Metabolomics in NEC: An Updated Review
Abstract
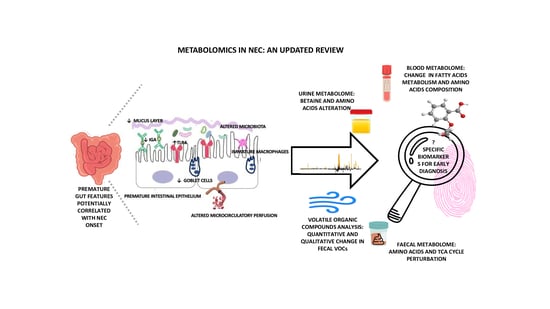
:1. Introduction
2. Intestine and the Microbiota of Preterm Newborns
| HIGHLIGHT: PREMATURE GUT FEATURES |
|
| HIGHLIGHT: IMMUNE ALTERATIONS CHARACTERISTIC OF THE PREMATURE INFANT’S GUT |
|
3. The Microbiota in Necrotizing Enterocolitis
4. Supporting Early Diagnosis of NEC: Potential Biomarkers
5. NEC Metabolomics Biomarkers
| HIGHLIGHT: MAIN METABOLOMIC ALTERATIONS |
|
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Meister, A.L.; Doheny, K.K.; Travagli, R.A. Necrotizing enterocolitis: It’s not all in the gut. Exp. Biol. Med. 2020, 245, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Warner, B.B.; Deych, E.; Zhou, Y.; Hall-Moore, C.; Weinstock, G.M.; Sodergren, E.; Shaikh, N.; Hoffmann, J.A.; Linneman, L.A.; Hamvas, A.; et al. Gut bacteria dysbiosis and necrotising enterocolitis in very low birthweight infants: A prospective case-control study. Lancet 2016, 387, 1928–1936. [Google Scholar] [CrossRef] [PubMed]
- Nanthakumar, N.; Meng, D.; Goldstein, A.M.; Zhu, W.; Lu, L.; Uauy, R.; Llanos, A.; Claud, E.C.; Walker, W.A. The mechanism of excessive intestinal inflammation in necrotizing enterocolitis: An immature innate immune response. PLoS ONE 2011, 6, e17776. [Google Scholar] [CrossRef] [PubMed]
- Rose, A.T.; Patel, R.M. A critical analysis of risk factors for necrotizing enterocolitis. Semin. Fetal Neonatal Med. 2018, 23, 374–379. [Google Scholar] [CrossRef] [PubMed]
- Gephart, S.M.; Effken, J.A.; McGrath, J.M.; Reed, P.G. Expert consensus building using e-Delphi for necrotizing enterocolitis risk assessment. J. Obstet. Gynecol. Neonatal Nurs. 2013, 42, 332–347. [Google Scholar] [CrossRef] [PubMed]
- Agakidou, E.; Agakidis, C.; Gika, H.; Sarafidis, K. Emerging Biomarkers for Prediction and Early Diagnosis of Necrotizing Enterocolitis in the Era of Metabolomics and Proteomics. Front. Pediatr. 2020, 8, 602255. [Google Scholar] [CrossRef] [PubMed]
- Samuels, N.; van de Graaf, R.A.; de Jonge, R.C.J.; Reiss, I.K.M.; Vermeulen, M.J. Risk factors for necrotizing enterocolitis in neonates: A systematic review of prognostic studies. BMC Pediatr. 2017, 17, 105. [Google Scholar] [CrossRef]
- Yu, Y.; Klemann, C.; Feng, X.; Ginzel, M.; Vieten, G.; Lacher, M.; Ure, B.; Kuebler, J.F. Increased inflammatory reaction to intestinal ischemia-reperfusion in neonatal versus adult mice. Eur. J. Pediatr. Surg. 2015, 25, 46–50. [Google Scholar] [CrossRef]
- Eaton, S.; Rees, C.M.; Hall, N.J. Current Research on the Epidemiology, Pathogenesis, and Management of Necrotizing Enterocolitis. Neonatology 2017, 111, 423–430. [Google Scholar] [CrossRef]
- Cotton, C.M. Modifiable Risk Factors in Necrotizing Enterocolitis. Clin. Perinatol. 2019, 46, 129–143. [Google Scholar] [CrossRef]
- Alexander, V.N.; Northrup, V.; Bizzarro, M.J. Antibiotic exposure in the newborn intensive care unit and the risk of necrotizing enterocolitis. J. Pediatr. 2011, 159, 392–397. [Google Scholar] [CrossRef] [PubMed]
