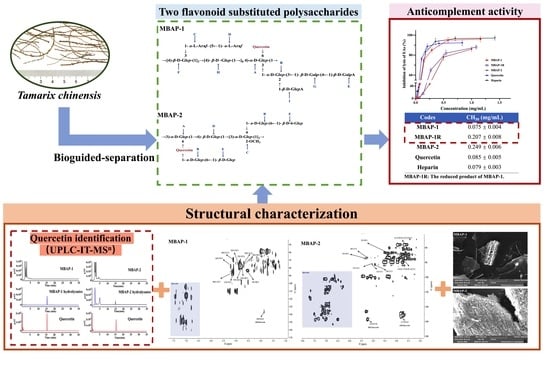
Two Natural Flavonoid Substituted Polysaccharides from Tamarix chinensis: Structural Characterization and Anticomplement Activities
Abstract
:1. Introduction
2. Results
2.1. Isolation and Purification of MBAP-1 and MBAP-2
2.2. Homogeneity and Molecular Weight Assessment
2.3. Monosaccharide Composition and Absolute Configuration Analysis
2.4. FT-IR Spectroscopy Assessments
2.5. Methylation Analysis
2.6. Identification of Substituted Flavonoid with UPLC-IT-MSn and NMR Analysis
2.7. NMR Analysis of Glycosidic Residues
2.8. NMR Analysis of Linkages and Sequences
2.9. Morphological Analysis
2.10. AFM Analysis
2.11. Anticomplement Activity
2.12. Antioxidant Activity
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Activity-Guided Extraction, Isolation and Purification
4.3. Homogeneity and Molecular Weight Determination
4.4. Monosaccharide Composition and Absolute Configuration Analysis
4.5. Reduction, Methylation and GC-MS Analysis
4.6. Qualitative and Quantitative Analysis of Flavonoid Component
4.7. NMR and FT-IR Spectroscopy Analysis
4.8. Microstructure and Atomic Force Microscopy (AFM) Analysis
4.9. Anticomplement Activity Evaluation
4.10. Antioxidant Activity Evaluation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Zoltán, P.; Michael, K.; Ashley, F.A. Complement analysis in the era of targeted therapeutics. Mol. Immunol. 2018, 102, 84–88. [Google Scholar]
- Zhu, H.Y.; Lu, X.X.; Ling, L.J.; Li, H.; Ou, Y.Y.; Shi, X.L.; Lu, Y.; Zhang, Y.Y.; Chen, D.F. Houttuynia cordata polysaccharides ameliorate pneumonia severity and intestinal injury in mice with influenza virus infection. J. Ethnopharmacol. 2018, 218, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Risitano, A.M.; Mastellos, D.C.; Huber-Lang, M.; Yancopoulou, D.; Lambris, J.D. Complement as a target in COVID-19? Nat. Rev. Immunol. 2020, 20, 343–344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polycarpou, A.; Howard, M.; Farrar, C.A.; Greenlaw, R.; Fanelli, G.; Wallis, R.; Klavinskis, L.S.; Sacks, S. Rationale for targeting complement in COVID-19. EMBO Mol. Med. 2020, 12, e12642. [Google Scholar] [CrossRef] [PubMed]
- Mastaglio, S.; Ruggeri, A.; Risitano, A.M.; Angelillo, P.; Yancopoulou, D.; Mastellos, D.C.; Markus, H.L.; Piemontese, S.; Assanelli, A.; Garlanda, C.; et al. The first case of COVID-19 treated with the complement C3 inhibitor AMY-101. Clin. Immunol. 2020, 215, 108450. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.L.; Xia, L.; De, J.; Zhu, M.X.; Li, H.; Lu, Y.; Chen, D.F. Beneficial effects on H1N1-induced acute lung injury and structure characterization of anti-complementary acidic polysaccharides from Juniperus pingii var. wilsonii. Int. J. Biol. Macro. 2019, 129, 246–253. [Google Scholar] [CrossRef] [PubMed]
- Ling, L.J.; Lu, Y.; Zhang, Y.Y.; Zhu, H.Y.; Tu, P.; Li, H.; Chen, D.F. Flavonoids from Houttuynia cordata attenuate H1N1-induced acute lung injury in mice via inhibition of influenza virus and Toll-like receptor signaling. Phytomedicine 2020, 67, 153150. [Google Scholar] [CrossRef]
