Screening and Comparison of Lignin Degradation Microbial Consortia from Wooden Antiques
Abstract
:1. Introduction
2. Results
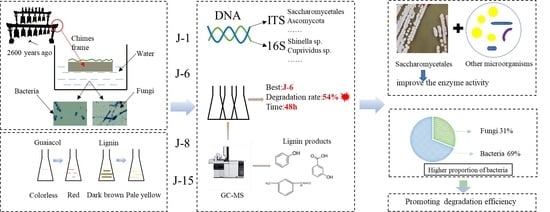
2.1. Screening of Lignin-Degrading Microbial Consortia
2.2. Comparative Study of the Lignin Degradation Performance of Different Microbial Consortia
2.3. Comparative Study of the Consortium Composition
2.3.1. Comparison of the Fungal Consortium Composition
2.3.2. Comparison of the Bacterial Consortium Composition
2.4. Comparison of the Biomass of Fungi and Bacteria in the Microbial Consortia
2.5. Comparison of the Lignin Degradation Products
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Culture Conditions
4.3. Screening of Microbial Consortia
4.4. Lignin Degradation Experiments
4.5. Analytical Methods
4.6. Statistical Methods
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Pu, W.F.; Shen, C.; Wei, B.; Yang, Y.; Li, Y.B. A comprehensive review of polysaccharide biopolymers for enhanced oil recovery (EOR) from flask to field. J. Ind. Eng. Chem. 2018, 61, 1–11. [Google Scholar] [CrossRef]
- Lubbers, R.J.; Dilokpimol, A.; Visser, J.; Mäkelä, M.R.; Hildén, K.S.; de Vries, R.P. A comparison between the homocyclic aromatic metabolic pathways from plant-derived compounds by bacteria and fungi. Biotechnol. Adv. 2019, 37, 107396. [Google Scholar] [CrossRef] [PubMed]
- Houtman, C.J.; Maligaspe, E.; Hunt, C.G.; Fernández-Fueyo, E.; Martínez, A.T.; Hammel, K.E. Fungal lignin peroxidase does not produce the veratryl alcohol cation radical as a diffusible ligninolytic oxidant. J. Biol. Chem. 2018, 293, 4702–4712. [Google Scholar] [CrossRef] [Green Version]
- Kumar, M.; Oyedun, A.O.; Kumar, A. A review on the current status of various hydrothermal technologies on biomass feedstock. Renew. Sustain. Energy Rev. 2018, 81, 1742–1770. [Google Scholar] [CrossRef]
- Asina, F.; Brzonova, I.; Kozliak, E.; Kubátová, A.; Ji, Y. Microbial treatment of industrial lignin: Successes, problems and challenges. Renew. Sustain. Energy Rev. 2017, 77, 1179–1205. [Google Scholar] [CrossRef]
- Kamimura, N.; Sakamoto, S.; Mitsuda, N.; Masai, E.; Kajita, S. Advances in microbial lignin degradation and its applications. Curr. Opin. Biotechnol. 2019, 56, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Priyadarshinee, R.; Singha, S.; Sengupta, B.; Roy, A.; Dasgupta, D.; Mandal, T. Biodegradation of alkali lignin by Bacillus flexus RMWW II: Analyzing performance for abatement of rice mill wastewater. Water Sci. Technol. 2019, 80, 1623–1632. [Google Scholar] [CrossRef]
- Zhang, Y.; Zang, G.-Q.; Tang, Z.-H.; Chen, X.-H.; Yu, Y.-S. Burning straw, air pollution, and respiratory infections in China. Am. J. Infect. Control 2014, 42, 815. [Google Scholar] [CrossRef]
- Arun, A.; Eyini, M. Comparative studies on lignin and polycyclic aromatic hydrocarbons degradation by basidiomycetes fungi. Bioresour. Technol. 2011, 102, 8063–8070. [Google Scholar] [CrossRef]
- Bugg, T.D.; Williamson, J.J.; Rashid, G.M. Bacterial enzymes for lignin depolymerisation: New biocatalysts for generation of renewable chemicals from biomass. Curr. Opin. Chem. Biol. 2020, 55, 26–33. [Google Scholar] [CrossRef]
- Ho, J.C.H.; Pawar, S.V.; Hallam, S.J.; Yadav, V.G. An Improved Whole-Cell Biosensor for the Discovery of Lignin-Transforming Enzymes in Functional Metagenomic Screens. ACS Synth. Biol. 2018, 7, 392–398. [Google Scholar] [CrossRef]
