The Rapid Non-Destructive Differentiation of Different Varieties of Rice by Fluorescence Hyperspectral Technology Combined with Machine Learning
Abstract
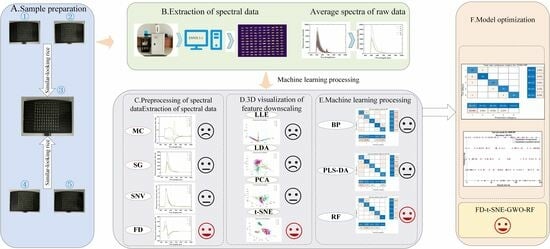
:1. Introduction
2. Results and Discussion
2.1. Characterization of Fluorescence Hyperspectral Imaging
2.2. Fluorescence Hyperspectral Data Preprocessing
2.3. Feature Downscaling and Selection
2.4. Modeling Analysis after Preprocessing and Feature Dimensionality Reduction Processing
- (a)
- In this study, a classification model was used to test the accuracy of the best rice model. After preprocessing and feature dimensionality reduction, the classification model accuracy was higher than that modeled using raw fluorescence hyperspectral data. This is because, by preprocessing, the spectral noise can be removed from the spectral curve as much as possible, highlighting the valuable information of the spectrum [31]. Then, after processing by feature dimensionality reduction, the spectral data dimensions are reduced to reduce further the influence of spectral noise, which reduces the amount of data and the influence of useless data [32]. After preprocessing and feature dimensionality reduction processing, the modeling accuracy is somewhat improved, and the robustness is enhanced.
- (b)
- Among all the feature dimensionality reduction processing methods, the accuracy of t-SNE in the RF modeling was dramatically improved. Theoretically, PCA is a matrix decomposition technique involving multiple conditional probabilities and gradient descent calculations [33], while t-SNE is a probabilistic method [34]. In other classification models, t-SNE also has a higher accuracy than PCA.
- (c)
- The accuracy of the experimental results showed significant differences in the effects of the different models. The accuracy of RF was improved on t-SNE and PCA and was not affected by its underlying evaluator. When the data were reduced to three dimensions by FD-t-SNE-RF, the model was more accurate than the other preprocessing methods. RF had a higher classification accuracy than BP and PLS-DA. RF had a significant advantage in the classification of rice in this study. After using RF, the accuracy of different preprocessing and feature processing methods was higher than the other two classification models. Especially in FD-t-SNE-RF, the accuracies of the training and test sets were 99.7% and 93.3%, respectively.
2.5. RF Classification Model Optimization
3. Materials and Methods
3.1. Sample
3.2. Fluorescence Hyperspectral Image Acquisition
3.3. Fluorescence Hyperspectral Data Extraction
3.4. Fluorescence Hyperspectral Data Preprocessing
- (1)
- For the rice sample set’s spectral matrix (), the average fluorescence hyperspectrum of the rice sample was calculated.
- (2)
- We used spectrum for the rice samples to obtain fluorescence hyperspectral data for the MC-treated rice.
3.5. Feature Downscaling and Feature Selection
- (1)
- Normalize the rice fluorescence hyperspectral dataset.
- (2)
- Calculate the covariance matrix of the standardized fluorescence hyperspectral data.
- (3)
- Based on the covariance matrix, the principal component eigenvalues, principal component contribution rates, and cumulative contribution rates of the rice samples were calculated, and the principal component loadings were calculated.
- (1)
- Input a fluorescence hyperspectral dataset of rice samples, where an arbitrary sample is an -dimensional vector downscaled to dimension .
- (2)
- Calculate the intra-class scatter matrix of the dataset .
- (3)
- Compute the interclass scatter matrix for dataset .
- (4)
- Compute the new matrix: multiply the inverse of (2) by (3).
- (5)
- Calculate the eigenvalues and eigenvectors of the matrix obtained from (4) and select the first eigenvalues and the corresponding eigenvectors in the order from smallest to largest to obtain the projection matrix .
- (6)
- New sample .
- (7)
- Output the fluorescence hyperspectral dataset of the rice samples .
- (1)
- Assume first that the expression for the neighborhood linear relationship of the high-dimensional fluorescence hyperspectral data is expressed by.where , , and are the weighting coefficients. Let the weighting coefficient be , which can be obtained by the following equation.where denotes the high-dimensional fluorescence hyperspectral data set of the neighborhood data points and denotes the number of rice fluorescence hyperspectral samples.