- Zhang, Z.L. Clinical summary of 41 cases of measles complicated with pneumonia Pract. Clin. J. Integr. Tradit. Chin. West Med. 1992, 2, 41–42. [Google Scholar]
- Wang, Z.D.; Zhang, Y.; Dai, Y.D.; Ren, K.; Han, C.; Wang, H.X.; Yi, S.Q. Tamarix chinensis Lour. inhibits chronic ethanol-induced liver injury in mice. World J. Gastroenterol. 2020, 26, 1286–1297. [Google Scholar] [CrossRef]
- Liu, J.; Wang, X.; Yong, H.; Kan, J.; Jin, C. Recent advances in flavonoid-grafted polysaccharides: Synthesis, structural characterization, bioactivities and potential applications. Int. J. Biol. Macro. 2018, 116, 1011–1025. [Google Scholar] [CrossRef]
- Wang, X.Y.; Chen, Y.H.; Dahmani, F.Z.; Yin, L.F.; Zhou, J.P.; Yao, J. Amphiphilic carboxymethyl chitosan-quercetin conjugate with P-gp inhibitory properties for oral delivery of paclitaxel. Biomaterials 2014, 35, 7654–7665. [Google Scholar] [CrossRef] [PubMed]
- Pang, X.; Lu, Z.; Du, H.J.; Yang, X.Y.; Zhai, G.X. Hyaluronic acid-quercetin conjugate micelles: Synthesis, characterization, in vitro and in vivo evaluation. Colloids Surf. B. 2014, 123, 778–786. [Google Scholar] [CrossRef] [PubMed]
- Izadi, M.; Cegolon, L.; Javanbakht, M.; Sarafzadeh, A.; Abolghasemi, H.; Alishiri, G.; Zhao, S.; Einollahi, B.; Kashaki, M.; Jonaidi-Jafari, N.; et al. Ozone therapy for the treatment of COVID-19 pneumonia: A scoping review. Int. Immunopharmacol. 2021, 92, 107307. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Qu, H.; Shan, S.; Song, C.; Baranenko, D.; Li, Y.Z.; Lu, W.H. A novel polysaccharide isolated from Ulva Pertusa: Structure and physicochemical property. Carbohyd. Polym. 2020, 223, 115849. [Google Scholar] [CrossRef]
- Zhang, S.J.; Zhang, Q.; An, L.J.; Zhang, J.J.; Li, Z.G.; Zhang, J.; Li, Y.H.; Tuerhong, M.; Ohizumi, Y.; Jin, J.; et al. A fructan from Anemarrhena asphodeloides Bunge showing neuroprotective and immunoregulatory effects. Carbohyd. Polym. 2020, 229, 115477. [Google Scholar] [CrossRef]
- Xia, L.; Deji; Zhu, M.X.; Chen, D.F.; Lu, Y. Juniperus pingii var. wilsonii acidic polysaccharide: Extraction, characterization and anticomplement activity. Carbohyd. Polym. 2020, 231, 115728. [Google Scholar]
- Martinez-Busi, M.; Arredondo, F.; González, D.; Echeverry, C.; Vega-Teijido, M.A.; Carvalho, D.; Rodríguez-Haralambides, A.; Rivera, F.; Dajas, D.; Abin-Carriquiry, J.A. Purification, structural elucidation, antioxidant capacity and neuroprotective potential of the main polyphenolic compounds contained in Achyrocline satureioides (Lam) D.C. (Compositae). Bioorgan. Med. Chem. 2019, 27, 2579–2591. [Google Scholar] [CrossRef]
- Zahmanov, G.; Alipieva, K.; Denev, P.; Todorov, D.; Hinkov, A.; Shishkov, S.; Simova, S. Flavonoid glycosides profiling in dwarf elder fruits (Sambucus ebulus L.) and evaluation of their antioxidant and anti-herpes simplex activities. Ind. Crop. Prod. 2015, 63, 58–64. [Google Scholar] [CrossRef]