- Li, C.; Chen, C.; Wu, X.; Tsang, C.-W.; Mou, J.; Yan, J.; Liu, Y.; Lin, C.S.K. Recent advancement in lignin biorefinery: With special focus on enzymatic degradation and valorization. Bioresour. Technol. 2019, 291, 121898. [Google Scholar] [CrossRef]
- Chen, Z.; Wan, C. Biological valorization strategies for converting lignin into fuels and chemicals. Renew. Sustain. Energy Rev. 2017, 73, 610–621. [Google Scholar] [CrossRef]
- Becker, J.; Kuhl, M.; Kohlstedt, M.; Starck, S.; Wittmann, C. Metabolic engineering of Corynebacterium glutamicum for the production of cis, cis-muconic acid from lignin. Microb. Cell Fact. 2018, 17, 115. [Google Scholar] [CrossRef]
- Xu, W.; Fu, S.; Yang, Z.; Lu, J.; Guo, R. Improved methane production from corn straw by microaerobic pretreatment with a pure bacteria system. Bioresour. Technol. 2018, 259, 18–23. [Google Scholar] [CrossRef]
- Lv, Y.; Chen, Y.; Sun, S.; Hu, Y. Interaction among multiple microorganisms and effects of nitrogen and carbon supplementations on lignin degradation. Bioresour. Technol. 2014, 155, 144–151. [Google Scholar] [CrossRef]
- Wei, Y.Q.; Wu, D.; Wei, D.; Zhao, Y.; Wu, J.Q.; Xie, X.Y.; Zhang, R.J.; Wei, Z.M. Improved lignocellulose-degrading performance during straw composting from diverse sources with actinomycetes inoculation by regulating the key enzyme activities. Bioresour. Technol. 2019, 27, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Rahmanpour, R.; Rea, D.; Jamshidi, S.; Fülop, V.; Bugg, T.D.H. Structure of Thermobifida fusca DyP-type peroxidase and activity towards Kraft lignin and lignin model compounds. Arch. Biochem. Biophys. 2016, 594, 54–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Xiao, J.; Wang, G.; Chen, G. Enzymatic hydrolysis of lignin by ligninolytic enzymes and analysis of the hydrolyzed lignin products. Bioresour. Technol. 2020, 304, 122975. [Google Scholar] [CrossRef]
- Chauhan, P.S. Role of various bacterial enzymes in complete depolymerization of lignin: A review. Biocatal. Agric. Biotechnol. 2020, 23, 101498. [Google Scholar] [CrossRef]
- Wilhelm, R.C.; Singh, R.; Eltis, L.D.; Mohn, W.W. Bacterial contributions to delignification and lignocellulose degradation in forest soils with metagenomic and quantitative stable isotope probing. ISME J. 2019, 13, 413–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, S.S.; Abomohra, A.E.; Sun, J.Z. Effective bio-pretreatment of sawdust waste with a novel microbial consortium for enhanced biomethanation. Bioresour Technol. 2017, 238, 425–432. [Google Scholar] [CrossRef]
- Wang, Y.X.; Liu, Q.; Yan, L.; Gao, Y.M.; Wang, Y.J.; Wang, W.D. A novel lignin degradation bacterial consortium for efficient pulping. Bioresour. Technol. 2013, 139, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.C.; Li, X.K.; Wang, X.W.; Liu, G.G.; Zuo, J.L.; Wang, S.T.; Wang, K. Impact of salinity on anaerobic microbial consortium structure in high organic loading purified terephthalic acid wastewater treatment system. J. Hazard. Mater. 2020, 383, 121132. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Singh, A.P.; Nilsson, T. Bacteria as Important Degraders in Waterlogged Archaeological Woods. Holzforschung 1996, 50, 389–392. [Google Scholar] [CrossRef]
- Guo, J.; Zhou, H.; Stevanic, J.S.; Dong, M.; Min, Y.; Lennart, S.; Yin, Y.F. Effects of ageing on the cell wall and its hygroscopicity of wood in ancient timber construction. Wood Sci. Technol. 2017, 52, 131–147. [Google Scholar] [CrossRef]