- (2)
- Keeping Equation (7) unchanged, the low-dimensional space data point A can be obtained by Equation (8).
- (1)
- Input the rice sample fluorescence hyperspectral dataset.
- (2)
- The conditional probability distributions and between the two data points and are calculated using Equation (9), and the location information of the rice samples is represented by a Gaussian probability distribution.In Equations (10) and (11), is the variance of the Gaussian distribution corresponding to the rice sample data point .
- (3)
- Setting up the joint probability substep , obtain a low-dimensional sample random initial solution .
- (4)
- Calculate the joint distribution of the low-dimensional sample space points in the -distribution.
- (5)
- Calculate the optimized gradient .
- (6)
- Loop iterations of (14) and (15) until the fluorescence hyperspectral low-dimensional data of the rice samples are obtained.
3.6. Machine Learning Classification Models
3.6.1. Partial Least Squares Discriminant Analysis (PLS-DA)
3.6.2. BP Neural Network (BP)
- (1)
- The five rice samples were divided into a training set and a test set according to the randomized division method; the training set was used for training the BP models, and the test set was used to test the trained BP models.
- (2)
- The data in the training set were normalized so as to reduce the adverse effects caused by singular sample data in the original rice sample data.
- (3)
- The purelin transfer function was selected, and the gradient descent method was used for training; the network parameters, such as hidden layers, number of training times, learning rate, learning accuracy, and weight threshold, were configured.
- (4)
- The BP model was trained with the normalized training rice sample set until the BP model was output when the set parameters were met (the number of training times was reached or the learning accuracy was satisfied).
- (5)
- The trained BP model was tested with the test set after the normalization process. If the predicted values of rice were within the allowed error range (0.001) from the ideal output, it meant that this rice classification BP model was valid; then, proceed to step (6), or otherwise return to step (3) for parameter modification and continue training.
- (6)
- The test set was classified with the trained rice classification BP model and the results were output.
3.6.3. Random Forest (RF)
- (1)
- Absolute majority voting method that restricts the number of labeled votes to be greater than half of the votes.
- (2)
- Relative majority voting method that decides the value simply based on the number of votes received.
- (3)
- The weighted voting method is similar to the weighted average method. A suitable voting mechanism can lead to more accurate prediction results.
- (1)
- A subset of samples was extracted from the raw rice fluorescence hyperspectral data.
- (2)
- A decision tree was generated using each sample subset, and at each node of the tree, variables were randomly selected to be split. The tree was grown continuously so that the number of nodes at each terminal node was not lower than the size of the node.
- (3)
- A classification was developed using a voting mechanism to count the results of the decision tree.
3.7. Model Optimization Algorithm
- (1)
- Input the rice sample data and normalize the data.
- (2)
- Set the optimization range of rice classification RF model parameters and initialize the wolf pack and GWO parameters.
- (3)
- Calculate the fitness value of the gray wolves and classify the wolves into four tiers, , , , and , by taking the root-mean-square error of the classification result as the fitness value.
- (4)
- Update the positions of the wolves, recalculate the fitness values at the new positions, and re-elect the new , , and .
- (5)
- When the number of iterations reaches the set maximum number of iterations, it indicates the end of training, and the optimal (the maximum growth in depth of the tree) and (the number of over-parameter subtrees) are output; otherwise, continue the parameter optimization.
- (6)
- Use the optimal and to establish a rice classification model, test the test set, and output the results of the inverse normalization process.
3.8. Confusion Matrix
3.9. Evaluation Indicators
4. Conclusions
- (1)
- FD preprocessing can effectively reduce the effects of sample background and baseline drift, improve the resolution and sensitivity of overlapping peaks, and improve the accuracy of the rice classification model.
- (2)
- Feature dimensionality reduction and feature selection can reduce data redundancy and improve the prediction accuracy and robustness of the model. In comparing the four feature dimensionality reduction methods, selecting t-SNE had the best effect. t-SNE can retain most of the information in the rice spectral curve, effectively reduce the dimensionality of the spectral data of rice samples, and improve the robustness and generalization of the rice classification model.