- Lian, Y.; Zhu, M.; Chen, J.; Yang, B.; Lv, Q.; Wang, L.; Guo, S.C.; Tan, X.B.; Li, C.; Bu, W.Q.; et al. Characterization of a novel polysaccharide from Moutan Cortex and its ameliorative effect on AGEs-induced diabetic nephropathy. Int. J. Biol. Macro. 2021, 176, 589–600. [Google Scholar] [CrossRef]
- Huang, F.; Liu, H.; Zhang, R.; Dong, L.; Liu, L.; Ma, Y.; Jia, X.C.; Wang, G.J.; Zhang, M.W. Physicochemical properties and prebiotic activities of polysaccharides from longan pulp based on different extraction techniques. Carbohyd. Polym. 2019, 206, 344–351. [Google Scholar] [CrossRef]
- Chang, X.; Shen, C.Y.; Jiang, J.G. Structural characterization of novel arabinoxylan and galactoarabinan from citron with potential antitumor and immunostimulatory activities. Carbohyd. Polym. 2021, 269, 118331. [Google Scholar] [CrossRef] [PubMed]
- Ye, G.; Li, J.; Zhang, J.; Liu, H.; Ye, Q.; Wang, Z. Structural characterization and antitumor activity of a polysaccharide from Dendrobium wardianum. Carbohyd. Polym. 2021, 269, 118253. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Xu, Z.; Wu, S.; Li, X.; Li, J.; Hu, H.; Wu, Y.; Ai, L.Z. Molecular properties and structural characterization of an alkaline extractable arabinoxylan from hull-less barley bran. Carbohyd. Polym. 2019, 218, 250–260. [Google Scholar] [CrossRef]
- Zhang, H.; Zou, P.; Zhao, H.; Qiu, J.; Regenstein, J.M.; Yang, X. Isolation, purification, structure and antioxidant activity of polysaccharide from pinecones of Pinus koraiensis. Carbohyd. Polym. 2021, 251, 117078. [Google Scholar] [CrossRef]
- Dong, Z.; Zhang, M.; Li, H.; Zhan, Q.; Lai, F.; Wu, H. Structural characterization and immunomodulatory activity of a novel polysaccharide from Pueraria lobata (Willd.) Ohwi root. Int. J. Biol. Macro. 2020, 154, 1556–1564. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.P.; Guo, M.R. Purification and structural characterization of polysaccharides isolated from Auricularia cornea var. Li. Carbohyd. Polym. 2020, 230, 115680. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Qi, J.; Ho, C.T.; Li, B.; Mu, J.; Zhang, Y.; Hu, H.P.; Mo, W.P.; Chen, Z.Z.; Xie, Y.Z. Structural characterization and immunomodulatory activity of a water-soluble polysaccharide from Ganoderma leucocontextum fruiting bodies. Carbohyd. Polym. 2020, 249, 116874. [Google Scholar] [CrossRef]
- Zhang, Y.; Pan, X.; Ran, S.; Wang, K. Purification, structural elucidation and anti-inflammatory activity in vitro of polysaccharides from Smilax china L. Int. J. Biol. Macro. 2019, 139, 233–243. [Google Scholar] [CrossRef]
- Xu, S.; Xu, X.; Zhang, L. Branching structure and chain conformation of water-soluble glucan extracted from Auricularia auricula-judae. J. Agr. Food. Chem. 2012, 60, 3498–3506. [Google Scholar] [CrossRef]
