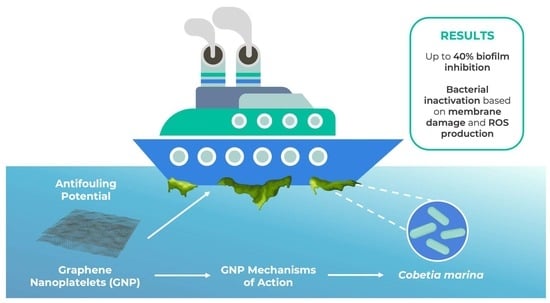
Graphene-Based Coating to Mitigate Biofilm Development in Marine Environments
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strain and Culture Conditions
2.2. Determination of GNP Minimal Inhibitory Concentration
2.3. Characterization of GNP Mechanisms of Action
2.3.1. Flow Cytometry Analysis
2.3.2. Epifluorescence Microscopy
2.3.3. Scanning Electron Microscopy (SEM)
2.4. Surface Preparation
2.5. Surface Characterization
2.5.1. Atomic Force Microscopy (AFM)
2.5.2. Scanning Electron Microscopy
2.5.3. Thermodynamic Analysis
2.6. Initial Bacterial Adhesion Assay
2.7. Biofilm Formation Assays
2.8. Biofilm Analysis
2.8.1. Biofilm Total Cell Count
2.8.2. Biofilm Thickness and Structure
2.9. Statistical Analysis
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Callow, M.E.; Callow, J.E. Marine Biofouling: A Sticky Problem. Biologist 2002, 49, 10–14. [Google Scholar] [PubMed]
- De Carvalho, C.C.C.R. Marine Biofilms: A Successful Microbial Strategy with Economic Implications. Front. Mar. Sci. 2018, 5, 126. [Google Scholar] [CrossRef] [Green Version]
- Schultz, M.P.; Bendick, J.A.; Holm, E.R.; Hertel, W.M. Economic Impact of Biofouling on a Naval Surface Ship. Biofouling 2011, 27, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Edyvean, R. Consequences of Fouling on Shipping. In Biofouling; Wiley-Blackwell: Oxford, UK, 2010; pp. 217–225. ISBN 9781444315462. [Google Scholar]
- Farkas, A.; Degiuli, N.; Martić, I.; Vujanović, M. Greenhouse Gas Emissions Reduction Potential by Using Antifouling Coatings in a Maritime Transport Industry. J. Clean Prod. 2021, 295, 126428. [Google Scholar] [CrossRef]
- Davidson, I.C.; Brown, C.W.; Sytsma, M.D.; Ruiz, G.M. The Role of Containerships as Transfer Mechanisms of Marine Biofouling Species. Biofouling 2009, 25, 645–655. [Google Scholar] [CrossRef]
- Schwindt, E.; López Gappa, J.; Raffo, M.P.; Tatián, M.; Bortolus, A.; Orensanz, J.M.; Alonso, G.; Diez, M.E.; Doti, B.; Genzano, G.; et al. Marine Fouling Invasions in Ports of Patagonia (Argentina) with Implications for Legislation and Monitoring Programs. Mar. Environ. Res. 2014, 99, 60–68. [Google Scholar] [CrossRef] [Green Version]
- Bannister, J.; Sievers, M.; Bush, F.; Bloecher, N. Biofouling in Marine Aquaculture: A Review of Recent Research and Developments. Biofouling 2019, 35, 631–648. [Google Scholar] [CrossRef] [Green Version]
- Lindholdt, A.; Dam-Johansen, K.; Olsen, S.M.; Yebra, D.M.; Kiil, S. Effects of Biofouling Development on Drag Forces of Hull Coatings for Ocean-Going Ships: A Review. J. Coat. Technol. Res. 2015, 12, 415–444. [Google Scholar] [CrossRef]
- Delauney, L.; Compare, C.; Lehaitre, M. Biofouling Protection for Marine Environmental Sensors. Ocean. Sci. 2010, 6, 503–511. [Google Scholar] [CrossRef] [Green Version]
- Deng, R.; Shen, T.; Chen, H.; Lu, J.; Yang, H.C.; Li, W. Slippery Liquid-Infused Porous Surfaces (SLIPSs): A Perfect Solution to Both Marine Fouling and Corrosion? J. Mater. Chem. A Mater. 2020, 8, 7536–7547. [Google Scholar] [CrossRef]
- Buskens, P.; Wouters, M.; Rentrop, C.; Vroon, Z. A Brief Review of Environmentally Benign Antifouling and Foul-Release Coatings for Marine Applications. J. Coat. Technol. Res. 2013, 10, 29–36. [Google Scholar] [CrossRef]