- Esaiassen, E.; Fjalstad, J.W.; Juvet, L.K.; van den Anker, J.N.; Klingenberg, C. Antibiotic exposure in neonates and early adverse outcomes: A systematic review and meta-analysis. J. Antimicrob. Chemother. 2017, 72, 1858–1870. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, G.P.; Sylvester, K.G. Biomarker Discovery and Utility in Necrotizing Enterocolitis. Clin. Perinatol. 2019, 46, 1–17. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo, G.; Impellizzeri, P.; Marseglia, L.; Montalto, A.S.; Russo, T.; Salamone, I.; Falsaperla, R.; Corsello, G.; Romeo, C.; Gitto, E. Current status of laboratory and imaging diagnosis of neonatal necrotizing enterocolitis. Ital. J. Pediatr. 2018, 44, 84. [Google Scholar] [CrossRef] [PubMed]
- Thomaidou, A.; Chatziioannou, A.C.; Deda, O.; Benaki, D.; Gika, H.; Mikros, E.; Agakidis, C.; Raikos, N.; Theodoridis, G.; Sarafidis, K. A pilot case-control study of urine metabolomics in preterm neonates with necrotizing enterocolitis. J. Chromatogr. B 2019, 1117, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Gordon, P.V.; Swanson, J.R.; MacQueen, B.C.; Christensen, R.D. A critical question for NEC researchers: Can we create a consensus definition of NEC that facilitates research progress? Semin. Perinatol. 2017, 41, 7–14. [Google Scholar] [CrossRef]
- Gordon, P.V.; Swanson, J.R.; Attridge, J.T.; Clark, R. Emerging trends in acquired neonatal intestinal disease: Is. it time to abandon bell’s criteria? J. Perinatol. 2007, 27, 661–671. [Google Scholar] [CrossRef]
- Wang, K.; Tao, G.; Sun, Z.; Sylvester, K.G. Recent Potential Noninvasive Biomarkers in Necrotizing Enterocolitis. Gastroenterol. Res. Pract. 2019, 2019, 8413698. [Google Scholar] [CrossRef]
- Dessì, A.; Pintus, R.; Marras, S.; Cesare Marincola, F.; De Magistris, A.; Fanos, V. Metabolomics in necrotizing enterocolitis: The state of the art. Expert. Rev. Mol. Diagn. 2016, 16, 1053–1058. [Google Scholar] [CrossRef]
- Moschino, L.; Verlato, G.; Duci, M.; Cavicchiolo, M.E.; Guiducci, S.; Stocchero, M.; Giordano, G.; Fascetti Leon, F.; Baraldi, E. The Metabolome and the Gut Microbiota for the Prediction of Necrotizing Enterocolitis and Spontaneous Intestinal Perforation: A Systematic Review. Nutrients 2022, 14, 3859. [Google Scholar] [CrossRef]
- Tarracchini, C.; Milani, C.; Longhi, G.; Fontana, F.; Mancabelli, L.; Pintus, R.; Lugli, G.A.; Alessandri, G.; Anzalone, R.; Viappiani, A.; et al. Unraveling the Microbiome of Necrotizing Enterocolitis: Insights in Novel Microbial and Metabolomic Biomarkers. Microbiol. Spectr. 2021, 9, e0117621. [Google Scholar] [CrossRef] [PubMed]
- Hackam, D.J.; Afrazi, A.; Good, M.; Sodhi, C.P. Innate immune signaling in the pathogenesis of necrotizing enterocolitis. J. Immunol. Res. 2013, 2013, 475415. [Google Scholar] [CrossRef] [PubMed]
- Hackam, D.J.; Sodhi, C.P. Toll like receptor mediated intestinal inflammatory imbalance in the pathogenesis of necrotizing enterocolitis. Cell. Mol. Gastroenterol. Hepatol. 2018, 6, 229–238. [Google Scholar] [CrossRef]
- Niño, D.F.; Sodhi, C.P.; Hackam, D.J. Necrotizing enterocolitis: New insights into pathogenesis and mechanisms. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 590–600. [Google Scholar] [CrossRef] [PubMed]