- Hu, D. The antisepsis of the bamboo slips and lacquer wares in water. Sci. Conserv. Archaeol. 2003, 15, 14–19. [Google Scholar]
- Wang, W.D.; Yan, L.; Cui, Z.J.; Gao, Y.M.; Wang, Y.J.; Jing, R.Y. Characterization of a microbial consortium capable of degrading lignocellulose. Bioresour. Technol. 2011, 102, 9321–9324. [Google Scholar] [CrossRef]
- Fang, X.; Li, Q.; Lin, Y.; Lin, X.; Dai, Y.; Guo, Z.; Pan, D. Screening of a microbial consortium for selective degradation of lignin from tree trimmings. Bioresour. Technol. 2018, 254, 247–255. [Google Scholar] [CrossRef]
- Zhu, D.; Zhang, P.; Xie, C.; Zhang, W.; Sun, J.; Qian, W.-J.; Yang, B. Biodegradation of alkaline lignin by Bacillus ligniniphilus L1. Biotechnol. Biofuels 2017, 10, 44. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Xiao, Y.-Z.; Yu, H.-Q. Degradation of lignin in pulp mill wastewaters by white-rot fungi on biofilm. Bioresour. Technol. 2005, 96, 1357–1363. [Google Scholar] [CrossRef] [PubMed]
- Raj, A.; Chandra, R.; Reddy, M.; Purohit, H.J.; Kapley, A. Biodegradation of kraft lignin by a newly isolated bacterial strain, Aneurinibacillus aneurinilyticus from the sludge of a pulp paper mill. World J. Microbiol. Biotechnol. 2006, 23, 793–799. [Google Scholar] [CrossRef]
- Chen, Y.H.; Chai, L.Y.; Zhu, Y.H.; Yang, Z.H.; Zheng, Y.; Zhang, H. Biodegradation of kraft lignin by a bacterial strain Comamonas sp. B-9 isolated from eroded bamboo slips. J. Appl. Microbiol. 2012, 112, 900–906. [Google Scholar] [CrossRef]
- Christopher, L.P.; Yao, B.; Ji, Y. Lignin biodegradation with laccase-mediator systems. Front. Energy Res. 2014, 2, 12. [Google Scholar] [CrossRef]
- Mate, D.M.; Alcalde, M. Laccase: A multi-purpose biocatalyst at the forefront of biotechnology. Microb. Biotechnol. 2017, 10, 1457–1467. [Google Scholar] [CrossRef]
- Xie, X.-G.; Dai, C.-C. Degradation of a model pollutant ferulic acid by the endophytic fungus Phomopsis liquidambari. Bioresour. Technol. 2015, 179, 35–42. [Google Scholar] [CrossRef]
- Shi, Y.; Chai, L.Y.; Tang, C.J.; Yang, Z.H.; Zheng, Y.; Chen, Y.H.; Jing, Q.X. Biochemical investigation of kraft lignin degradation by Pandoraea sp. B-6 isolated from bamboo slips. Bioproc. Biosyst. Eng. 2013, 36, 1957–1965. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Ren, X.H.; He, J.; Zhang, Q.R.; Qiu, C.; Fan, B.M. Application of natural mixed bacteria immobilized carriers to different kinds of organic wastewater treatment and microbial consortium comparison. J. Hazard. Mater. 2019, 377, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Chandra, R.; Bharagava, R.N. Bacterial degradation of synthetic and kraft lignin by axenic and mixed culture and their metabolic products. J. Environ. Biol. 2013, 34, 991–999. [Google Scholar]
- Jin, R. Identification and characterization of a fungal strain with lignin and cellulose hydrolysis activities. Afr. J. Microbiol. Res. 2012, 6, 6545–6550. [Google Scholar] [CrossRef]
- Rodríguez, A.; Carnicero, A.; Perestelo, F.; Fuente, D.G.; Milstein, O.; Falcón, M.A. Effect of Penicillium chrysogenum on lignin transformation. Appl. Environ. Microbiol. 1994, 60, 2971–2976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, N.; Xu, H.C.; Yan, Z.S.; Yang, T.; Wang, C.H.; Jiang, H.L. Improved lignin degradation through distinct microbial consortium in subsurface sediments of one eutrophic lake. Renew. Energy 2019, 138, 861–869. [Google Scholar] [CrossRef]