- Gao, Y.; Guo, Q.; Zhang, K.; Wang, N.; Li, C.; Li, Z.; Zhang, A.L.; Wang, C.L. Polysaccharide from Pleurotus nebrodensis: Physicochemical, structural characterization and in vitro fermentation characteristics. Int. J. Biol. Macro. 2020, 165, 1960–1969. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, M.; Zhang, D.; Wang, S.; Yan, C. An O-acetyl-glucomannan from the rhizomes of Curculigo orchioides: Structural characterization and anti-osteoporosis activity in vitro. Carbohyd. Polym. 2017, 174, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.L.; Wang, B.; Hu, R.; Zhao, X.; Li, H.; Zhou, G.; Song, L.L.; Wu, A.M. Characterization of hemicelluloses in Phyllostachys edulis (moso bamboo) culm during xylogenesis. Carbohyd. Polym. 2019, 221, 127–136. [Google Scholar] [CrossRef]
- Marszalek, P.E.; Dufrene, Y.F. Stretching single polysaccharides and proteins using atomic force microscopy. Chem. Soc. Rev. 2012, 41, 3523–3534. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Delbianco, M.; Anggara, K.; Michnowicz, T.; Pardo-Vargas, A.; Bharate, P.; Sen, S.; Pristl, M.; Rauschenbach, S.; Schlickum, U.; et al. Imaging single glycans. Nature 2020, 582, 375–378. [Google Scholar] [CrossRef] [PubMed]
- Wolosiak, R.; Drużyńska, B.; Derewiaka, D.; Piecyk, M.; Majewska, E.; Ciecierska, M.; Worobiej, E.; Pakosz, P. Verification of the conditions for determination of antioxidant activity by ABTS and DPPH assays—A practical approach. Molecules 2021, 27, 50. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.L.; Huang, P.; Zhang, L.; Qiu, Y.; Qi, H.; Leng, A.J.; Shang, D. Hepatoprotective effect of plant polysaccharides from natural resources: A review of the mechanisms and structure-activity relationship. Int. J. Biol. Macro. 2020, 161, 24–34. [Google Scholar] [CrossRef]
- Lu, Y.; Zhang, J.J.; Huo, J.Y.; Chen, D.F. Structural characterization and anti-complementary activities of two polysaccharides from Houttuynia cordata. Planta Med. 2019, 85, 108–1106. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.W.; Li, N.; Zhu, C.; Shi, S.S.; Jin, H.; Wang, S.C. Anti-complementary activity of two homogeneous polysaccharides from Eclipta prostrata. Biochem. Bioph. Res. Commun. 2017, 393, 887–893. [Google Scholar] [CrossRef]
- Zeng, F.K.; Chen, W.B.; He, P.; Zhan, Q.P.; Wang, Q.; Wu, H.; Zhang, M.M. Structural characterization of polysaccharides with potential antioxidant and immunomodulatory activities from Chinese water chestnut peels. Carbohyd. Polym. 2020, 246, 116551. [Google Scholar] [CrossRef]
- Zhu, W.L.; Zhang, Z.J. Preparation and characterization of catechin-grafted chitosan with antioxidant and antidiabetic potential. Int. J. Biol. Macro. 2014, 70, 150–155. [Google Scholar] [CrossRef]
- Ahn, S.; Halake, K.; Lee, J. Antioxidant and ion-induced gelation functions of pectins enabled by polyphenol conjugation. Int. J. Biol. Macro. 2017, 101, 776–782. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.K.; Hua, D.H.; Huang, D.; Zhang, Q.; Yan, C.Y. Characterization of a new heteropolysaccharide from green guava and its application as an α-glucosidase inhibitor for the treatment of type II diabetes. Food. Funct. 2018, 9, 3997–4007. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.X.; Li, T.Y.; Qing, D.G.; Chen, J.; Zhang, Q.; Yan, C.Y. Structural characterization and osteogenic bioactivities of a novel Humulus lupulus polysaccharide. Food. Funct. 2020, 11, 1165–1175. [Google Scholar] [CrossRef] [PubMed]