- Sodhi, C.P.; Shi, X.H.; Richardson, W.M.; Grant, Z.S.; Shapiro, R.A.; Prindle, T., Jr.; Branca, M.; Russo, A.; Gribar, S.C.; Ma, C.; et al. Toll-like receptor-4 inhibits enterocyte proliferation via impaired beta-catenin signaling in necrotizing enterocolitis. Gastroenterology 2010, 138, 185–196. [Google Scholar] [CrossRef] [PubMed]
- Deplancke, B.; Gaskins, H.R. Microbial modulation of innate defense: Goblet cells and the intestinal mucus layer. Am. J. Clin. Nutr. 2001, 73, 1131S–1141S. [Google Scholar] [CrossRef] [PubMed]
- Ou, J.; Courtney, C.M.; Steinberger, A.E.; Tecos, M.E.; Warner, B.W. Nutrition in Necrotizing Enterocolitis and Following Intestinal Resection. Nutrients 2020, 12, 520. [Google Scholar] [CrossRef]
- Hackam, D.; Caplan, M. Necrotizing enterocolitis: Pathophysiology from a historical context. Semin. Pediatr. Surg. 2018, 27, 11–18. [Google Scholar] [CrossRef]
- Sodhi, C.P.; Wipf, P.; Yamaguchi, Y.; Fulton, W.B.; Kovler, M.; Niño, D.F.; Zhou, Q.; Banfield, E.; Werts, A.D.; Ladd, M.R.; et al. The human milk oligosaccharides 2’-fucosyllactose and 6’-sialyllactose protect against the development of necrotizing enterocolitis by inhibiting toll-like receptor 4 signaling. Pediatr. Res. 2021, 89, 91–101. [Google Scholar] [CrossRef]
- Soliman, A.; Michelsen, K.S.; Karahashi, H.; Lu, J.; Meng, F.J.; Qu, X.; Crother, T.R.; Rabizadeh, S.; Chen, S.; Caplan, M.S.; et al. Platelet-activating factor induces TLR4 expression in intestinal epithelial cells: Implication for the pathogenesis of necrotizing enterocolitis. PLoS ONE 2010, 5, e15044. [Google Scholar] [CrossRef]
- Caplan, M.S.; Sun, X.M.; Hseuh, W.; Hageman, J.R. Role of platelet activating factor and tumor necrosis factor-alpha in neonatal necrotizing enterocolitis. J. Pediatr. 1990, 116, 960–964. [Google Scholar] [CrossRef] [PubMed]
- Maheshwari, A.; Kelly, D.R.; Nicola, T.; Ambalavanan, N.; Jain, S.K.; Murphy-Ullrich, J.; Athar, M.; Shimamura, M.; Bhandari, V.; Aprahamian, C.; et al. TGF-β2 suppresses macrophage cytokine production and mucosal inflammatory responses in the developing intestine. Gastroenterology 2011, 140, 242–253. [Google Scholar] [CrossRef]
- Moles, L.; Gómez, M.; Heilig, H.; Bustos, G.; Fuentes, S.; de Vos, W.; Fernández, L.; Rodríguez, J.M.; Jiménez, E. Bacterial diversity in meconium of preterm neonates and evolution of their fecal microbiota during the first month of life. PLoS ONE 2013, 8, e66986. [Google Scholar] [CrossRef] [PubMed]
- Dahl, C.; Stigum, H.; Valeur, J.; Iszatt, N.; Lenters, V.; Peddada, S.; Bjørnholt, J.V.; Midtvedt, T.; Mandal, S.; Eggesbø, M. Preterm infants have distinct microbiomes not explained by mode of delivery, breastfeeding duration or antibiotic exposure. Int. J. Epidemiol. 2018, 47, 1658–1669. [Google Scholar] [CrossRef] [PubMed]
- Claud, E.C.; Keegan, K.P.; Brulc, J.M.; Lu, L.; Bartels, D.; Glass, E.; Chang, E.B.; Meyer, F.; Antonopoulos, D.A. Bacterial community structure and functional contributions to emergence of health or necrotizing enterocolitis in preterm infants. Microbiome 2013, 1, 20. [Google Scholar] [CrossRef] [PubMed]
