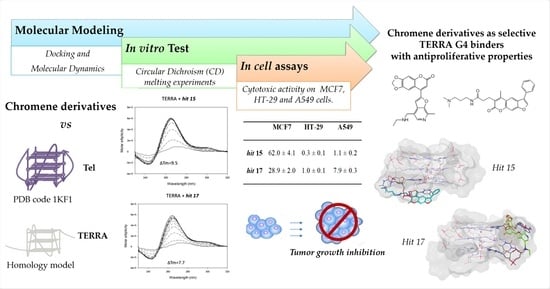
Chromene Derivatives as Selective TERRA G-Quadruplex RNA Binders with Antiproliferative Properties
Abstract
:1. Introduction
2. Results and Discussion
2.1. Computational Studies
2.2. In Vitro Analysis
2.3. In Cell Assays
3. Materials and Methods
3.1. Target Preparation for In Silico Analysis
3.2. Molecular Docking Protocol
3.3. Molecular Dynamics of the Thermodynamically Best Complexes
3.4. Circular Dichroism
3.5. Binding Affinity
3.6. Compounds’ Cytotoxicity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gordon, K.E.; Parkinson, E.K. Analysis of telomerase activity and telomere function in cancer. Methods Mol. Biol. 2004, 281, 333–348. [Google Scholar] [CrossRef] [PubMed]
- Parrotta, L.; Ortuso, F.; Moraca, F.; Rocca, R.; Costa, G.; Alcaro, S.; Artese, A. Targeting unimolecular G-quadruplex nucleic acids: A new paradigm for the drug discovery? Expert Opin. Drug Discov. 2014, 9, 1167–1187. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Carver, M.; Yang, D. Polymorphism of human telomeric quadruplex structures. Biochimie 2008, 90, 1172–1183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parkinson, G.N.; Lee, M.P.; Neidle, S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 2002, 417, 876–880. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Punchihewa, C.; Ambrus, A.; Chen, D.; Jones, R.A.; Yang, D. Structure of the intramolecular human telomeric G-quadruplex in potassium solution: A novel adenine triple formation. Nucleic Acids Res. 2007, 35, 2440–2450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, J.; Carver, M.; Punchihewa, C.; Jones, R.A.; Yang, D. Structure of the Hybrid-2 type intramolecular human telomeric G-quadruplex in K+ solution: Insights into structure polymorphism of the human telomeric sequence. Nucleic Acids Res. 2007, 35, 4927–4940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Patel, D.J. Solution structure of the human telomeric repeat d [AG3 (T2AG3) 3] G-tetraplex. Structure 1993, 1, 263–282. [Google Scholar] [CrossRef]
- Campbell, N.H.; Parkinson, G.N.; Reszka, A.P.; Neidle, S. Structural basis of DNA quadruplex recognition by an acridine drug. J. Am. Chem. Soc. 2008, 130, 6722–6724. [Google Scholar] [CrossRef]
- Hounsou, C.; Guittat, L.; Monchaud, D.; Jourdan, M.; Saettel, N.; Mergny, J.L.; Teulade-Fichou, M.P. G-quadruplex recognition by quinacridines: A SAR, NMR, and biological study. ChemMedChem 2007, 2, 655–666. [Google Scholar] [CrossRef]
- Phan, A.T.; Patel, D.J. Two-repeat human telomeric d(TAGGGTTAGGGT) sequence forms interconverting parallel and antiparallel G-quadruplexes in solution: Distinct topologies, thermodynamic properties, and folding/unfolding kinetics. J. Am. Chem. Soc. 2003, 125, 15021–15027. [Google Scholar] [CrossRef] [Green Version]
- Azzalin, C.M.; Reichenbach, P.; Khoriauli, L.; Giulotto, E.; Lingner, J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 2007, 318, 798–801. [Google Scholar] [CrossRef] [PubMed]
- Feuerhahn, S.; Iglesias, N.; Panza, A.; Porro, A.; Lingner, J. TERRA biogenesis, turnover and implications for function. FEBS Lett. 2010, 584, 3812–3818. [Google Scholar] [CrossRef] [Green Version]