- Zhang, W.; Ding, W.; Li, Y.X.; Tam, C.; Bougouffa, S.; Wang, R.; Pei, B.; Chiang, H.; Leung, P.; Lu, Y.; et al. Marine Biofilms Constitute a Bank of Hidden Microbial Diversity and Functional Potential. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, P.-Y.; Lau, S.C.K.; Dahms, H.-U.; Dobretsov, S.; Harder, T. Marine Biofilms as Mediators of Colonization by Marine Macroorganisms: Implications for Antifouling and Aquaculture. Mar. Biotechnol. 2007, 9, 399–410. [Google Scholar] [CrossRef]
- Salta, M.; Wharton, J.A.; Blache, Y.; Stokes, K.R.; Briand, J.-F. Marine Biofilms on Artificial Surfaces: Structure and Dynamics. Environ. Microbiol. 2013, 15, 2879–2893. [Google Scholar] [CrossRef] [PubMed]
- Lima, M.; Gomes, L.C.; Teixeira-Santos, R.; Romeu, M.J.; Valcarcel, J.; Vázquez, J.A.; Cerqueira, M.A.; Pastrana, L.; Bourbon, A.I.; de Jong, E.D.; et al. Assessment of the Antibiofilm Performance of Chitosan-Based Surfaces in Marine Environments. Int. J. Mol. Sci. 2022, 23, 14647. [Google Scholar] [CrossRef] [PubMed]
- Ista, L.K.; Fan, H.; Baca, O.; López, G.P. Attachment of Bacteria to Model Solid Surfaces: Oligo(Ethylene Glycol) Surfaces Inhibit Bacterial Attachment. FEMS Microbiol. Lett. 1996, 142, 59–63. [Google Scholar] [CrossRef]
- Ista, L.K.; Callow, M.E.; Finlay, J.A.; Coleman, S.E.; Nolasco, A.C.; Simons, R.H.; Callow, J.A.; Lopez, G.P. Effect of Substratum Surface Chemistry and Surface Energy on Attachment of Marine Bacteria and Algal Spores. Appl. Environ. Microbiol. 2004, 70, 4151–4157. [Google Scholar] [CrossRef] [Green Version]
- Schwarze, J.; Wanka, R.; Rosenhahn, A. Microfluidic Accumulation Assay to Quantify the Attachment of the Marine Bacterium Cobetia Marina on Fouling-Release Coatings. Biointerphases 2020, 15, 31014. [Google Scholar] [CrossRef]
- Akuzov, D.; Franca, L.; Grunwald, I.; Vladkova, T. Sharply Reduced Biofilm Formation from Cobetia marina and in Black Sea Water on Modified Siloxane Coatings. Coatings 2018, 8, 136. [Google Scholar] [CrossRef] [Green Version]
- Abouelkheir, S.S.; Abdelghany, E.A.; Sabry, S.A.; Ghozlan, H.A. Biofilm Formation by Marine Cobetia Marina Alex and Pseudoalteromonas Spp: Development and Detection of Quorum Sensing N-Acyl Homoserine Lactones (AHLs) Molecules. J. Mar. Sci. 2021, 3. [Google Scholar] [CrossRef]
- Shea, C.; Lovelace, L.J.; Smith-Somerville, H.E. Deleya Marina as a Model Organism for Studies of Bacterial Colonization and Biofilm Formation. J. Ind. Microbiol 1995, 15, 290–296. [Google Scholar] [CrossRef]
- Rossi, F.; de Philippis, R. Role of Cyanobacterial Exopolysaccharides in Phototrophic Biofilms and in Complex Microbial Mats. Life 2015, 5, 1218–1238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, R.; Jing, Y.; Ni, Y.; Jiang, Q. Effects of Chitin Nanocrystals on Coverage of Coating Layers and Water Retention of Coating Color. J. Bioresour. Bioprod. 2022, 7, 201–210. [Google Scholar] [CrossRef]
- Oyeoka, H.C.; Ewulonu, C.M.; Nwuzor, I.C.; Obele, C.M.; Nwabanne, J.T. Packaging and Degradability Properties of Polyvinyl Alcohol/Gelatin Nanocomposite Films Filled Water Hyacinth Cellulose Nanocrystals. J. Bioresour. Bioprod. 2021, 6, 168–185. [Google Scholar] [CrossRef]
