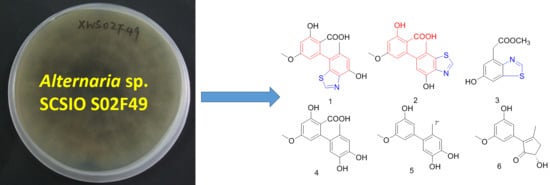
Two New Altenusin/Thiazole Hybrids and a New Benzothiazole Derivative from the Marine Sponge-Derived Fungus Alternaria sp. SCSIOS02F49
Abstract
:1. Introduction
2. Results and Discussion
2.1. Structural Elucidation
2.2. Biological Activities
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Fungal Materials
3.3. ITS Region Sequence and Phylogenetic Analysis
3.4. Fermentation and Extraction
3.5. Bioassay Protocols
3.5.1. DPPH Radical Scavenging Activity
3.5.2. COX-2 Inhibitory Activity Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Aly, A.H.; Debbab, A.; Proksch, P. Fungal endophytes: Unique plant inhabitants with great promises. Appl. J. Microbiol. Biotechnol. 2011, 90, 1829–1845. [Google Scholar] [CrossRef] [PubMed]
- Moreno, M.A.; Alonso, I.G.; De Santos, R.M.; Lacarra, T.G. Importancia del género Alternaria como productor de micotoxinas y agente causal de enfermedades humanas. Nutr. Hosp. 2012, 27, 1772–1781. [Google Scholar] [CrossRef]
- Chua, S.W.; Cornejo, A.; Van Eersel, J.; Stevens, C.H.; Vaca, I.; Mercedes, C.; Kassiou, M.; Gladbach, A.; Macmillan, A.; Lewis, L.; et al. The polyphenol altenusin inhibits in vitro fibrillization of tau and reduces induced tau pathology in primary neurons. ACS Chem. Neurosci. 2017, 8, 743–751. [Google Scholar] [CrossRef] [PubMed]
- Askin, S.; Bond, T.E.H.; Sorenson, A.E.; Moreau, M.J.J.; Antony, H.; Davis, R.A.; Schaeffer, P.M. Selective protein unfolding: A universal mechanism of action for the development of irreversible inhibitors. Chem. Commun. 2018, 54, 1738–1741. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.H.; Zhao, Z.M.; Li, S.Q.; Lu, X.H.; Jiang, M.X.; Lin, J.; An, Y.Q.; Xie, Y.; Xu, M.S.; Shen, W.B.; et al. Altenusin, a non-steroidal microbial metabolite, attenuates non-alcoholic fatty liver disease by activating the farnesoid X receptor. Mol. Pharmacol. 2017, 92, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Johann, S.; Rosa, L.H.; Rosa, C.A.; Perez, P.; Cisalpino, P.S.; Zani, C.L.; Cota, B.B. Antifungal activity of altenusin isolated from the endophytic fungus Alternaria sp. against the pathogenic fungus Paracoccidioides brasiliensis. Rev. Iberom. Micol. 2012, 29, 205–209. [Google Scholar] [CrossRef] [PubMed]
