic Structure System. J. Chem. Phys. 2020, 152, 154102. [Google Scholar] [CrossRef]
- Machado, M.R.; Pantano, S. Split the Charge Difference in Two! A Rule of Thumb for Adding Proper Amounts of Ions in MD Simulations. J. Chem. Theory Comput. 2020, 16, 1367–1372. [Google Scholar] [CrossRef]














| Inhibition (%) | |||
|---|---|---|---|
| Extracts | Spike:ACE2 | 3CLpro | PLpro |
| Eugenia brasiliensis | 12 | 69 | 34 |
| Myrcia splendens | 81 | 58 | 52 |
| Eugenia mosenii | 84 | 100 | 72 |
| Myrcia strigipes | 79 | 46 | 43 |
| Eugenia prasina | 96 | 53 | 57 |
| E. mosenii | E. prasina | M. splendens | M. strigipes | E. brasiliensis | |
|---|---|---|---|---|---|
| EC50 (μg·mL−1) | <25 | 115 | 47 | 45 | 65 |
| Compound | Rt (min) | Molecular Formula | [M − H]− (m/z) | MS/MS (MS2) | Proposed/Annotated Compound | Relative Content (%) | Ref. |
|---|---|---|---|---|---|---|---|
| Network 1 | |||||||
| 1 | 21.86 | C25H34O7 | 445.1 | 235, 209, 181 | semimyrtucommulone | E. mosenii (6.2) | [17,28] |
| 2 | 23.78 | C24H32O7 | 431.1 | 221, 209, 165, 151 | nor-semimyrtucommulone | E. mosenii (7.9) | [17,29] |
| 3 | 25.24 | C27H38O7 | 473.2 | 263, 209, 151 | undescribed | E. mosenii (4.2) | - |
| 4 | 27.76 | C36H42O1 | 633.3 | 423, 219, 205, 209, 165 | undescribed | E. mosenii (0.9) | - |
| 5 | 28.54 | C47H54O1 | 841.3 | 631, 577, 421, 367 | undescribed | E. mosenii (26) | - |
| E. mosenii | E. prasina | M. splendens | M. strigipes | E. brasiliensis | |
|---|---|---|---|---|---|
| EC50 (µg·mL−1) | 8.14 ± 0.6 | 2.57 ± 0.5 | 8.15 ± 0.7 | 6.75 ± 0.4 | 3.03 ± 0.3 |
| Ligand Name | ACE2 (Fitness Score) | Spike (Fitness Score) |
|---|---|---|
| compound 3 (m/z 473 [M − H]−) | 45.41 | 19.18 |
| compound 4 (m/z 633 [M − H]−) | 56.37 | 18.24 |
| compound 5 (m/z 841 [M − H]−) | 67.74 | 20.2 |
| nor-semimyrtucommulone 2 | 42.18 | 14.15 |
| semimyrtucommulone 1 | 44.65 | 19.17 |
| Compounds | MW (Da) | nHA | nHD | TPSA (Å) | nRot | LogP |
|---|---|---|---|---|---|---|
| compound 3 (m/z 473 [M − H]−) | 474.26 | 7 | 3 | 128.97 | 7 | 4.706 |
| compound 4 (m/z 633 [M − H]−) | 634.28 | 10 | 5 | 178.66 | 6 | 7.036 |
| compound 5 (m/z 841 [M − H]−) | 842.35 | 14 | 7 | 245.42 | 10 | 8.359 |
| nor-semimyrtucommulone 2 | 432.21 | 7 | 3 | 128.97 | 5 | 3.845 |
| semimyrtucommulone 1 | 446.23 | 7 | 3 | 128.97 | 5 | 4.04 |
| Compounds | H-HT | DILI | AMES | Carc. | R.T. | PAINS |
|---|---|---|---|---|---|---|
| compound 3 | ||||||
| compound 4 | ||||||
| compound 5 | ||||||
| nor-semimyrtucommulone 2 | ||||||
| semimyrtucommulone 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mendonça, S.C.; Gomes, B.A.; Campos, M.F.; da Fonseca, T.S.; Esteves, M.E.A.; Andriolo, B.V.; Cheohen, C.F.d.A.R.; Constant, L.E.C.; da Silva Costa, S.; Calil, P.T.; et al. Myrtucommulones and Related Acylphloroglucinols from Myrtaceae as a Promising Source of Multitarget SARS-CoV-2 Cycle Inhibitors. Pharmaceuticals 2024, 17, 436. https://doi.org/10.3390/ph17040436
Mendonça SC, Gomes BA, Campos MF, da Fonseca TS, Esteves MEA, Andriolo BV, Cheohen CFdAR, Constant LEC, da Silva Costa S, Calil PT, et al. Myrtucommulones and Related Acylphloroglucinols from Myrtaceae as a Promising Source of Multitarget SARS-CoV-2 Cycle Inhibitors. Pharmaceuticals. 2024; 17(4):436. https://doi.org/10.3390/ph17040436
Chicago/Turabian StyleMendonça, Simony Carvalho, Brendo Araujo Gomes, Mariana Freire Campos, Thamirys Silva da Fonseca, Maria Eduarda Alves Esteves, Bruce Veiga Andriolo, Caio Felipe de Araujo Ribas Cheohen, Larissa Esteves Carvalho Constant, Stephany da Silva Costa, Pedro Telles Calil, and et al. 2024. "Myrtucommulones and Related Acylphloroglucinols from Myrtaceae as a Promising Source of Multitarget SARS-CoV-2 Cycle Inhibitors" Pharmaceuticals 17, no. 4: 436. https://doi.org/10.3390/ph17040436