- Parra, C.; Dorta, F.; Jimenez, E.; Henríquez, R.; Ramírez, C.; Rojas, R.; Villalobos, P. A Nanomolecular Approach to Decrease Adhesion of Biofouling-Producing Bacteria to Graphene-Coated Material. J. Nanobiotechnol. 2015, 13, 82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Ye, F.; Dobretsov, S.; Dutta, J. Nanocoating Is a New Way for Biofouling Prevention. Front. Nanotechnol. 2021, 3, 771098. [Google Scholar] [CrossRef]
- Scardino, A.J.; Guenther, J.; de Nys, R. Attachment Point Theory Revisited: The Fouling Response to a Microtextured Matrix. Biofouling 2008, 24, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Callow, J.A.; Callow, M.E. Trends in the Development of Environmentally Friendly Fouling-Resistant Marine Coatings. Nat. Commun. 2011, 2, 244. [Google Scholar] [CrossRef] [Green Version]
- Sousa-Cardoso, F.; Teixeira-Santos, R.; Mergulhão, F.J.M. Antifouling Performance of Carbon-Based Coatings for Marine Applications: A Systematic Review. Antibiotics 2022, 11, 1102. [Google Scholar] [CrossRef]
- Romeu, M.J.; Lima, M.; Gomes, L.C.; de Jong, E.D.; Morais, J.; Vasconcelos, V.; Pereira, M.F.R.; Soares, O.S.G.P.; Sjollema, J.; Mergulhão, F.J. The Use of 3D Optical Coherence Tomography to Analyze the Architecture of Cyanobacterial Biofilms Formed on a Carbon Nanotube Composite. Polymers 2022, 14, 4410. [Google Scholar] [CrossRef]
- Romeu, M.J.; Gomes, L.C.; Sousa-Cardoso, F.; Morais, J.; Vasconcelos, V.; Whitehead, K.A.; Pereira, M.F.R.; Soares, O.S.G.P.; Mergulhão, F.J. How Do Graphene Composite Surfaces Affect the Development and Structure of Marine Cyanobacterial Biofilms? Coatings 2022, 12, 1775. [Google Scholar] [CrossRef]
- Sattar, T. Current Review on Synthesis, Composites and Multifunctional Properties of Graphene. Top Curr. Chem. 2019, 377, 10. [Google Scholar] [CrossRef] [PubMed]
- Idumah, C.I.; Hassan, A. Emerging Trends in Graphene Carbon Based Polymer Nanocomposites and Applications. Rev. Chem. Eng. 2016, 32, 223–264. [Google Scholar] [CrossRef]
- Kumar, A.; Sharma, K.; Dixit, A.R. A Review of the Mechanical and Thermal Properties of Graphene and Its Hybrid Polymer Nanocomposites for Structural Applications. J. Mater. Sci. 2019, 54, 5992–6026. [Google Scholar] [CrossRef]
- Teixeira-Santos, R.; Gomes, M.; Gomes, L.C.; Mergulhão, F.J. Antimicrobial and Anti-Adhesive Properties of Carbon Nanotube-Based Surfaces for Medical Applications: A Systematic Review. iScience 2021, 24, 102001. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Huo, P.; Zhang, R.; Liu, B. Antibacterial Properties of Graphene-Based Nanomaterials. Nanomaterials 2019, 9, 737. [Google Scholar] [CrossRef] [Green Version]
- Radhi, A.; Mohamad, D.; Abdul Rahman, F.S.; Abdullah, A.M.; Hasan, H. Mechanism and Factors Influence of Graphene-Based Nanomaterials Antimicrobial Activities and Application in Dentistry. J. Mater. Res. Technol. 2021, 11, 1290–1307. [Google Scholar] [CrossRef]
- Jin, H.; Tian, L.; Bing, W.; Zhao, J.; Ren, L. Toward the Application of Graphene for Combating Marine Biofouling. Adv. Sustain. Syst. 2021, 5, 2000076. [Google Scholar] [CrossRef]
- Arahal, D.R.; Castillo, A.M.; Ludwig, W.; Schleifer, K.H.; Ventosa, A. Proposal of Cobetia marina Gen. Nov., Comb. Nov., within the Family Halomonadaceae, to Include the Species Halomonas Marina. Syst. Appl. Microbiol. 2002, 25, 207–211. [Google Scholar] [CrossRef]