- Bresesti, I.; Salvatore, S.; Valetti, G.; Baj, A.; Giaroni, C.; Agosti, M. The Microbiota-Gut Axis in Premature Infants: Physio-Pathological Implications. Cells 2022, 11, 379. [Google Scholar] [CrossRef] [PubMed]
- Arboleya, S.; Saturio, S.; Suárez, M.; Fernández, N.; Mancabelli, L.; de Los Reyes-Gavilán, C.G.; Ventura, M.; Solís, G.; Gueimonde, M. Donated Human Milk as a Determinant Factor for the Gut Bifidobacterial Ecology in Premature Babies. Microorganisms 2020, 8, 760. [Google Scholar] [CrossRef]
- La Rosa, P.S.; Warner, B.B.; Zhou, Y.; Weinstock, G.M.; Sodergren, E.; Hall-Moore, C.M.; Stevens, H.J.; Bennett, W.E., Jr.; Shaikh, N.; Linneman, L.A.; et al. Patterned progression of bacterial populations in the premature infant gut. Proc. Natl. Acad. Sci. USA 2014, 111, 12522–12527. [Google Scholar] [CrossRef]
- Chu, D.M.; Ma, J.; Prince, A.L.; Antony, K.M.; Seferovic, M.D.; Aagaard, K.M. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat. Med. 2017, 23, 314–326. [Google Scholar] [CrossRef]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef]
- Arboleya, S.; Sánchez, B.; Milani, C.; Duranti, S.; Solís, G.; Fernández, N.; de los Reyes-Gavilán, C.G.; Ventura, M.; Margolles, A.; Gueimonde, M. Intestinal microbiota development in preterm neonates and effect of perinatal antibiotics. J. Pediatr. 2015, 166, 538–544. [Google Scholar] [CrossRef] [PubMed]
- Chernikova, D.A.; Madan, J.C.; Housman, M.L.; Zain-Ul-Abideen, M.; Lundgren, S.N.; Morrison, H.G.; Sogin, M.L.; Williams, S.M.; Moore, J.H.; Karagas, M.R.; et al. The premature infant gut microbiome during the first 6 weeks of life differs based on gestational maturity at birth. Pediatr. Res. 2018, 84, 71–79. [Google Scholar] [CrossRef]
- Korpela, K.; Blakstad, E.W.; Moltu, S.J.; Strømmen, K.; Nakstad, B.; Rønnestad, A.E.; Brække, K.; Iversen, P.O.; Drevon, C.A.; de Vos, W. Intestinal microbiota development and gestational age in preterm neonates. Sci. Rep. 2018, 8, 2453. [Google Scholar] [CrossRef] [PubMed]
- Gregory, K.E.; Samuel, B.S.; Houghteling, P.; Shan, G.; Ausubel, F.M.; Sadreyev, R.I.; Walker, W.A. Influence of maternal breast milk ingestion on acquisition of the intestinal microbiome in preterm infants. Microbiome 2016, 4, 68. [Google Scholar] [CrossRef] [PubMed]
- Cong, X.; Judge, M.; Xu, W.; Diallo, A.; Janton, S.; Brownell, E.A.; Maas, K.; Graf, J. Influence of Feeding Type on Gut Microbiome Development in Hospitalized Preterm Infants. Nurs. Res. 2017, 66, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Ford, S.L.; Lohmann, P.; Preidis, G.A.; Gordon, P.S.; O’Donnell, A.; Hagan, J.; Venkatachalam, A.; Balderas, M.; Luna, R.A.; Hair, A.B. Improved feeding tolerance and growth are linked to increased gut microbial community diversity in very-low-birth-weight infants fed mother’s own milk compared with donor breast milk. Am. J. Clin. Nutr. 2019, 109, 1088–1097. [Google Scholar] [CrossRef] [PubMed]
- Parra-Llorca, A.; Gormaz, M.; Alcántara, C.; Cernada, M.; Nuñez-Ramiro, A.; Vento, M.; Collado, M.C. Preterm Gut Microbiome Depending on Feeding Type: Significance of Donor Human Milk. Front. Microbiol. 2018, 9, 1376. [Google Scholar] [CrossRef]