- (3)
- Among the three classification models, RF provided the best results. Combining RF with preprocessing and feature dimensionality reduction processing provided the best performance. Among them, the FD-t-SNE-RF method achieved 99.7% and 93.3% accuracy values for the training and test sets, respectively, and the classification model accuracy was higher than that of other spectroscopic and chemical methods. The RF classification model effectively identifies rice varieties accurately and has a superior reliability compared to other classification models.
- (4)
- After the classification modeling of the data after preprocessing and feature dimensionality reduction, the parameters of the RF classification model were optimization-seeking after the introduction of the GWO optimization algorithm, and the optimal model for rice classification and identification was finally determined. Comparing the modeling results, it can be seen that the GWO optimization algorithm played a positive role in the classification of the RF model and improved the classification accuracy. The optimal classification model was FD-t-SNE-GWO-RF, and the training set accuracy and test set accuracy were 99.8% and 95.3%, respectively. This study demonstrates the superiority of the GWO optimization algorithm in rice classification model identification.
- (5)
- The confusion matrix of the classification model shows that rice varieties with similar fluorescence hyperspectral information have specific errors in identification. Further machine-learning data processing is needed for rice variety identification analysis.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- D’Amour, C.B.; Anderson, W. International trade and the stability of food supplies in the Global South. Environ. Res. Lett. 2020, 15, 074005. [Google Scholar] [CrossRef]
- Watson, C.; Gustave, W. Prevalence of arsenic contamination in rice and the potential health risks to the Baham-ian population-A preliminary study. Front. Environ. Sci. 2022, 10, 1011785. [Google Scholar] [CrossRef]
- Das, G.; Patra, J.K.; Choi, J.; Baek, K.H. RICE GRAIN, A RICH SOURCE OF NATURAL BIOACTIVE COMPOUNDS. Pak. J. Agric. Sci. 2017, 54, 671–682. [Google Scholar] [CrossRef]
- Zafar, S.; Xu, J.L. Recent Advances to Enhance Nutritional Quality of Rice. Rice Sci. 2023, 30, 523–536. [Google Scholar] [CrossRef]
- Yu, J.; Chen, H.; Zhang, X.; Cui, X.; Zhao, Z. A Rice Hazards Risk Assessment Method for a Rice Processing Chain Based on a Multidimensional Trapezoidal Cloud Model. Foods 2023, 12, 1203. [Google Scholar] [CrossRef] [PubMed]
- Pezzotti, G.; Zhu, W.; Hashimoto, Y.; Marin, E.; Masumura, T.; Sato, Y.-I.; Nakazaki, T. Raman Fingerprints of Rice Nutritional Quality: A Comparison between Japanese Koshihikari and Internationally Renowned Cultivars. Foods 2021, 10, 2936. [Google Scholar] [CrossRef]
- Sokudlor, N.; Laloon, K.; Junsiri, C.; Sudajan, S. Enhancing milled rice qualitative classification with machine learning techniques using morphological features of binary images. Int. J. Food Prop. 2023, 26, 2978–2992. [Google Scholar] [CrossRef]
- Liu, W.; Xu, X.; Liu, C.; Zheng, L. Nondestructive Detection of Authenticity of Thai Jasmine Rice Using Multispectral Imaging. J. Food Qual. 2021, 2021, 6642220. [Google Scholar] [CrossRef]
- Li, C.; Li, B.; Ye, D. Analysis and Identification of Rice Adulteration Using Terahertz Spectroscopy and Pattern Recognition Algorithms. IEEE Access 2020, 8, 26839–26850. [Google Scholar] [CrossRef]