- Holmström, C.; James, S.; Neilan, B.A.; White, D.C.; Kjelleberg, S. Pseudoalteromonas tunicata Sp. Nov., a Bacterium That Produces Antifouling Agents. Int. J. Syst. Bacteriol. 1998, 48, 1205–1212. [Google Scholar] [CrossRef]
- D’Souza, F.; Bruin, A.; Biersteker, R.; Donnelly, G.; Klijnstra, J.; Rentrop, C.; Willemsen, P. Bacterial Assay for the Rapid Assessment of Antifouling and Fouling Release Properties of Coatings and Materials. J. Ind. Microbiol. Biotechnol. 2010, 37, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Mortimer, M.; Petersen, E.; Buchholz, B.; Holden, P. Separation of Bacteria, Protozoa and Carbon Nanotubes by Density Gradient Centrifugation. Nanomaterials 2016, 6, 181. [Google Scholar] [CrossRef] [PubMed]
- Kramer, B.; Muranyi, P. Effect of Pulsed Light on Structural and Physiological Properties of Listeria innocua and Escherichia coli. J. Appl. Microbiol. 2014, 116, 596–611. [Google Scholar] [CrossRef]
- Li, J.; Ma, L.; Liao, X.; Liu, D.; Lu, X.; Chen, S.; Ye, X.; Ding, T. Ultrasound-Induced Escherichia coli O157:H7 Cell Death Exhibits Physical Disruption and Biochemical Apoptosis. Front. Microbiol. 2018, 9, 2486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenkranz, A.R.; Schmaldienst, S.; Stuhlmeier, K.M.; Chen, W.; Knapp, W.; Zlabinger, G.J. A Microplate Assay for the Detection of Oxidative Products Using 2′,7′-Dichlorofluorescin-Diacetate. J. Immunol. Methods 1992, 156, 39–45. [Google Scholar] [CrossRef]
- Gomes, L.C.; Silva, L.N.; Simões, M.; Melo, L.F.; Mergulhão, F.J. Escherichia coli Adhesion, Biofilm Development and Antibiotic Susceptibility on Biomedical Materials. J. Biomed. Mater. Res. A 2015, 103, 1414–1423. [Google Scholar] [CrossRef] [Green Version]
- Vorobii, M.; Teixeira-Santos, R.; Gomes, L.C.; Garay-Sarmiento, M.; Wagner, A.M.; Mergulhão, F.J.; Rodriguez-Emmenegger, C. Oriented Immobilization of Pep19-2.5 on Antifouling Brushes Suppresses the Development of Staphylococcus aureus Biofilms. Prog. Org. Coat. 2022, 163, 106609. [Google Scholar] [CrossRef]
- Faria, S.I.; Teixeira-Santos, R.; Romeu, M.J.; Morais, J.; Vasconcelos, V.; Mergulhão, F.J. The Relative Importance of Shear Forces and Surface Hydrophobicity on Biofilm Formation by Coccoid Cyanobacteria. Polymers 2020, 12, 653. [Google Scholar] [CrossRef] [Green Version]
- Vagos, M.R.; Moreira, J.M.R.; Soares, O.S.G.P.; Pereira, M.F.R.; Mergulhão, F.J. Incorporation of Carbon Nanotubes in Polydimethylsiloxane to Control Escherichia coli Adhesion. Polym. Compos. 2019, 40, E1697–E1704. [Google Scholar] [CrossRef]
- Oliveira, I.M.; Gomes, M.; Gomes, L.C.; Pereira, M.F.R.; Soares, O.S.G.P.; Mergulhão, F.J. Performance of Graphene/Polydimethylsiloxane Surfaces against S. aureus and P. aeruginosa Single- and Dual-Species Biofilms. Nanomaterials 2022, 12, 355. [Google Scholar] [CrossRef]
- Gomes, M.; Gomes, L.C.; Teixeira-Santos, R.; Pereira, M.F.R.; Soares, O.S.G.P.; Mergulhão, F.J. Optimizing CNT Loading in Antimicrobial Composites for Urinary Tract Application. Appl. Sci. 2021, 11, 4038. [Google Scholar] [CrossRef]