- Gong, C.J.; Su, D.; Wang, X.; Pu, Y.; Wang, T.J. Impacts of cold-resistant mixed strains immobilized by different carrier materials on remediation of PAHs polluted soils. Chin. J. Ecol. 2018, 37, 3713–3720. [Google Scholar]
- Madadi, M.; Hano, A. Lignin Degradation by Fungal Pretreatment: A Review. J. Plant Pathol. Microbiol. 2017, 8, 1–6. [Google Scholar]
- Bugg, T.D.H.; Ahmad, M.; Hardiman, E.; Rahman, P.R. ChemInform Abstract: Pathways for Degradation of Lignin in Bacteria and Fungi. Nat. Prod. Rep. 2011, 28, 1883–1896. [Google Scholar] [CrossRef]
- Young, D.; Dollhofer, V.; Callaghan, T.M.; Reitberger, S.; Lebuhn, M.; Benz, J.P. Isolation, identification and characterization of lignocellulolytic aerobic and anaerobic fungi in one- and two-phase biogas plants. Bioresour. Technol. 2018, 268, 470–479. [Google Scholar] [CrossRef] [PubMed]
- Šlosarčíková, P.; Novotný, Č.; Malachová, K.; Válková, H.; Fojtík, J. Effect of yeasts on biodegradation potential of immobilized cultures of white rot fungi. Sci. Total Environ. 2017, 589, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Yediler, A.; Yang, M.; Kettrup, A. Decolorization of an azo dye, Reactive Black 5 and MnP production by yeast isolate: Debaryomyces polymorphus. Biochem. Eng. J. 2005, 24, 249–253. [Google Scholar] [CrossRef]
- Moraes, E.C.; Alvarez, T.M.; Persinoti, G.F.; Tomazetto, G.; Brenelli, L.B.; Paixão, D.A.A.; Ematsu, G.C.; Aricetti, J.A. Lignolytic-consortium omics analyses reveal novel genomes and pathways involved in lignin modification and valorization. Biotechnol. Biofuels 2018, 11, 75. [Google Scholar] [CrossRef]
- Shi, Y.; Yan, X.; Li, Q.; Wang, X.; Liu, M.; Xie, S.S.; Chai, L.Y.; Yuan, J.S. Directed bioconversion of Kraft lignin to polyhydroxyalkanoate by Cupriavidus basilensis B-8 without any pretreatment. Process Biochem. 2017, 52, 238–242. [Google Scholar] [CrossRef]
- Singh, A.K.; Yadav, P.; Bharagava, R.N.; Saratale, G.D.; Raj, A. Biotransformation and Cytotoxicity Evaluation of Kraft Lignin Degraded by Ligninolytic Serratia liquefaciens. Front. Microbiol. 2019, 10, 2364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Nie, Y.; Tang, Y.-Q.; Song, X.-M.; Cao, K.; Sun, L.-Z.; Wang, Z.-J.; Wu, X.-L. Diverse Bacteria with Lignin Degrading Potentials Isolated from Two Ranks of Coal. Front. Microbiol. 2016, 7, 1428. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.-H.; Pourcher, A.-M.; Bouchez, T.; Gelhaye, E.; Peu, P. Occurrence of lignin degradation genotypes and phenotypes among prokaryotes. Appl. Microbiol. Biotechnol. 2014, 98, 9527–9544. [Google Scholar] [CrossRef]
- Brown, M.E.; Chang, M.C. Exploring bacterial lignin degradation. Curr. Opin. Chem. Biol. 2014, 19, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Azadi, P.; Inderwildi, O.R.; Farnood, R.; King, D.A. Liquid fuels, hydrogen and chemicals from lignin: A critical review. Renew. Sustain. Energy Rev. 2013, 21, 506–523. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Magoč, T.; Magoč, M.; Salzberg, S.L. FLASH: Fast Length Adjustment of Short Reads to Improve Genome Assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Chang, Y.-C.; Choi, D.; Takamizawa, K.; Kikuchi, S. Isolation of Bacillus sp. strains capable of decomposing alkali lignin and their application in combination with lactic acid bacteria for enhancing cellulase performance. Bioresour. Technol. 2014, 152, 429–436. [Google Scholar] [CrossRef] [Green Version]