- Sliwinska-Bartel, M.; Burns, D.T.; Elliott, C. Rice fraud a global problem: A review of analytical tools to detect species, country of origin and adulterations. Trends Food Sci. Technol. 2021, 116, 36–46. [Google Scholar] [CrossRef]
- Zhang, Y.; Jiang, L.; Chen, Y.; Zhang, Q.; Kang, C.; Chen, D. Detection of chlorpyrifos residue in apple and rice samples based on aptamer sensor: Improving quantitative accuracy with partial least squares model. Microchem. J. 2023, 194, 109352. [Google Scholar] [CrossRef]
- Wadood, S.A.; Nie, J.; Li, C.L.; Rogers, K.M.; Khan, A.; Khan, W.A.; Qamar, A.; Zhang, Y.Z.; Yuwei, Y. Rice authentication: An overview of different analytical techniques combined with multivariate analysis. J. Food Compos. Anal. 2022, 112, 104677. [Google Scholar] [CrossRef]
- Park, M.S.; Kim, H.N.; Bahk, G.J. The analysis of food safety incidents in South Korea, 1998–2016. Food Control 2017, 81, 196–199. [Google Scholar] [CrossRef]
- King, T.; Cole, M.; Farber, J.M.; Eisenbrand, G.; Zabaras, D.; Fox, E.M.; Hill, J.P. Food safety for food security: Relationship between global megatrends and developments in food safety. Trends Food Sci. Technol. 2017, 68, 160–175. [Google Scholar] [CrossRef]
- Uawisetwathana, U.; Karoonuthaisiri, N. Metabolomics for rice quality and traceability: Feasibility and future aspects. Curr. Opin. Food Sci. 2019, 28, 58–66. [Google Scholar] [CrossRef]
- Daygon, V.D.; Prakash, S.; Calingacion, M.; Riedel, A.; Ovenden, B.; Snell, P.; Mitchell, J.; Fitzgerald, M. Understanding the Jasmine phenotype of rice through metabolite profiling and sensory evaluation. Metabolomics 2016, 12, 63. [Google Scholar] [CrossRef]
- Lu, L.; Hu, Z.; Fang, C.; Hu, X.; Tian, S. Improvement on the Identification and Discrimination Ability for Rice of Electronic Tongue Multi-Sensor Array Based on Information Entropy. J. Electrochem. Soc. 2022, 169, 037524. [Google Scholar] [CrossRef]
- Yan, C.; Lu, A. A deep learning method combined with electronic nose to identify the rice origin. J. Instrum. 2022, 17, 08016. [Google Scholar] [CrossRef]
- Shi, S.J.; Tang, Z.H.; Ma, Y.Y.; Cao, C.G.; Jiang, Y. Application of spectroscopic techniques combined with chemometrics to the authenticity and quality attributes of rice. Crit. Rev. Food Sci. Nutr. 2023. [Google Scholar] [CrossRef]
- Hu, Y.; Xu, L.; Huang, P.; Luo, X.; Wang, P.; Kang, Z. Reliable Identification of Oolong Tea Species: Nondestructive Testing Classification Based on Fluorescence Hyperspectral Technology and Machine Learning. Agriculture 2021, 11, 1106. [Google Scholar] [CrossRef]
- Hu, Y.; Kang, Z. The Rapid Non-Destructive Detection of Adulteration and Its Degree of Tieguanyin by Fluorescence Hyperspectral Technology. Molecules 2022, 27, 1196. [Google Scholar] [CrossRef]
- Wang, J.; Lin, T.; Ma, S.; Ju, J.; Wang, R.; Chen, G.; Jiang, R.; Wang, Z. The qualitative and quantitative analysis of industrial paraffin contamination levels in rice using spectral pretreatment combined with machine learning models. J. Food Compos. Anal. 2023, 121, 105430. [Google Scholar] [CrossRef]
- Kim, M.-J.; Lim, J.; Kwon, S.W.; Kim, G.; Kim, M.S.; Cho, B.-K.; Baek, I.; Lee, S.H.; Seo, Y.; Mo, C. Geographical Origin Discrimination of White Rice Based on Image Pixel Size Using Hyperspectral Fluorescence Imaging Analysis. Appl. Sci. 2020, 10, 5794. [Google Scholar] [CrossRef]
- Zhang, P.; Liu, B.; Mu, X.; Xu, J.; Du, B.; Wang, J.; Liu, Z.; Tong, Z. Performance of Classification Models of Toxins Based on Raman Spectroscopy Using Machine Learning Algorithms. Molecules 2024, 29, 197. [Google Scholar] [CrossRef]