- Alves, P.; Gomes, L.; Vorobii, M.; Rodriguez-Emmenegger, C.; Mergulhão, F. The Potential Advantages of Using a Poly(HPMA) Brush in Urinary Catheters: Effects on Biofilm Cells and Architecture. Colloids Surf. B Biointerfaces 2020, 191, 110976. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Cao, X.; Feng, X.; Ma, Y.; Zou, H. Fabrication of Super-Hydrophobic Film from PMMA with Intrinsic Water Contact Angle below 90°. Polymer 2007, 48, 7455–7460. [Google Scholar] [CrossRef]
- van Oss, C.J. Interfacial Forces in Aqueous Media; CRC Press: Boca Raton, FL, USA, 2006; ISBN 9780429134418. [Google Scholar]
- Busscher, H.J.; Weerkamp, A.H.; van Der Mei, H.C.; van Pelt, A.W.; de Jong, H.P.; Arends, J. Measurement of the Surface Free Energy of Bacterial Cell Surfaces and Its Relevance for Adhesion. Appl. Environ. Microbiol. 1984, 48, 980–983. [Google Scholar] [CrossRef] [Green Version]
- Bayoudh, S.; Othmane, A.; Bettaieb, F.; Bakhrouf, A.; Ouada, H.B.; Ponsonnet, L. Quantification of the Adhesion Free Energy between Bacteria and Hydrophobic and Hydrophilic Substrata. Mater. Sci. Eng. C 2006, 26, 300–305. [Google Scholar] [CrossRef]
- Faria, S.I.; Teixeira-Santos, R.; Romeu, M.J.; Morais, J.; de Jong, E.; Sjollema, J.; Vasconcelos, V.; Mergulhão, F.J. Unveiling the Antifouling Performance of Different Marine Surfaces and Their Effect on the Development and Structure of Cyanobacterial Biofilms. Microorganisms 2021, 9, 1102. [Google Scholar] [CrossRef] [PubMed]
- Romeu, M.J.; Alves, P.; Morais, J.; Miranda, J.M.; Jong, E.D.; Sjollema, J.; Ramos, V.; Vasconcelos, V.; Mergulhão, F.J.M. Biofilm Formation Behaviour of Marine Filamentous Cyanobacterial Strains in Controlled Hydrodynamic Conditions. Environ. Microbiol. 2019, 21, 4411–4424. [Google Scholar] [CrossRef] [PubMed]
- Bakker, D.P.; Van der Plaats, A.; Verkerke, G.J.; Busscher, H.J.; Van der Mei, H.C. Comparison of Velocity Profiles for Different Flow Chamber Designs Used in Studies of Microbial Adhesion to Surfaces. Appl. Environ. Microbiol. 2003, 69, 6280–6287. [Google Scholar] [CrossRef] [Green Version]
- Amara, I.; Miled, W.; Slama, R.B.; Ladhari, N. Antifouling Processes and Toxicity Effects of Antifouling Paints on Marine Environment. A Review. Environ. Toxicol. Pharmacol. 2018, 57, 115–130. [Google Scholar] [CrossRef]
- Faria, S.I.; Teixeira-Santos, R.; Morais, J.; Vasconcelos, V.; Mergulhão, F.J. The Association between Initial Adhesion and Cyanobacterial Biofilm Development. FEMS Microbiol. Ecol. 2021, 97, 52. [Google Scholar] [CrossRef]
- Romeu, M.J.; Domínguez-Pérez, D.; Almeida, D.; Morais, J.; Araújo, M.J.; Osório, H.; Campos, A.; Vasconcelos, V.; Mergulhão, F.J. Quantitative Proteomic Analysis of Marine Biofilms Formed by Filamentous Cyanobacterium. Environ. Res. 2021, 201, 111566. [Google Scholar] [CrossRef] [PubMed]
- Faria, S.I.; Gomes, L.C.; Teixeira-Santos, R.; Morais, J.; Vasconcelos, V.; Mergulhão, F.J.M. Developing New Marine Antifouling Surfaces: Learning from Single-Strain Laboratory Tests. Coatings 2021, 11, 90. [Google Scholar] [CrossRef]
- Dobretsov, S. Marine Biofilms. In Biofouling; Wiley-Blackwell: Oxford, UK, 2010; pp. 123–136. ISBN 9781444315462. [Google Scholar]
- Otsu, N. Threshold Selection Method From Gray-Level Histograms. IEEE Trans. Syst. Man Cybern. 1979, SMC-9, 62–66. [Google Scholar] [CrossRef] [Green Version]
