Isolation and Biological Activity of Iezoside and Iezoside B, SERCA Inhibitors from Floridian Marine Cyanobacteria
Abstract
:1. Introduction
2. Results
2.1. Isolation and Structure Elucidation
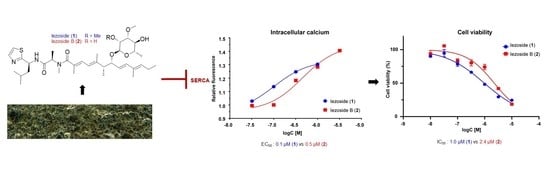
2.2. Cytotoxicity and Effect on Intracellular Calcium
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Biological Material
3.3. Extraction and Isolation
3.4. Cell Viability Assay
3.5. HeLa Cell Morphology Study
3.6. Intracellular Calcium Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carafoli, E. Intracellular Calcium Homeostasis. Annu. Rev. Biochem. 1987, 56, 395–433. [Google Scholar] [CrossRef] [PubMed]
- Carafoli, E. Calcium signaling: A tale for all seasons. Proc. Natl. Acad. Sci. USA 2002, 99, 1115–1122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blaustein, M.P.; Lederer, W.J. Sodium/calcium exchange: Its physiological implications. Physiol. Rev. 1999, 79, 763–854. [Google Scholar] [CrossRef] [PubMed]
- Denmeade, S.R.; Isaacs, J.T. The SERCA pump as a therapeutic target: Making a “smart bomb” for prostate cancer. Cancer Biol. Ther. 2005, 4, 21–29. [Google Scholar] [CrossRef] [Green Version]
- Chemaly, E.R.; Troncone, L.; Lebeche, D. SERCA control of cell death and survival. Cell Calcium 2018, 69, 46–61. [Google Scholar] [CrossRef] [PubMed]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2021, 38, 362–413. [Google Scholar] [CrossRef]
- Salvador-Reyes, L.A.; Luesch, H. Biological targets and mechanisms of action of natural products from marine cyanobacteria. Nat. Prod. Rep. 2015, 32, 478–503. [Google Scholar] [CrossRef] [Green Version]
- Mi, Y.; Zhang, J.; He, S.; Yan, X. New peptides isolated from marine cyanobacteria, an overview over the past decade. Mar. Drugs 2017, 15, 132. [Google Scholar] [CrossRef] [Green Version]
- Teruya, T.; Sasaki, H.; Kitamura, K.; Nakayama, T.; Suenaga, K. Biselyngbyaside, a macrolide glycoside from the marine Cyanobacterium Lyngbya sp. Org. Lett. 2009, 11, 2421–2424. [Google Scholar] [CrossRef]
- Morita, M.; Ohno, O.; Teruya, T.; Yamori, T.; Inuzuka, T.; Suenaga, K. Isolation and structures of biselyngbyasides B, C, and D from the marine cyanobacterium Lyngbya sp., and the biological activities of biselyngbyasides. Tetrahedron 2012, 68, 5984–5990. [Google Scholar] [CrossRef]
- Iwasaki, A.; Ohno, O.; Sumimoto, S.; Suda, S.; Suenaga, K. Kurahyne, an acetylene-containing lipopeptide from a marine cyanobacterial assemblage of Lyngbya sp. RSC Adv. 2014, 4, 12840–12843. [Google Scholar] [CrossRef] [Green Version]
- Iwasaki, A.; Ohno, O.; Katsuyama, S.; Morita, M.; Sasazawa, Y.; Dan, S.; Simizu, S.; Yamori, T.; Suenaga, K. Identification of a molecular target of kurahyne, an apoptosis-inducing lipopeptide from marine cyanobacterial assemblages. Bioorganic Med. Chem. Lett. 2015, 25, 5295–5298. [Google Scholar] [CrossRef] [PubMed]
- Kurisawa, N.; Iwasaki, A.; Teranuma, K.; Dan, S.; Toyoshima, C.; Hashimoto, M.; Suenaga, K. Structural Determination, Total Synthesis, and Biological Activity of Iezoside, a Highly Potent Ca2+-ATPase Inhibitor from the Marine Cyanobacterium Leptochromothrix valpauliae. J. Am. Chem. Soc. 2022, 144, 11019–11032. [Google Scholar] [CrossRef] [PubMed]