- Vasat, R.; Kodesova, R.; Klement, A.; Boruvka, L. Simple but efficient signal pre-processing in soil organic carbon spectroscopic estimation. Geoderma 2017, 298, 46–53. [Google Scholar] [CrossRef]
- Xie, S.G.; Ding, F.J.; Chen, S.G.; Wang, X.; Li, Y.H.; Ma, K. Prediction of soil organic matter content based on characteristic band selection method. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 273, 120949. [Google Scholar] [CrossRef]
- Rezvantalab, S.; Mihandoost, S.; Rezaiee, M. Machine learning assisted exploration of the influential parameters on the PLGA nanoparticles. Sci. Rep. 2024, 14, 1114. [Google Scholar] [CrossRef] [PubMed]
- Silva, R.; Melo-Pinto, P. A review of different dimensionality reduction methods for the prediction of sugar cont-ent from hyperspectral images of wine grape berries. Appl. Soft Comput. 2021, 113, 107889. [Google Scholar] [CrossRef]
- Hajderanj, L.; Chen, D.; Grisan, E.; Dudley, S. Single- and Multi-Distribution Dimensionality Reduction Approaches for a Better Data Structure Capturing. IEEE Access 2020, 8, 207141–207155. [Google Scholar] [CrossRef]
- Liu, W.; Liu, C.H.; Hu, X.H.; Yang, J.B.; Zheng, L. Application of terahertz spectroscopy imaging for discrimi-nation of transgenic rice seeds with chemometrics. Food Chem. 2016, 210, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Miao, X.; Miao, Y.; Tao, S.; Liu, D.; Chen, Z.; Wang, J.; Huang, W.; Yu, Y. Classification of rice based on storag-e time by using near infrared spectroscopy and chemometric methods. Microchem. J. 2021, 171, 106841. [Google Scholar] [CrossRef]
- Li, H.D.; Cui, J.; Zhang, X.L.; Han, Y.Q.; Cao, L.Y. Dimensionality Reduction and Classification of Hyperspectral Remote Sensing Image Feature Extraction. Remote Sens. 2022, 14, 4579. [Google Scholar] [CrossRef]
- Zare, A.; Ozdemir, A.; Iwen, M.A.; Aviyente, S. Extension of PCA to Higher Order Data Structures: An Introduction to Tensors, Tensor Decompositions, and Tensor PCA. Proc. IEEE 2018, 106, 1341–1358. [Google Scholar] [CrossRef]
- Xu, X.; Xie, Z.; Yang, Z.; Li, D.; Xu, X. A t-SNE Based Classification Approach to Compositional Microbiome Data. Front. Genet. 2020, 11, 620143. [Google Scholar] [CrossRef]
- Hu, J.C.; Szymczak, S. A review on longitudinal data analysis with random forest. Brief. Bioinform. 2023, 24, bbad002. [Google Scholar] [CrossRef]
- Jiang, X.; Xu, C.H. Deep Learning and Machine Learning with Grid Search to Predict Later Occurrence of Breast Cancer Metastasis Using Clinical Data. J. Clin. Med. 2022, 11, 5772. [Google Scholar] [CrossRef] [PubMed]
- Peng, D.; Xu, R.; Zhou, Q.; Yue, J.; Su, M.; Zheng, S.; Li, J. Discrimination of Milk Freshness Based on Synchronous Two-Dimensional Visible/Near-Infrared Correlation Spectroscopy Coupled with Chemometrics. Molecules 2023, 28, 5728. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.L.; Zheng, J.D.; Huang, S.Q.; Pan, H.Y.; Liu, Q.Y. Time-Shift Multi-scale Weighted Permutation Ent-ropy and GWO-SVM Based Fault Diagnosis Approach for Rolling Bearing. Entropy 2019, 21, 621. [Google Scholar] [CrossRef]
- Mirjalili, S.; Mirjalili, S.M.; Lewis, A. Grey Wolf Optimizer. Adv. Eng. Softw. 2014, 69, 46–61. [Google Scholar] [CrossRef]
- Cao, Q.; Zhao, C.J.; Bai, B.N.; Cai, J.; Chen, L.Y.; Wang, F.; Xu, B.; Duan, D.D.; Jiang, P.; Meng, X.Y.; et al. Oolong tea cultivars categorization and germination period classification based on multispectral information. Front. Plant Sci. 2023, 14, 1251418. [Google Scholar] [CrossRef]