- Quigley, M.; Embleton, N.D.; McGuire, W. Formula versus donor breast milk for feeding preterm or low birth weight infants. Cochrane Database Syst. Rev. 2019, 7, CD002971. [Google Scholar] [CrossRef]
- Altobelli, E.; Angeletti, P.M.; Verrotti, A.; Petrocelli, R. The Impact of Human Milk on Necrotizing Enterocolitis: A Systematic Review and Meta-Analysis. Nutrients 2020, 12, 1322. [Google Scholar] [CrossRef]
- Mestecky, J. Homeostasis of the mucosal immune system: Human milk and lactation. Adv. Exp. Med. Biol. 2001, 501, 197–205. [Google Scholar]
- Lin, P.W.; Stoll, B.J. Necrotising enterocolitis. Lancet 2006, 368, 1271–1283. [Google Scholar] [CrossRef] [PubMed]
- Vongbhavit, K.; Underwood, M.A. Prevention of Necrotizing Enterocolitis Through Manipulation of the Intestinal Microbiota of the Premature Infant. Clin. Ther. 2016, 38, 716–732. [Google Scholar] [CrossRef] [PubMed]
- Pammi, M.; Cope, J.; Tarr, P.I.; Warner, B.B.; Morrow, A.L.; Mai, V.; Gregory, K.E.; Kroll, J.S.; McMurtry, V.; Ferris, M.J.; et al. Intestinal dysbiosis in preterm infants preceding necrotizing enterocolitis: A systematic review and meta-analysis. Microbiome 2017, 5, 31. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, R.; Rani, A.; Metwally, A.; McGee, H.S.; Perkins, D.L. Analysis of the microbiome: Advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem. Biophys. Res. Commun. 2016, 469, 967–977. [Google Scholar] [CrossRef] [PubMed]
- Dobbler, P.T.; Procianoy, R.S.; Mai, V.; Silveira, R.C.; Corso, A.L.; Rojas, B.S.; Roesch, L.F.W. Low Microbial Diversity and Abnormal Microbial Succession Is Associated with Necrotizing Enterocolitis in Preterm Infants. Front. Microbiol. 2017, 8, 2243. [Google Scholar] [CrossRef]
- Zhou, Y.; Shan, G.; Sodergren, E.; Weinstock, G.; Walker, W.A.; Gregory, K.E. Longitudinal analysis of the premature infant intestinal microbiome prior to necrotizing enterocolitis: A case-control study. PLoS ONE 2015, 10, e0118632. [Google Scholar] [CrossRef] [PubMed]
- Ward, D.V.; Scholz, M.; Zolfo, M.; Taft, D.H.; Schibler, K.R.; Tett, A.; Segata, N.; Morrow, A.L. Metagenomic Sequencing with Strain-Level Resolution Implicates Uropathogenic E. coli in Necrotizing Enterocolitis and Mortality in Preterm Infants. Cell Rep. 2016, 14, 2912–2924. [Google Scholar] [CrossRef]
- Brehin, C.; Dubois, D.; Dicky, O.; Breinig, S.; Oswald, E.; Serino, M. Evolution of Gut Microbiome and Metabolome in Suspected Necrotizing Enterocolitis: A Case-Control Study. J. Clin. Med. 2020, 9, 2278. [Google Scholar] [CrossRef]
- McMurtry, V.E.; Gupta, R.W.; Tran, L.; Blanchard, E.E., IV; Penn, D.; Taylor, C.M.; Ferris, M.J. Bacterial diversity and Clostridia abundance decrease with increasing severity of necrotizing enterocolitis. Microbiome 2015, 3, 11. [Google Scholar] [CrossRef]
- Patel, R.M.; Ferguson, J.; McElroy, S.J.; Khashu, M.; Caplan, M.S. Defining necrotizing enterocolitis: Current difficulties and future opportunities. Pediatr. Res. 2020, 88 (Suppl. S1), 10–15. [Google Scholar] [CrossRef]
- Garstin, W.I.; Boston, V.E. Sequential assay of expired breath hydrogen as a means of predicting necrotizing enterocolitis in susceptible infants. J. Pediatr. Surg. 1987, 22, 208–210. [Google Scholar] [PubMed]