- Kostalova, D.; Tekel’ová, D.; Czigle, S.; Tóth, J.; Lalkovi, S. The content of flavonoids in selected drug formulations containing Ginkgo biloba. Farmaceutický. Obzor. 2005, 74, 119–123. [Google Scholar]
- Chen, X.; Cao, D.; Zhou, L.; Jin, H.; Dong, J.; Yao, J.; Ding, K. Structure of a polysaccharide from Gastrodia elata Bl., and oligosaccharides prepared thereof with anti-pancreatic cancer cell growth activities. Carbohyd. Polym. 2020, 86, 1300–1305. [Google Scholar] [CrossRef]
- Huo, J.; Lu, Y.; Jiao, Y.; Chen, D. Structural characterization and anticomplement activity of an acidic polysaccharide from Hedyotis diffusa. Int. J. Biol. Macromol. 2020, 155, 1553–1560. [Google Scholar] [CrossRef]








| Molecular Characteristics | Parameter | Detection Results | |
|---|---|---|---|
| MBAP-1 | MBAP-2 | ||
| Polydispersity | Mw/Mn | 1.01 | 1.07 |
| Mz/Mn | 1.04 | 1.05 | |
| Molar mass moments (g/mol) | Mw | 2.693 × 105 | 4.650 × 104 |
| Mn | 2.537 × 105 | 4.611 × 104 | |
| Mz | 2.501 × 105 | 4.520 × 104 | |
| Mp | 2.51 × 105 | 3.91 × 104 | |
| Rms radius moments (nm) | Rz | 1.5 nm | 1.5 nm |
| Sample | Yield (%) | Total Sugar Content (%) | Uronic Acid (%) | Protein (%) | Flavonoids (%) | Monosaccharide Composition (w/w) | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Glc | GlcA | GalA | Gal | Ara | ||||||
| MBAP-1 | 0.14 | 86.06 ± 2.76 | 9.64 ± 0.56 | 1.90 ± 0.15 | 12.03 ± 1.20 | 65.17 (54.54 #) | 6.27 (4.87 #) | 5.42 (4.21 #) | 5.03 (4.21 #) | 18.10 (18.18 #) |
| MBAP-2 | 0.16 | 82.03 ± 1.77 | - | 2.06 ± 0.13 | 15.96 ± 1.36 | 88.18 (88.41 #) | - | - | - | - |
| PMAAs | Linkages | Molar Ratio | Major Mass Fragments (m/z) | ||
|---|---|---|---|---|---|
| MBAP-1 | MBAP-1R | MBAP-2 | |||
| 1,3,4,5,6-Penta-O-acetyl-1-deuterio-2-O-methyl-d-glucitol | 1,3,4,6-linked-Glcp | 1.00 | 1.00 | 43, 59, 87, 118, 139, 333 | |
| 1,2,3,5-Tetra-O-acetyl-1-deuterio-4,6-di-O-methyl-d-glucitol | 1,2,3-linked-Glcp | 1.03 | 0.95 | 43, 59, 71, 87, 101, 129, 161, 202, 262 | |
| 1,4,5-Tri-O-acetyl-1-deuterio-2,3-di-O-methyl-l-arabinitol | 1,5-linked-Araf | 1.87 | 1.95 | 43, 59, 87, 102, 118, 129, 189 | |
| 1,4-Di-O-acetyl-1-deuterio-2,3,5-tri-O-methyl-l-arabinitol | 1-linked-Araf | 2.21 | 2.24 | 43, 59, 71, 87, 102, 118, 129, 161, 162 | |
| 1,5-Di-O-acetyl-1-deuterio-2,3,4,6-tetra-O-methyl-d-galactitol | 1-linked-Galp | n.d. | 1.12 | 43, 71, 87, 102, 118, 129, 145, 161, 162, 205 | |
| 1,3,4,5-Tetra-O-acetyl-1-deuterio-2,6-di-O-methyl-d-glucitol | 1,3,4-linked-Glcp | 1.98 | 2.01 | 0.89 | 43, 59, 87, 118, 129, 160, 185, 305 |
| 1,4,5-Tri-O-acetyl-1-deuterio-2,3,6-tri-O-methyl-d-galactitol | 1,4-linked-Galp | 1.03 | 0.99 | 43, 59, 71, 87, 102, 118, 129, 162, 233 | |