- Jairin, J.; Teangdeerith, S.; Leelagud, P.; Kothcharerk, J.; Sansen, K.; Yi, M.; Vanavichit, A.; Toojinda, T. Development of rice introgression lines with brown planthopper resistance and KDML105 grain quality characteristics through marker-assisted selection. Field Crops Res. 2009, 110, 263–271. [Google Scholar] [CrossRef]
- Sun, J.; Lu, X.Z.; Mao, H.P.; Jin, X.M.; Wu, X.H. A Method for Rapid Identification of Rice Origin by Hyperspectral Imaging Technology. J. Food Process Eng. 2017, 40, 12297. [Google Scholar] [CrossRef]
- Zou, Z.; Wu, Q.; Wang, J.; Xu, L.; Zhou, M.; Lu, Z.; He, Y.; Wang, Y.; Liu, B.; Zhao, Y. Research on non-destructive testing of hotpot oil quality by fluorescence hyperspectral technology combined with machine learning. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2023, 284, 121785. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xu, L.; Chen, H.; Zou, Z.; Huang, P.; Xin, B. Non-Destructive Detection of pH Value of Kiwifruit Bas-ed on Hyperspectral Fluorescence Imaging Technology. Agriculture 2022, 12, 208. [Google Scholar] [CrossRef]
- Wei, D.; Huang, Y.B.; Zhao, C.J.; Xiu, W. Identification of seedling cabbages and weeds using hyperspectral i-maging. Int. J. Agric. Biol. Eng. 2015, 8, 65–72. [Google Scholar] [CrossRef]
- Li, X.L.; Li, Z.X.; Yang, X.F.; He, Y. Boosting the generalization ability of Vis-NIR-spectroscopy-based regression models through dimension reduction and transfer learning. Comput. Electron. Agric. 2021, 186, 106157. [Google Scholar] [CrossRef]
- Dong, C.W.; Liu, Z.Y.; Yang, C.S.; An, T.; Hu, B.; Luo, X.; Jin, J.; Li, Y. Rapid detection of exogenous sucros-e in black tea samples based on near-infrared spectroscopy. Infrared Phys. Technol. 2021, 119, 103934. [Google Scholar] [CrossRef]
- Jiao, Y.P.; Li, Z.C.; Chen, X.S.; Fei, S.M. Preprocessing methods for near-infrared spectrum calibration. J. Chemom. 2020, 34, e3306. [Google Scholar] [CrossRef]
- Pu, Y.-Y.; Sun, D.-W.; Buccheri, M.; Grassi, M.; Cattaneo, T.M.P.; Gowen, A. Ripeness Classification of Bananito Fruit (Musa acuminata, AA): A Comparison Study of Visible Spectroscopy and Hyperspectral Imaging. Food Anal. Methods 2019, 12, 1693–1704. [Google Scholar] [CrossRef]
- Ozbekova, Z.; Kulmyrzaev, A. Study of moisture content and water activity of rice using fluorescence spectroscopy and multivariate analysis. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2019, 223, 117357. [Google Scholar] [CrossRef]
- Jayaprakash, C.; Damodaran, B.B.; Viswanathan, S.; Soman, K.P. Randomized independent component analysis and linear discriminant analysis dimensionality reduction methods for hyperspectral image classification. J. Appl. Remote Sens. 2020, 14, 036507. [Google Scholar] [CrossRef]
- Pu, H.; Wang, L.; Sun, D.-W.; Cheng, J.-H. Comparing Four Dimension Reduction Algorithms to Classify Algae Concentration Levels in Water Samples Using Hyperspectral Imaging. Water Air Soil Pollut. 2016, 227, 315. [Google Scholar] [CrossRef]
- He, Z.; Wu, M.; Zhao, X.; Zhang, S.; Tan, J. Representative null space LDA for discriminative dimensionality reduction. Pattern Recognit. 2021, 111, 107664. [Google Scholar] [CrossRef]
- Yu, W.; Zhang, M.; Shen, Y. Learning a local manifold representation based on improved neighborhood rough s-et and LLE for hyperspectral dimensionality reduction. Signal Process. 2019, 164, 20–29. [Google Scholar] [CrossRef]