- Porro, A.; Feuerhahn, S.; Delafontaine, J.; Riethman, H.; Rougemont, J.; Lingner, J. Functional characterization of the TERRA transcriptome at damaged telomeres. Nat. Commun. 2014, 5, 5379. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Kaminaga, K.; Komiyama, M. G-quadruplex formation by human telomeric repeats-containing RNA in Na+ solution. J. Am. Chem. Soc. 2008, 130, 11179–11184. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Norseen, J.; Wiedmer, A.; Riethman, H.; Lieberman, P.M. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell 2009, 35, 403–413. [Google Scholar] [CrossRef] [Green Version]
- Biffi, G.; Tannahill, D.; Balasubramanian, S. An intramolecular G-quadruplex structure is required for binding of telomeric repeat-containing RNA to the telomeric protein TRF2. J. Am. Chem. Soc. 2012, 134, 11974–11976. [Google Scholar] [CrossRef] [PubMed]
- Baker, A.M.; Fu, Q.; Hayward, W.; Victoria, S.; Pedroso, I.M.; Lindsay, S.M.; Fletcher, T.M. The telomere binding protein TRF2 induces chromatin compaction. PLoS ONE 2011, 6, e19124. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.U.; Park, W.J.; Jeong, W.K.; Baek, S.K.; Lee, H.W.; Lee, J.H. Prognostic impact of telomeric repeat-containing RNA expression on long-term oncologic outcomes in colorectal cancer. Medicine 2019, 98, e14932. [Google Scholar] [CrossRef] [PubMed]
- Marzano, S.; Pagano, B.; Iaccarino, N.; Di Porzio, A.; De Tito, S.; Vertecchi, E.; Salvati, E.; Randazzo, A.; Amato, J. Targeting of Telomeric Repeat-Containing RNA G-Quadruplexes: From Screening to Biophysical and Biological Characterization of a New Hit Compound. Int. J. Mol. Sci. 2021, 22, 10315. [Google Scholar] [CrossRef] [PubMed]
- Bryan, T.M.; Englezou, A.; Gupta, J.; Bacchetti, S.; Reddel, R.R. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 1995, 14, 4240–4248. [Google Scholar] [CrossRef]
- Collie, G.W.; Haider, S.M.; Neidle, S.; Parkinson, G.N. A crystallographic and modelling study of a human telomeric RNA (TERRA) quadruplex. Nucleic Acids Res. 2010, 38, 5569–5580. [Google Scholar] [CrossRef]
- Lim, K.W.; Amrane, S.; Bouaziz, S.; Xu, W.; Mu, Y.; Patel, D.J.; Luu, K.N.; Phan, A.T. Structure of the human telomere in K+ solution: A stable basket-type G-quadruplex with only two G-tetrad layers. J. Am. Chem. Soc. 2009, 131, 4301–4309. [Google Scholar] [CrossRef] [Green Version]
- Phan, A.T.; Kuryavyi, V.; Luu, K.N.; Patel, D.J. Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Res. 2007, 35, 6517–6525. [Google Scholar] [CrossRef] [Green Version]
- Martadinata, H.; Phan, A.T. Structure of human telomeric RNA (TERRA): Stacking of two G-quadruplex blocks in K(+) solution. Biochemistry 2013, 52, 2176–2183. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Tan, J.; Ou, T.; Huang, Z.; Gu, L. DNA G-quadruplex binders: A patent review. Expert Opin. Ther. Pat. 2013, 23, 1495–1509. [Google Scholar] [CrossRef] [PubMed]
- Rocca, R.; Moraca, F.; Costa, G.; Alcaro, S.; Distinto, S.; Maccioni, E.; Ortuso, F.; Artese, A.; Parrotta, L. Structure-based virtual screening of novel natural alkaloid derivatives as potential binders of h-telo and c-myc DNA G-quadruplex conformations. Molecules 2014, 20, 206–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rocca, R.; Costa, G.; Artese, A.; Parrotta, L.; Ortuso, F.; Maccioni, E.; Pinato, O.; Greco, M.L.; Sissi, C.; Alcaro, S. Hit Identification of a Novel Dual Binder for h-telo/c-myc G-Quadruplex by a Combination of Pharmacophore Structure-Based Virtual Screening and Docking Refinement. ChemMedChem 2016, 11, 1721–1733. [Google Scholar] [CrossRef] [PubMed]