- Cheu, H.W.; Brown, D.R.; Rowe, M.I. Breath hydrogen excretion as a screening test for the early diagnosis of necrotizing enterocolitis. Am. J. Dis. Child. 1989, 143, 156–159. [Google Scholar] [CrossRef] [PubMed]
- Lin, J. Too much short chain fatty acids cause neonatal necrotizing enterocolitis. Med. Hypotheses 2004, 62, 291–293. [Google Scholar] [CrossRef]
- Bardanzellu, F.; Fanos, V. How could metabolomics change pediatric health? Ital. J. Pediatr. 2020, 46, 37. [Google Scholar] [CrossRef] [PubMed]
- Garner, C.E.; Ewer, A.K.; Elasouad, K.; Power, F.; Greenwood, R.; Ratcliffe, N.M.; Costello, B.d.L.; Probert, C.S. Analysis of faecal volatile organic compounds in preterm infants who develop necrotising enterocolitis: A pilot study. J. Pediatr. Gastroenterol. Nutr. 2009, 49, 559–565. [Google Scholar] [CrossRef] [PubMed]
- Morrow, A.L.; Lagomarcino, A.J.; Schibler, K.R.; Taft, D.H.; Yu, Z.; Wang, B.; Altaye, M.; Wagner, M.; Gevers, D.; Ward, D.V.; et al. Early microbial and metabolomic signatures predict later onset of necrotizing enterocolitis in preterm infants. Microbiome 2013, 1, 13. [Google Scholar] [CrossRef] [PubMed]
- Wilcock, A.; Begley, P.; Stevens, A.; Whatmore, A.; Victor, S. The metabolomics of necrotising enterocolitis in preterm babies: An exploratory study. J. Matern. Fetal Neonatal Med. 2016, 29, 758–762. [Google Scholar] [CrossRef]
- de Meij, T.G.; van der Schee, M.P.; Berkhout, D.J.; van de Velde, M.E.; Jansen, A.E.; Kramer, B.W.; van Weissenbruch, M.M.; van Kaam, A.H.; Andriessen, P.; van Goudoever, J.B.; et al. Early Detection of Necrotizing Enterocolitis by Fecal Volatile Organic Compounds Analysis. J. Pediatr. 2015, 167, 562–567.e1. [Google Scholar] [CrossRef]
- Stewart, C.J.; Nelson, A.; Treumann, A.; Skeath, T.; Cummings, S.P.; Embleton, N.D.; Berrington, J.E. Metabolomic and proteomic analysis of serum from preterm infants with necrotising entercolitis and late-onset sepsis. Pediatr. Res. 2016, 79, 425–431. [Google Scholar] [CrossRef]
- Stewart, C.J.; Embleton, N.D.; Marrs, E.C.; Smith, D.P.; Nelson, A.; Abdulkadir, B.; Skeath, T.; Petrosino, J.F.; Perry, J.D.; Berrington, J.E.; et al. Temporal bacterial and metabolic development of the preterm gut reveals specific signatures in health and disease. Microbiome 2016, 4, 67. [Google Scholar] [CrossRef]
- Sylvester, K.G.; Kastenberg, Z.J.; Moss, R.L.; Enns, G.M.; Cowan, T.M.; Shaw, G.M.; Stevenson, D.K.; Sinclair, T.J.; Scharfe, C.; Ryckman, K.K.; et al. Acylcarnitine Profiles Reflect Metabolic Vulnerability for Necrotizing Enterocolitis in Newborns Born Premature. J. Pediatr. 2017, 181, 80–85.e1. [Google Scholar] [CrossRef] [PubMed]
- Wandro, S.; Osborne, S.; Enriquez, C.; Bixby, C.; Arrieta, A.; Whiteson, K. The Microbiome and Metabolome of Preterm Infant Stool Are Personalized and Not Driven by Health Outcomes, Including Necrotizing Enterocolitis and Late-Onset Sepsis. mSphere 2018, 3, e00104–e00118. [Google Scholar] [CrossRef]
- Rusconi, B.; Jiang, X.; Sidhu, R.; Ory, D.S.; Warner, B.B.; Tarr, P.I. Gut Sphingolipid Composition as a Prelude to Necrotizing Enterocolitis. Sci. Rep. 2018, 8, 10984. [Google Scholar] [CrossRef]