- Gao, T.; Ma, Z.; Gao, W.; Liu, S. Dimensionality reduction of tensor data based on local linear embedding and mode product. J. Intell. Fuzzy Syst. 2021, 41, 2779–2796. [Google Scholar] [CrossRef]
- Traven, G.; Matijevic, G.; Zwitter, T.; Zerjal, M.; Kos, J.; Asplund, M.; Bland-Hawthorn, J.; Casey, A.R.; De Silva, G.; Freeman, K.; et al. The Galah Survey: Classification and Diagnostics with t-SNE Reduction of Spectral Information. Astrophys. J. Suppl. Ser. 2017, 228, 24. [Google Scholar] [CrossRef]
- van der Maaten, L.; Hinton, G. Visualizing Data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Sampaio, P.S.; Castanho, A.; Almeida, A.S.; Oliveira, J.; Brites, C. Identification of rice flour types with near-infrared spectroscopy associated with PLS-DA and SVM methods. Eur. Food Res. Technol. 2020, 246, 527–537. [Google Scholar] [CrossRef]
- Wang, H.; Zhu, H.; Zhao, Z.; Zhao, Y.; Wang, J. The study on increasing the identification accuracy of waxed a-pples by hyperspectral imaging technology. Multimed. Tools Appl. 2018, 77, 27505–27516. [Google Scholar] [CrossRef]
- Amjad, A.; Ullah, R.; Khan, S.; Bilal, M.; Khan, A. Raman spectroscopy based analysis of milk using random forest classification. Vib. Spectrosc. 2018, 99, 124–129. [Google Scholar] [CrossRef]
- Amaratunga, D.; Cabrera, J.; Lee, Y.S. Enriched random forests. Bioinformatics 2008, 24, 2010–2014. [Google Scholar] [CrossRef] [PubMed]
- Brereton, R.G. Contingency tables, confusion matrices, classifiers and quality of prediction. J. Chemom. 2021, 35, e3331. [Google Scholar] [CrossRef]
- Agarwal, S.; Zubair, M. Classification of Alcoholic and Non-Alcoholic EEG Signals Based on Sliding-SSA and Independent Component Analysis. IEEE Sens. J. 2021, 21, 26198–26206. [Google Scholar] [CrossRef]











| Methods | Models of Classification | ||
|---|---|---|---|
| BP | PLS-DA | RF | |
| RAW | 78% | 79% | 82% |
| MC | 77% | 78% | 81% |
| SG | 80% | 79% | 85% |
| SNV | 82% | 83% | 88% |
| FD | 84% | 85% | 91% |
| Models | Methods | Training Accuracy | Test Accuracy |
|---|---|---|---|
| RF | FD-TSNE | 99.7% | 93.3% |
| FD-PCA | 96.2% | 90.2% | |
| SNV-TSNE | 90.7% | 86.6% | |
| SNV-PCA | 90.2% | 88.5% | |
| PLS-DA | FD-TSNE | 91.2% | 90.6% |
| FD-PCA | 90.5% | 90.2% | |
| SNV-TSNE | 90.2% | 90.1% | |
| SNV-PCA | 90.1% | 90.0% | |
| BP | FD-TSNE | 95.5% | 86.6% |
| FD-PCA | 90.2% | 84.5% | |
| SNV-TSNE | 90.1% | 84.2% | |
| SNV-PCA | 89.5% | 82.2% |
| Methods | Training Accuracy | Test Accuracy |
|---|---|---|
| RF | 99.7% | 93.3% |
| GWO-RF | 99.8% | 95.3% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, Z.; Fan, R.; Zhan, C.; Wu, Y.; Lin, Y.; Li, K.; Qing, R.; Xu, L. The Rapid Non-Destructive Differentiation of Different Varieties of Rice by Fluorescence Hyperspectral Technology Combined with Machine Learning. Molecules 2024, 29, 682. https://doi.org/10.3390/molecules29030682
Kang Z, Fan R, Zhan C, Wu Y, Lin Y, Li K, Qing R, Xu L. The Rapid Non-Destructive Differentiation of Different Varieties of Rice by Fluorescence Hyperspectral Technology Combined with Machine Learning. Molecules. 2024; 29(3):682. https://doi.org/10.3390/molecules29030682
Chicago/Turabian StyleKang, Zhiliang, Rongsheng Fan, Chunyi Zhan, Youli Wu, Yi Lin, Kunyu Li, Rui Qing, and Lijia Xu. 2024. "The Rapid Non-Destructive Differentiation of Different Varieties of Rice by Fluorescence Hyperspectral Technology Combined with Machine Learning" Molecules 29, no. 3: 682. https://doi.org/10.3390/molecules29030682