- Alcaro, S.; Musetti, C.; Distinto, S.; Casatti, M.; Zagotto, G.; Artese, A.; Parrotta, L.; Moraca, F.; Costa, G.; Ortuso, F.; et al. Identification and characterization of new DNA G-quadruplex binders selected by a combination of ligand and structure-based virtual screening approaches. J. Med. Chem. 2013, 56, 843–855. [Google Scholar] [CrossRef]
- Costa, G.; Rocca, R.; Moraca, F.; Talarico, C.; Romeo, I.; Ortuso, F.; Alcaro, S.; Artese, A. A Comparative Docking Strategy to Identify Polyphenolic Derivatives as Promising Antineoplastic Binders of G-quadruplex DNA c-myc and bcl-2 Sequences. Mol. Inf. 2016, 35, 391–402. [Google Scholar] [CrossRef] [PubMed]
- Catalano, R.; Moraca, F.; Amato, J.; Cristofari, C.; Rigo, R.; Via, L.D.; Rocca, R.; Lupia, A.; Maruca, A.; Costa, G.; et al. Targeting multiple G-quadruplex-forming DNA sequences: Design, biophysical and biological evaluations of indolo-naphthyridine scaffold derivatives. Eur. J. Med. Chem. 2019, 182, 111627. [Google Scholar] [CrossRef] [PubMed]
- Collie, G.; Reszka, A.P.; Haider, S.M.; Gabelica, V.; Parkinson, G.N.; Neidle, S. Selectivity in small molecule binding to human telomeric RNA and DNA quadruplexes. Chem. Commun. 2009, 7482–7484. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Komiyama, M. Structure, function and targeting of human telomere RNA. Methods 2012, 57, 100–105. [Google Scholar] [CrossRef] [PubMed]
- Di Antonio, M.; Biffi, G.; Mariani, A.; Raiber, E.A.; Rodriguez, R.; Balasubramanian, S. Selective RNA versus DNA G-quadruplex targeting by in situ click chemistry. Angew. Chem. Int. Ed. Engl. 2012, 51, 11073–11078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rocca, R.; Talarico, C.; Moraca, F.; Costa, G.; Romeo, I.; Ortuso, F.; Alcaro, S.; Artese, A. Molecular recognition of a carboxy pyridostatin toward G-quadruplex structures: Why does it prefer RNA? Chem. Biol. Drug Des. 2017, 90, 919–925. [Google Scholar] [CrossRef] [PubMed]
- Rocca, R.; Moraca, F.; Costa, G.; Nadai, M.; Scalabrin, M.; Talarico, C.; Distinto, S.; Maccioni, E.; Ortuso, F.; Artese, A.; et al. Identification of G-quadruplex DNA/RNA binders: Structure-based virtual screening and biophysical characterization. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1329–1340. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y. Chemistry in human telomere biology: Structure, function and targeting of telomere DNA/RNA. Chem. Soc. Rev. 2011, 40, 2719–2740. [Google Scholar] [CrossRef]
- Xu, Y.; Ishizuka, T.; Kimura, T.; Komiyama, M. A U-tetrad stabilizes human telomeric RNA G-quadruplex structure. J. Am. Chem. Soc. 2010, 132, 7231–7233. [Google Scholar] [CrossRef] [PubMed]
- Alcaro, S.; Costa, G.; Distinto, S.; Moraca, F.; Ortuso, F.; Parrotta, L.; Artese, A. The polymorphisms of DNA G-quadruplex investigated by docking experiments with telomestatin enantiomers. Curr. Pharm. Des. 2012, 18, 1873–1879. [Google Scholar] [CrossRef]
- Li, J.; Abel, R.; Zhu, K.; Cao, Y.; Zhao, S.; Friesner, R.A. The VSGB 2.0 model: A next generation energy model for high resolution protein structure modeling. Proteins 2011, 79, 2794–2812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lombardo, C.M.; Sánchez Martínez, I.; Haider, S.; Gabelica, V.; De Pauw, E.; Moses, J.E.; Neidle, S. Structure-based design of selective high-affinity telomeric quadruplex-binding ligands. Chem. Commun. 2010, 46, 9116–9118. [Google Scholar] [CrossRef] [Green Version]