- Probert, C.; Greenwood, R.; Mayor, A.; Hughes, D.; Aggio, R.; Jackson, R.E.; Simcox, L.; Barrow, H.; García-Finana, M.; Ewer, A.K. Faecal volatile organic compounds in preterm babies at risk of necrotising enterocolitis: The DOVE study. Arch. Dis. Child. Fetal Neonatal Ed. 2020, 105, 474–479. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, T.J.; Ye, C.; Chen, Y.; Zhang, D.; Li, T.; Ling, X.B.; Cohen, H.J.; Shaw, G.M.; Stevenson, D.K.; Chace, D.; et al. Progressive Metabolic Dysfunction and Nutritional Variability Precedes Necrotizing Enterocolitis. Nutrients 2020, 12, 1275. [Google Scholar] [CrossRef] [PubMed]
- Picaud, J.C.; De Magistris, A.; Mussap, M.; Corbu, S.; Dessì, A.; Noto, A.; Fanos, V.; Cesare Marincola, F. Urine NMR Metabolomics Profile of Preterm Infants with Necrotizing Enterocolitis Over the First Two Months of Life: A Pilot Longitudinal Case-Control Study. Front. Mol. Biosci. 2021, 8, 680159. [Google Scholar] [CrossRef]
- Thomaidou, A.; Deda, O.; Begou, O.; Lioupi, A.; Kontou, A.; Gika, H.; Agakidou, E.; Theodoridis, G.; Sarafidis, K. A Prospective, Case-Control Study of Serum Metabolomics in Neonates with Late-Onset Sepsis and Necrotizing Enterocolitis. J. Clin. Med. 2022, 11, 5270. [Google Scholar] [CrossRef]
- Deianova, N.; El Manouni El Hassani, S.; Struijs, E.A.; Jansen, E.E.W.; Bakkali, A.; van de Wiel, M.A.; de Boode, W.P.; Hulzebos, C.V.; van Kaam, A.H.; Kramer, B.W.; et al. Fecal amine metabolite analysis before onset of severe necrotizing enterocolitis in preterm infants: A prospective case-control study. Sci. Rep. 2022, 12, 12310. [Google Scholar] [CrossRef]
- Du, T.T.; Liu, X.C.; He, Y.; Gao, X.; Liu, Z.Z.; Wang, Z.L.; Li, L.Q. Changes of gut microbiota and tricarboxylic acid metabolites may be helpful in early diagnosis of necrotizing enterocolitis: A pilot study. Front. Microbiol. 2023, 14, 1119981. [Google Scholar] [CrossRef]


| Authors | Patients | Samples | Technique | Results |
|---|---|---|---|---|
| Garner et al. [65] 2009 | 6 NEC and 7 matched controls | Faecal VOCs | SPME GC/MS | ↓ VOCs in the days before and after the NEC diagnosis 2-ethylhexyl acetic ester, decanoic acid ethyl ester, dodecanoic acid ethyl ester, and hexadecanoic acid ethyl ester consistently absent from all NEC faecal samples in the 4 days before the onset of the diseases |
| Morrow et al. [66] (2013) | 11 cases vs. 21 controls | Urine | 1H-NMR | High urinary ratio alanine/histidine provided a good prediction of NEC Alanine and histidine: discriminant metabolites between NEC Bell Stage I and NEC Bell Stage II |
| Wilcock et al. [67] (2015) | 7 NEC vs. 8 preterms without NEC. | Serum | GC-MS | No metabolomics differences in NEC cases after complete enteral nutrition is introduced. ↑ ‘IL-1b before clinical diagnosis |
| De Meij et al. [68] 2015 | 13 NEC, 14 matched controls and 31 sepsis | Faecal VOCs | e-Nose | NEC fecal VOCs profiles could be discriminated from controls, from 2–3 days before clinical symptoms onset |
| Stewart et al. [69] (2015) | 6 NEC, 4 LOS vs. 9 controls | Serum | UPLC-MS | No significant protein among the groups |