- Mohammed, H.; Kumar, A.; Bekyarova, E.; Al-Hadeethi, Y.; Zhang, X.; Chen, M.; Ansari, M.S.; Cochis, A.; Rimondini, L. Antimicrobial Mechanisms and Effectiveness of Graphene and Graphene-Functionalized Biomaterials. A Scope Review. Front. Bioeng. Biotechnol. 2020, 8, 465. [Google Scholar] [CrossRef] [PubMed]
- Dawan, J.; Ahn, J. Bacterial Stress Responses as Potential Targets in Overcoming Antibiotic Resistance. Microorganisms 2022, 10, 1385. [Google Scholar] [CrossRef]
- McBee, M.E.; Chionh, Y.H.; Sharaf, M.L.; Ho, P.; Cai, M.W.L.; Dedon, P.C. Production of Superoxide in Bacteria Is Stress- and Cell State-Dependent: A Gating-Optimized Flow Cytometry Method That Minimizes ROS Measurement Artifacts with Fluorescent Dyes. Front. Microbiol. 2017, 8, 459. [Google Scholar] [CrossRef] [Green Version]
- Zecher, K.; Aitha, V.P.; Heuer, K.; Ahlers, H.; Roland, K.; Fiedel, M.; Philipp, B. A Multi-Step Approach for Testing Non-Toxic Amphiphilic Antifouling Coatings against Marine Microfouling at Different Levels of Biological Complexity. J. Microbiol. Methods 2018, 146, 104–114. [Google Scholar] [CrossRef]
- Chien, H.W.; Chen, X.Y.; Tsai, W.P.; Lee, M. Inhibition of Biofilm Formation by Rough Shark Skin-Patterned Surfaces. Colloids Surf. B Biointerfaces 2020, 186, 110738. [Google Scholar] [CrossRef]
- Krsmanovic, M.; Biswas, D.; Ali, H.; Kumar, A.; Ghosh, R.; Dickerson, A.K. Hydrodynamics and Surface Properties Influence Biofilm Proliferation. Adv. Colloid Interface Sci. 2021, 288, 102336. [Google Scholar] [CrossRef]
- Zhou, W.; Apkarian, R.; Wang, Z.L.; Joy, D. Fundamentals of Scanning Electron Microscopy (SEM). In Scanning Microscopy for Nanotechnology; Springer: New York, NY, USA, 2006; pp. 1–40. ISBN 0387333258. [Google Scholar]
- Chatterjee, S.; Nüesch, F.A.; Chu, B.T.T. Comparing Carbon Nanotubes and Graphene Nanoplatelets as Reinforcements in Polyamide 12 Composites. Nanotechnology 2011, 22, 275714. [Google Scholar] [CrossRef]
- Staneva, A.D.; Dimitrov, D.K.; Gospodinova, D.N.; Vladkova, T.G. Antibiofouling Activity of Graphene Materials and Graphene-Based Antimicrobial Coatings. Microorganisms 2021, 9, 1839. [Google Scholar] [CrossRef] [PubMed]
- Lukowiak, A.; Kedziora, A.; Strek, W. Antimicrobial Graphene Family Materials: Progress, Advances, Hopes and Fears. Adv. Colloid Interface Sci. 2016, 236, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Quan, K.; Hou, J.; Zhang, Z.; Ren, Y.; Peterson, B.W.; Flemming, H.-C.; Mayer, C.; Busscher, H.J.; van der Mei, H.C. Water in Bacterial Biofilms: Pores and Channels, Storage and Transport Functions. Crit. Rev. Microbiol. 2022, 48, 283–302. [Google Scholar] [CrossRef] [PubMed]
- Chang, I.; Gilbert, E.S.; Eliashberg, N.; Keasling, J.D. A Three-Dimensional, Stochastic Simulation of Biofilm Growth and Transport-Related Factors That Affect Structure. Microbiology 2003, 149, 2859–2871. [Google Scholar] [CrossRef]



 ) and G5/PDMS (
) and G5/PDMS ( ) throughout 42 days. The analyzed parameters refer to the number of biofilm cells (a) and biofilm thickness (b). Results are presented as mean ± SD. ns—p > 0.05; *—p ≤ 0.05; **—p ≤ 0.01; ***—p ≤ 0.001.
) throughout 42 days. The analyzed parameters refer to the number of biofilm cells (a) and biofilm thickness (b). Results are presented as mean ± SD. ns—p > 0.05; *—p ≤ 0.05; **—p ≤ 0.01; ***—p ≤ 0.001.