- Scalabrin, M.; Nadai, M.; Tassinari, M.; Lago, S.; Doria, F.; Frasson, I.; Freccero, M.; Richter, S.N. Selective Recognition of a Single HIV-1 G-Quadruplex by Ultrafast Small-Molecule Screening. Anal. Chem. 2021, 93, 15243–15252. [Google Scholar] [CrossRef]
- Hess, B.; Kutzner, C.; van der Spoel, D.; Lindahl, E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J. Chem. Theory Comput. 2008, 4, 435–447. [Google Scholar] [CrossRef] [Green Version]
- Weiner, S.J.; Kollman, P.A.; Case, D.A.; Singh, U.C.; Ghio, C.; Alagona, G.; Profeta, S.; Weiner, P. A new force field for molecular mechanical simulation of nucleic acids and proteins. J. Am. Chem. Soc. 1984, 106, 765–784. [Google Scholar]
- Cheatham, T.E.; Cieplak, P.; Kollman, P.A. A modified version of the Cornell et al. force field with improved sugar pucker phases and helical repeat. J. Biomol. Struct. Dyn. 1999, 16, 845–862. [Google Scholar] [CrossRef]
- Pérez, A.; Marchán, I.; Svozil, D.; Sponer, J.; Cheatham, T.E.; Laughton, C.A.; Orozco, M. Refinement of the AMBER force field for nucleic acids: Improving the description of alpha/gamma conformers. Biophys. J. 2007, 92, 3817–3829. [Google Scholar] [CrossRef] [Green Version]
- Zgarbová, M.; Luque, F.J.; Sponer, J.; Cheatham, T.E.; Otyepka, M.; Jurečka, P. Toward Improved Description of DNA Backbone: Revisiting Epsilon and Zeta Torsion Force Field Parameters. J. Chem. Theory Comput. 2013, 9, 2339–2354. [Google Scholar] [CrossRef]
- Krepl, M.; Zgarbová, M.; Stadlbauer, P.; Otyepka, M.; Banáš, P.; Koča, J.; Cheatham, T.E.; Jurečka, P.; Sponer, J. Reference simulations of noncanonical nucleic acids with different χ variants of the AMBER force field: Quadruplex DNA, quadruplex RNA and Z-DNA. J. Chem. Theory Comput. 2012, 8, 2506–2520. [Google Scholar] [CrossRef] [Green Version]
- Sousa da Silva, A.W.; Vranken, W.F. ACPYPE—AnteChamber PYthon Parser interfacE. BMC Res. Notes 2012, 5, 367. [Google Scholar] [CrossRef] [Green Version]
- Jorgensen, W.L.; Madura, J.D. Quantum and statistical mechanical studies of liquids. 25. Solvation and conformation of methanol in water. J. Am. Chem. Soc. 1983, 105, 1407–1413. [Google Scholar]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef] [Green Version]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar]
- Daura, X.; Gademann, K.; Jaun, B.; Seebach, D.; Van Gunsteren, W.F.; Mark, A.E. Peptide folding: When simulation meets experiment. Angew. Chem. Int. Ed. 1999, 38, 236–240. [Google Scholar]
- Schrödinger. Glide; Schrödinger, LLC.: New York, NY, USA, 2018. [Google Scholar]
- Schrödinger. Maestro; Schrödinger, LLC.: New York, NY, USA, 2019. [Google Scholar]
- Schrödinger. LigPrep; Schrödinger, LLC.: New York, NY, USA, 2018. [Google Scholar]
- Kollman, P.A.; Massova, I.; Reyes, C.; Kuhn, B.; Huo, S.; Chong, L.; Lee, M.; Lee, T.; Duan, Y.; Wang, W.; et al. Calculating Structures and Free Energies of Complex Molecules: Combining Molecular Mechanics and Continuum Models. Acc. Chem. Res. 2000, 33, 889–897. [Google Scholar] [CrossRef]
- Prime; Schrodinger, LLC.: New York, NY, USA, 2018.