| 1,4,5-Tri-O-acetyl-1-deuterio-2,3,6-tri-O-methyl-d-glucitol | 1,4-linked-Glcp | 8.01 | 7.89 | 0.35 | 43, 59, 71, 87, 102, 118, 129, 162, 233 |
| 1,5-Di-O-acetyl-1-deuterio-2,3,4,6-tetra-O-methyl-d-glucitol | 1-linked-Glcp | n.d. | 1.10 | 1.20 | 43, 71, 87, 102, 118, 129, 145, 161, 162, 205 |
| 1,3,5,6-Tetra-O-acetyl-1-deuterio-2,4-di-O-methyl-d-glucitol | 1,3,6-linked-Glcp | 0.38 | 43, 59, 87, 118, 129, 139, 160, 189, 234, 305 | ||
| 1,5,6-Tri-O-acetyl-1-deuterio-2,3,4-tri-O-methyl-d-glucitol | 1,6-linked-Glcp | 1.01 | 43, 59, 87, 99, 101, 118, 129, 162, 189, 233 | ||
| Name | Code | Residues | Chemical Shifts (ppm) | |||||
|---|---|---|---|---|---|---|---|---|
| H1/C1 | H2/C2 | H3/C3 | H4/C4 | H5/C5 | H6/C6 | |||
| MBAP-1 | A | →3,4,6)-α-d-Glcp-(1→ | 5.26/101.76 | 3.36/72.47 | 4.01/86.21 | 3.50/77.49 | 3.98/71.22 | 3.85/69.46 |
| B | →2,3)-α-d-Glcp-(1→ | 5.21/94.86 | 4.33/84.89 | 4.00/80.62 | 3.88/72.97 | 3.92/73.13 | 3.61/61.54 | |
| C | →5)-α-l-Araf-(1→ | 5.13/109.84 | 4.11/79.08 | 3.99/79.21 | 4.11/83.30 | 3.78/68.53 | ||
| D | α-l-Araf-(1→ | 5.07/110.23 | 4.12/84.16 | 3.88/76.51 | 4.06/86.96 | 3.72/63.41 | ||
| E | β-d-GalpA-(1→ | 4.95/100.47 | 3.77/69.59 | 3.47/72.12 | 3.53/73.99 | 3.73/68.31 | -/175.87 | |
| F | →3,4)-β-d-Glcp-(1→ | 4.73/102.72 | 3.65/76.30 | 4.06/86.66 | 3.77/76.40 | 3.69/78.60 | 3.92/63.41 | |
| G | →4)-β-d-Galp-(1→ | 4.70/98.66 | 3.79/74.14 | 3.97/75.00 | 3.65/76.18 | 4.32/78.82 | 3.80/59.78 | |
| H | →4)-β-d-Glcp-(1→ | 4.62/98.70 | 3.30/72.40 | 3.55/74.20 | 3.92/78.55 | 3.63/71.68 | 3.73/58.92 | |
| I | β-d-GlcpA-(1→ | 4.48/105.32 | 3.50/72.33 | 3.60/76.54 | 3.62/75.58 | 3.59/77.17 | -/178.66 | |
| MBAP-2 | A | →3,6)-α-d-Glcp-(1→ | 5.32/98.91 | 4.40/80.62 | 4.31/81.47 | 3.76/70.88 | 3.84/73.76 | 3.71/69.56 |
| B | →6)-α-d-Glcp-(1→ | 5.23/98.68 | 4.25/79.52 | 3.93/73.76 | 3.88/70.28 | 3.90/74.01 | 3.62/68.56 | |
| C | →3,4)-α-d-2-OCH3-Glcp-(1→ | 5.08/100.60 | 3.67/75.51 | 4.39/82.41 | 3.76/74.51 | 3.78/74.77 | 3.53/60.32 | |
| D | →4)-β-d-Glcp-(1→ | 4.92/100.26 | 3.62/70.51 | 3.48/72.21 | 3.64/74.82 | 3.35/72.40 | 3.74/60.23 | |
| E | β-d-Glcp-(1→ | 4.37/102.53 | 3.75/73.10 | 3.56/73.52 | 3.40/70.22 | 3.46/75.83 | 3.68/60.48 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiao, Y.; Yang, Y.; Zhou, L.; Chen, D.; Lu, Y. Two Natural Flavonoid Substituted Polysaccharides from Tamarix chinensis: Structural Characterization and Anticomplement Activities. Molecules 2022, 27, 4532. https://doi.org/10.3390/molecules27144532
Jiao Y, Yang Y, Zhou L, Chen D, Lu Y. Two Natural Flavonoid Substituted Polysaccharides from Tamarix chinensis: Structural Characterization and Anticomplement Activities. Molecules. 2022; 27(14):4532. https://doi.org/10.3390/molecules27144532
Chicago/Turabian StyleJiao, Yukun, Yiting Yang, Lishuang Zhou, Daofeng Chen, and Yan Lu. 2022. "Two Natural Flavonoid Substituted Polysaccharides from Tamarix chinensis: Structural Characterization and Anticomplement Activities" Molecules 27, no. 14: 4532. https://doi.org/10.3390/molecules27144532