 ) and G5/PDMS (
) and G5/PDMS ( ) throughout 42 days. The analyzed parameters refer to the number of biofilm cells (a) and biofilm thickness (b). Results are presented as mean ± SD. ns—p > 0.05; *—p ≤ 0.05; **—p ≤ 0.01; ***—p ≤ 0.001.
) throughout 42 days. The analyzed parameters refer to the number of biofilm cells (a) and biofilm thickness (b). Results are presented as mean ± SD. ns—p > 0.05; *—p ≤ 0.05; **—p ≤ 0.01; ***—p ≤ 0.001.

 ) and G5/PDMS (
) and G5/PDMS ( ), determined at incubation day 42. **—p ≤ 0.01. (b) Porosity of biofilms formed on PDMS (
), determined at incubation day 42. **—p ≤ 0.01. (b) Porosity of biofilms formed on PDMS ( ) and G5/PDMS (
) and G5/PDMS ( ) determined on incubation day 42. ***—p ≤ 0.001. (c,d) Two-dimensional cross-sectional OCT images of C. marina DSM 4741 biofilms formed on PDMS (c) and G5/PDMS (d) acquired on incubation day 42. Empty spaces highlighted in light blue. White scale bars = 100 μm.
) determined on incubation day 42. ***—p ≤ 0.001. (c,d) Two-dimensional cross-sectional OCT images of C. marina DSM 4741 biofilms formed on PDMS (c) and G5/PDMS (d) acquired on incubation day 42. Empty spaces highlighted in light blue. White scale bars = 100 μm.
 ) and G5/PDMS (
) and G5/PDMS ( ), determined at incubation day 42. **—p ≤ 0.01. (b) Porosity of biofilms formed on PDMS (
), determined at incubation day 42. **—p ≤ 0.01. (b) Porosity of biofilms formed on PDMS ( ) and G5/PDMS (
) and G5/PDMS ( ) determined on incubation day 42. ***—p ≤ 0.001. (c,d) Two-dimensional cross-sectional OCT images of C. marina DSM 4741 biofilms formed on PDMS (c) and G5/PDMS (d) acquired on incubation day 42. Empty spaces highlighted in light blue. White scale bars = 100 μm.
) determined on incubation day 42. ***—p ≤ 0.001. (c,d) Two-dimensional cross-sectional OCT images of C. marina DSM 4741 biofilms formed on PDMS (c) and G5/PDMS (d) acquired on incubation day 42. Empty spaces highlighted in light blue. White scale bars = 100 μm.

| Sample | Contact Angle (°) | Hydrophobicity (mJ·m−2) | C. marina–Surface Interaction (mJ·m−2) | ||
|---|---|---|---|---|---|
| θW | θF | θB | ΔG | ΔGAdh | |
| Surface | |||||
| PDMS | 114.3 ± 1.4 | 111.8 ± 1.6 | 88.3 ± 3.8 | −63.1 | −5.5 |
| G5/PDMS | 115.8 ± 2.1 | 112.2 ± 1.7 | 89.8 ± 3.7 | −67.5 | −6.7 |
| Bacteria | |||||
| C. marina DSM 4741 | 37.4 ± 1.7 | 31.5 ± 2.3 | 55.0 ± 1.9 | 14.9 | n.a. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sousa-Cardoso, F.; Teixeira-Santos, R.; Campos, A.F.; Lima, M.; Gomes, L.C.; Soares, O.S.G.P.; Mergulhão, F.J. Graphene-Based Coating to Mitigate Biofilm Development in Marine Environments. Nanomaterials 2023, 13, 381. https://doi.org/10.3390/nano13030381
Sousa-Cardoso F, Teixeira-Santos R, Campos AF, Lima M, Gomes LC, Soares OSGP, Mergulhão FJ. Graphene-Based Coating to Mitigate Biofilm Development in Marine Environments. Nanomaterials. 2023; 13(3):381. https://doi.org/10.3390/nano13030381
Chicago/Turabian StyleSousa-Cardoso, Francisca, Rita Teixeira-Santos, Ana Francisca Campos, Marta Lima, Luciana C. Gomes, Olívia S. G. P. Soares, and Filipe J. Mergulhão. 2023. "Graphene-Based Coating to Mitigate Biofilm Development in Marine Environments" Nanomaterials 13, no. 3: 381. https://doi.org/10.3390/nano13030381