- Anderson, J.P.E.; Domsch, K.H. Measurement of bacterial and fungal contributions to respiration of selected agricultural and forest soils. Can. J. Microbiol. 1975, 21, 314–322. [Google Scholar] [CrossRef]













| No. | RT 1 | Compound | J-1 | J-6 | J-8 | J-15 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1d | 2d | 6d | 1d | 2d | 6d | 1d | 2d | 6d | 1d | 2d | 6d | |||
| 1 | 10.45 min | Phenol | + | |||||||||||
| 2 | 11.58 min | Ethyl 3-hydroxybutyrate | + | + | + | + | ||||||||
| 3 | 13.27 min | Benzaldehyde, 3-methyl | + | + | + | + | + | + | + | + | + | + | + | + |
| 4 | 13.91 min | Salicylic acid | + | |||||||||||
| 5 | 13.92 min | 3-Hydroxybenzoic acid | + | |||||||||||
| 6 | 14.27 min | 2,6-Dihydroxyacetophenone | + | |||||||||||
| 7 | 14.61 min | 2,6-Dihydroxybenzoic acid | + | + | + | |||||||||
| 8 | 14.84 min | 2-Ethylhexanoic acid | + | + | + | + | ||||||||
| 9 | 16.81 min | Benzeneacetic acid, 3-pentadecyl ester | + | |||||||||||
| 10 | 17.19 min | 2-Methoxy-4-vinylphenol | + | |||||||||||
| 11 | 18.32 min | 5,5′-Dimethoxy-3,3′,7,7′-tetramethyl-2,2′-binaphthalene-1,1′,4,4′-tetrone | + | |||||||||||
| 12 | 19.81 min | Butylated Hydroxytoluene | + | + | + | + | + | + | + | + | + | + | ||
| 13 | 20.42 min | Tyrosol, acetate | + | |||||||||||
| 14 | 22.12 min | Cinnamic acid, 4-hydroxy-3-methoxy-, (5-hydroxy-2-hydroxymethyl-6-[2-(4-hydroxy-3-methoxyphenyl)ethoxy | + | |||||||||||
| 15 | 22.79 min | Benzenebutyric acid, 2,3-dimethoxy- | + | |||||||||||
| 16 | 23.77 min | 1,2-Benzenedicarboxylic acid, butyl octyl ester | + | |||||||||||
| 17 | 23.77 min | Phthalic acid, hept-3-yl isobutyl ester | + | + | + | + | + | + | + | |||||
| 18 | 24.64 min | n-Hexadecanoic acid | + | + | + | + | + | + | + | + | + | + | + | + |
| 19 | 24.71min | Dibutyl phthalate | + | + | + | + | + | + | + | + | + | + | + | + |
| 20 | 26.51 min | Octadecanoic acid | + | + | + | + | + | + | + | + | + | + | + | + |
| 21 | 30.14 min | Phenol, 2,4-bis(1-phenylethyl)- | + | + | ||||||||||
| 22 | 31.79 min | Chromone,2-[2-[3,4,5-trimethoxyphenyl]ethenyl]-5,6,7,8-tetramethoxy-3-methyl- | + | |||||||||||
| 23 | 32.69 min | 1-Hydroxy-2-(2,3,4,6-tetra-O-acetyl-beta-D-glucopyranosyl)-9H-xanthene-3,6,7-triyl triacetate | + | + | + | + | + | + | ||||||
| Microbial Strains | Lignin Degradation% | Degradation Time | Lignin Load | |
|---|---|---|---|---|
| Bacteria | ||||
| Bacillus flexus [7] | 20% | 9 d | 0.4 g/L | |
| Cupriavidus basilensis B-8 [37] | 38% | 7 d | 0.5 g/L | |
| Citrobacter freundii [9] | 49% | 6 d | 600 ppm | |
| Fungi | ||||
| Cladosporium sp. Bio-1 [39] | 35% | 10 d | 2.0 g/L | |
| Penicillium chrysogenum [40] | 83.50% | 30 d | 1.0 g/L | |
| Phellinus sp. [41] | 36% | 10 d | 0.50% | |
| J-6 | 54% | 48 h | 0.5 g/L | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, W.; Ren, X.; Lei, Q.; Wang, L. Screening and Comparison of Lignin Degradation Microbial Consortia from Wooden Antiques. Molecules 2021, 26, 2862. https://doi.org/10.3390/molecules26102862
Zhang W, Ren X, Lei Q, Wang L. Screening and Comparison of Lignin Degradation Microbial Consortia from Wooden Antiques. Molecules. 2021; 26(10):2862. https://doi.org/10.3390/molecules26102862
Chicago/Turabian StyleZhang, Wen, Xueyan Ren, Qiong Lei, and Lei Wang. 2021. "Screening and Comparison of Lignin Degradation Microbial Consortia from Wooden Antiques" Molecules 26, no. 10: 2862. https://doi.org/10.3390/molecules26102862