- Phaopongthai, J.; Wiyakrutta, S.; Meksuriyen, D.; Sriubolmas, N.; Suwanborirux, K. Azole-synergistic anti-candidal activity of altenusin, a biphenyl metabolite of the endophytic fungus Alternaria alternata isolated from Terminalia chebula Retz. J. Microbiol. 2013, 51, 821–828. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.Y.; Awakawa, T.; Noguchi, H.; Abe, I. Induced production of mycotoxins in an endophytic fungus from the medicinal plant Datura stramonium L. Bioorg. Med. Chem. Lett. 2012, 22, 6397–6400. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.X.; Bao, L.; Yang, X.L.; Guo, H.; Yang, R.N.; Ren, B.; Zhang, L.X.; Dai, H.Q.; Guo, L.D.; Liu, H.W. Polyketides with antimicrobial activity from the solid culture of an endolichenic fungus Ulocladium sp. Fitoterapia 2012, 83, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.Y.; Wu, Y.N.; Zhai, R.; Liu, Z.M.; Huang, X.S.; She, Z.G. Altenusin derivatives from mangrove endophytic fungus Alternaria sp. SK6YW3L. RSC Adv. 2016, 6, 72127–72132. [Google Scholar] [CrossRef]
- Ye, F.; Chen, G.D.; He, J.W.; Li, X.X.; Sun, X.; Guo, L.D.; Li, Y.; Gao, H. Xinshengin, the first altenusin with tetracyclic skeleton core from Phialophora spp. Tetrahedron Lett. 2014, 44, 4551–4554. [Google Scholar] [CrossRef]
- Nakanishi, S.; Toki, S.; Saitoh, Y.; Tsukuda, E.; Kawahara, K.; Ando, K.; Matsuda, Y. Isolation of myosin light chain kinase inhibitors from microorganisms: Dehydroaltenusin, altenusin, atrovenetinone, and cyclooctasulfur. Biosci. Biotechnol. Biochem. 1995, 59, 1333–1335. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.Y.; Yang, Q.; Xia, G.P.; Huang, H.B.; Li, H.X.; Ma, L.; Lu, Y.J.; He, L.; Xia, X.K.; She, Z.G. Polyketides with α-glucosidase inhibitory activity from a mangrove endophytic fungus, Penicillium sp. hn29-3b1. J. Nat. Prod. 2015, 3864, 1816–1822. [Google Scholar] [CrossRef] [PubMed]
- Pang, X.Y.; Lin, X.P.; Wang, P.; Zhou, X.F.; Yang, B.; Wang, J.F.; Liu, Y.H. Perylenequione derivatives with anticancer activities isolated from the marine sponge-derived fungus, Alternaria sp. SCSIO 41014. Mar. Drugs 2018, 16, 280. [Google Scholar] [CrossRef] [PubMed]
- Kumagai, M.; Nishikawa, K.; Matsuura, H.; Umezawa, T.; Matsuda, F.; Okino, T. Antioxidants from the Brown Alga Dictyopteris undulate. Molecules 2018, 23, 1214. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.Q.; Qin, X.C.; Lin, X.P.; Kaliyaperumal, K.; Zhou, X.F.; Liu, J.; Ju, Z.R.; Tu, Z.C.; Liu, Y.H. Sydoxanthone C and acremolin B produced by deep-sea-derived fungus Aspergillus sp. SCSIO Ind09F01. J. Antibiot. 2015, 68, 703–706. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Abeydeera, N.D.; Bale, S.; Pai, P.J.; Dorrestein, P.C.; Russell, D.H.; Ealick, S.E.; Begley, T.P. Saccharomyces cerevisiae THI4p is a suicide thiamine thiazole synthase. Nature 2011, 7370, 542–546. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds 1–6 are available from the authors. |

 ) and 1D NOESY (
) and 1D NOESY (  ) correlations of compounds 1 and 2.
) correlations of compounds 1 and 2.