| Stewart et al. [70] (2016) | 16 preterm infants (6 NEC and 10 matched controls) | Feces | UPLC-MS | Discriminant metabolites involved into C-21 steroid hormones, linoleate, prostaglandines, leucotrienes pathways |
| Sylvester et al. [71] (2016) | 116 NEC vs. 22876 controls | Blood | MS/MS | Acylcarnitines associated at higher risk of NEC |
| Wandro et al. [72] (2018) | 32 VLBW: 3 NEC 8 OS 21 controls | Feces | GC-MS | No discriminant metabolites found |
| Rusconi et al. [73] (2018) | 9 NEC (stage II–III Bell) vs. 19 controls for broad range metabolomics 23 NEC (stage II–III Bell) vs. 46 controls for targeted metabolomics | Feces | UPLC-MS/MS | ↑ Sphingomyelins ↓ Ceramides |
| Thomaidou et al. [15] (2019) | 5 NEC stage I, 10 NEC stage II–III vs. 30 controls | Urine | 1H-NMR HILIC-UHPLC-MS/MS | 25 discriminant metabolites in NEC: ↓ alanine, tirosine, asparagine, proline, betaine, citrate, fumarate, riboflavine, polyols ↑ phenylalanine, arginine, pyridoxine |
| Probert et al. [74] 2019 | 32 NEC vs. frequency-matched controls without NEC | Faecal VOCs | SPME GC/MS | Change in fecal VOCs between NEC and controls up to 3–4 days before clinical diagnosis |
| Brehin et al. [58] (2020) | 32 preterms: 11 NEC stage I vs. 21 controls | Feces | 1H-NMR | ↓ serine in NEC-1 ↓ ethanol e leucine in NEC-1 in the 2°month of life. |
| Sinclair et al. [75] (2020) | 887 preterms: 73 NEC vs. 814 controlls | Blood | MS-MS | ↓ Citrulline/phenylalanine ratio, 3-idrossioleilcarnitine ↑ phenylalanine/tyrosine ratio and octanoilcarnitine/decanoilcarnitine ratio in cases. |
| Picaud et al. [76] (2021) | 18 VLBW: 6 NEC with food intolerance (group 1, NEC), 6 with food intolerance without NEC (group 2, FI) and 6 controls (group 3, GDT) | Urine | 1H-NMR | Discriminant metabolites: lattate, betaine, myo-inositol, urea, creatinine e N,N-dimetilglicine between LOS NEC and group 3 |
| Thomaidou et al., 2022 [77] | 17 NEC vs. 15 LOS vs. 16 controls | Blood | LC-QTOF-MS | L-carnitine discriminant metabolite in NEC and LOS NEC ↓ PC (16:0/0:0) o LysoPC (16:0/0:0), PC (18:1/0:0) o LysoPC (18:1/0:0) ↑ PC (20:4/0:0) |
| Deianova et al (2022) [78] | 31 preterms (<30 weeks) with severe NEC (15 NEC IIIA, 16 NEC IIIB) and 31 controls; | Feces | HPLC | ↑ isoleucine, leucine, methionine, phenylalanina and valine ↓ ratios of lisine and ethanolamine in preclinical samples of NEC newborns |
| Du et al (2023) [79] | 16 NEC vs. 16 controls | Feces | MRM UHPLC LC-MS | ↑ succinate, L-malic acid and oxalacetate in NEC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bosco, A.; Piu, C.; Picciau, M.E.; Pintus, R.; Fanos, V.; Dessì, A. Metabolomics in NEC: An Updated Review. Metabolites 2024, 14, 14. https://doi.org/10.3390/metabo14010014
Bosco A, Piu C, Picciau ME, Pintus R, Fanos V, Dessì A. Metabolomics in NEC: An Updated Review. Metabolites. 2024; 14(1):14. https://doi.org/10.3390/metabo14010014
Chicago/Turabian StyleBosco, Alice, Claudia Piu, Marta Emanuela Picciau, Roberta Pintus, Vassilios Fanos, and Angelica Dessì. 2024. "Metabolomics in NEC: An Updated Review" Metabolites 14, no. 1: 14. https://doi.org/10.3390/metabo14010014