- Bochevarov, A.D.; Harder, E.; Hughes, T.F.; Greenwood, J.R.; Braden, D.A.; Philipp, D.M.; Rinaldo, D.; Halls, M.D.; Zhang, J.; Friesner, R.A. Jaguar: A high-performance quantum chemistry software program with strengths in life and materials sciences. Int. J. Quantum Chem. 2013, 113, 2110–2142. [Google Scholar] [CrossRef]
- Cornell, W.D.; Cieplak, P.; Bayly, C.I.; Kollman, P.A. Application of RESP charges to calculate conformational energies, hydrogen bond energies, and free energies of solvation. J. Am. Chem. Soc. 2002, 115, 9620–9631. [Google Scholar] [CrossRef]
- Wang, J.; Wang, W.; Kollman, P.A.; Case, D.A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 2006, 25, 247–260. [Google Scholar] [CrossRef]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Greenfield, N.J. Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat. Protoc. 2006, 1, 2527–2535. [Google Scholar] [CrossRef]
- Fogh, J.; Trempe, G. New human tumor cell lines. In Human Tumor Cells in Vitro; Springer: Boston, MA, USA, 1975; pp. 115–159. [Google Scholar]





| MCF7 | HT-29 | A549 | |
|---|---|---|---|
| hit 7 | >50 | 1.9 ± 0.2 | 20.3 ± 0.4 |
| hit 15 | 62.0 ± 4.1 | 0.3 ± 0.1 | 1.1 ± 0.2 |
| hit 17 | 28.9 ± 2.0 | 1.0 ± 0.1 | 7.9 ± 0.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rocca, R.; Scionti, F.; Nadai, M.; Moraca, F.; Maruca, A.; Costa, G.; Catalano, R.; Juli, G.; Di Martino, M.T.; Ortuso, F.; et al. Chromene Derivatives as Selective TERRA G-Quadruplex RNA Binders with Antiproliferative Properties. Pharmaceuticals 2022, 15, 548. https://doi.org/10.3390/ph15050548
Rocca R, Scionti F, Nadai M, Moraca F, Maruca A, Costa G, Catalano R, Juli G, Di Martino MT, Ortuso F, et al. Chromene Derivatives as Selective TERRA G-Quadruplex RNA Binders with Antiproliferative Properties. Pharmaceuticals. 2022; 15(5):548. https://doi.org/10.3390/ph15050548
Chicago/Turabian StyleRocca, Roberta, Francesca Scionti, Matteo Nadai, Federica Moraca, Annalisa Maruca, Giosuè Costa, Raffaella Catalano, Giada Juli, Maria Teresa Di Martino, Francesco Ortuso, and et al. 2022. "Chromene Derivatives as Selective TERRA G-Quadruplex RNA Binders with Antiproliferative Properties" Pharmaceuticals 15, no. 5: 548. https://doi.org/10.3390/ph15050548