| No. | 1 | 2 | 4 | |||||
|---|---|---|---|---|---|---|---|---|
| δH (CD3OD) | δH (J in Hz) | δC | δH (CD3OD) | δH (J in Hz) | δC | δH (J in Hz) | δC | |
| 1 | 143.2, C | 144.0, C | 145.0, C | |||||
| 2 | 106.8, C | 108.2, C | 108.8, C | |||||
| 3 | 163.4, C | 162.1, C | 161.6, C | |||||
| 4 | 6.47, d (2.4) | 6.57, d (2.4) | 100.7, CH | 6.42, d (2.0) | 6.50, d (2.0) | 100.2, CH | 6.44, d (2.7) | 99.6, CH |
| 5 | 163.3, C | 162.6, C | 163.0, C | |||||
| 6 | 6.17, d (2.4) | 6.24, d (2.4) | 109.2, CH | 6.14, d (2.0) | 6.19, d (2.0) | 108.8, CH | 6.10, d (2.7) | 108.9, CH |
| 7 | 171.6, C | 171.5, C | 171.6, C | |||||
| 8 | 3.80, s | 3.79, s | 55.6, CH3 | 3.79, s | 3.78, s | 55.5, CH3 | 3.76, s | 55.3, CH3 |
| 1′ | 126.0, C | 139.0, C | 132.4, C | |||||
| 2′ | 132.9, C | 118.1, C | 124.9, C | |||||
| 3′ | 6.77, s | 6.78, s | 113.3, CH | 135.8, C | 6.54, s | 116.6, CH | ||
| 4′ | 150.1, C | 141.4, C | 143.9, C | |||||
| 5′ | 140.3, C | 148.6, C | 142.1, C | |||||
| 6′ | 136.1, C | 6.70, s | 6.64, s | 112.0, CH | 6.42, s | 115.9, CH | ||
| 7′ | 2.13, s | 2.07, s | 19.6, CH3 | 2.19, s | 2.13, s | 18.7, CH3 | 1.86, s | 18.8, CH3 |
| 8′ | 8.83, s | 9.02, s | 152.0, CH | 8.97, s | 9.20, s | 152.5, CH | ||
| 3-OH | ||||||||
| 4′-OH | 11.96, br.s | 8.70, s | ||||||
| 5′-OH | 10.04, s | 8.65, s | ||||||
| COOH | 10.11, br.s | 11.53, br.s | ||||||
| No. | δH (J in Hz) | δc | No. | δH (J in Hz) | δc |
|---|---|---|---|---|---|
| 1 | 4.07, s | 36.8, CH2 | 7 | 7.34, d (1.5) | 105.5, CH |
| 2 | 9.05, s | 151.7, CH | 8 | 134.8, C | |
| 3 | 145.8, C | 9 | 171.2, C | ||
| 4 | 129.6, C | 10 | 3.59, s | 51.7, CH3 | |
| 5 | 6.91, d (1.5) | 116.8, CH | 6-OH | 9.86, br.s | |
| 6 | 155.4, C |
| Compounds | DPPH Free Radical Scavenging Activity | COX-2 Inhibitory Activity |
|---|---|---|
| 1 | >1000 | >30 |
| 2 | >1000 | >30 |
| 3 | >1000 | >30 |
| 4 | 10.7 ± 0.09 | >30 |
| 5 | 100.6 ± 0.025 | 9.5 ± 0.08 |
| 6 | >1000 | >30 |
| BHT | 170.3 ± 0.06 | - |
| Celecoxib | - | 0.008 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Chen, R.; Xu, J.; Tian, Y.; Xu, J.; Liu, Y. Two New Altenusin/Thiazole Hybrids and a New Benzothiazole Derivative from the Marine Sponge-Derived Fungus Alternaria sp. SCSIOS02F49. Molecules 2018, 23, 2844. https://doi.org/10.3390/molecules23112844
Chen Y, Chen R, Xu J, Tian Y, Xu J, Liu Y. Two New Altenusin/Thiazole Hybrids and a New Benzothiazole Derivative from the Marine Sponge-Derived Fungus Alternaria sp. SCSIOS02F49. Molecules. 2018; 23(11):2844. https://doi.org/10.3390/molecules23112844
Chicago/Turabian StyleChen, Yaping, Ruyan Chen, Jinhuai Xu, Yongqi Tian, Jiangping Xu, and Yonghong Liu. 2018. "Two New Altenusin/Thiazole Hybrids and a New Benzothiazole Derivative from the Marine Sponge-Derived Fungus Alternaria sp. SCSIOS02F49" Molecules 23, no. 11: 2844. https://doi.org/10.3390/molecules23112844