- Lytton, J.; Westlin, M.; Hanley, M.R. Thapsigargin inhibits the sarcoplasmic or endoplasmic reticulum Ca-ATPase family of calcium pumps. J. Biol. Chem. 1991, 266, 17067–17071. [Google Scholar] [CrossRef]
- Sehgal, P.; Szalai, P.; Olesen, C.; Praetorius, H.A.; Nissen, P.; Christensen, S.B.; Engedal, N.; Møller, J.V. Inhibition of the sarco/endoplasmic reticulum (ER) Ca2-ATPase by thapsigargin analogs induces cell death via ER Ca2 depletion and the unfolded protein response. J. Biol. Chem. 2017, 292, 19656–19673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seidler, N.W.; Jona, I.; Vegh, M.; Martonosi, A. Cyclopiazonic acid is a specific inhibitor of the Ca2+-ATPase of sarcoplasmic reticulum. J. Biol. Chem. 1989, 264, 17816–17823. [Google Scholar] [CrossRef]
- Uyama, Y.; Imaizumi, Y.; Watanabe, M. Cyclopiazonic acid, an inhibitor of Ca2+-ATPase in sarcoplasmic reticulum, increases excitability in ileal smooth muscle. Br. J. Pharmacol. 1993, 110, 565–572. [Google Scholar] [CrossRef] [Green Version]
- Doan, N.T.Q.; Paulsen, E.S.; Sehgal, P.; Møller, J.V.; Nissen, P.; Denmeade, S.R.; Isaacs, J.T.; Dionne, C.A.; Christensen, S.B. Targeting thapsigargin towards tumors. Steroids 2015, 97, 2–7. [Google Scholar] [CrossRef] [Green Version]
- Jaskulska, A.; Janecka, A.E.; Gach-Janczak, K. Thapsigargin—From Traditional Medicine to Anticancer Drug. Int. J. Mol. Sci. 2020, 22, 4. [Google Scholar] [CrossRef]
- Isaacs, J.T.; Brennen, W.N.; Christensen, S.B.; Denmeade, S.R. Mipsagargin: The beginning—Not the end—Of thapsigargin prodrug-based cancer therapeutics. Molecules 2021, 26, 7469. [Google Scholar] [CrossRef]
- Andersen, T.; López, C.; Manczak, T.; Martinez, K.; Simonsen, H. Thapsigargin—From Thapsia L. to Mipsagargin. Molecules 2015, 20, 6113–6127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morita, M.; Ogawa, H.; Ohno, O.; Yamori, T.; Suenaga, K.; Toyoshima, C. Biselyngbyasides, cytotoxic marine macrolides, are novel and potent inhibitors of the Ca2+ pumps with a unique mode of binding. FEBS Lett. 2015, 589, 1406–1411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, D.; Putra, M.Y.; Ye, T.; Paul, V.J.; Luesch, H. Isolation, Structure Elucidation and Biological Evaluation of Lagunamide D: A New Cytotoxic Macrocyclic Depsipeptide from Marine Cyanobacteria. Mar. Drugs 2019, 17, 83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, D.; Ratnayake, R.; Atanasova, K.R.; Paul, V.J.; Luesch, H. Targeted and functional genomics approaches to the mechanism of action of lagunamide D, a mitochondrial cytotoxin from marine cyanobacteria. Biochem. Pharmacol. 2023, 213, 115608. [Google Scholar] [CrossRef]






| Iezoside (1) | Iezoside B (2) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Position | δH, mult., J (Hz) a | δC b | HMBC | COSY | NOESY | δH, mult., J (Hz) a | δC b | HMBC | COSY | NOESY |
| 1 | 7.70, d (3.3) | 142.1 | 2, 3 | 2 | 7.70, d (3.3) | 142.2 | 2, 3 | 2 | ||
| 2 | 7.58, d (3.3) | 119.1 | 1 | 1 | 7.58, d (3.3) | 119.4 | 1 | |||
| 3 | 173.6 | 173.6 | ||||||||
| 4 | 5.19, m | 49.2 | 3, 5 | 5, 8-NH | 5.19, m | 49.2 | 3, 5 | 5, 8-NH | ||
| 5 | 1.77, m | 43.2 | 4 | 4, 6 | 1.77, m | 43.2 | 3, 4, 6, 7, 8 | 4, 6 | ||
| 6 | 1.65, m | 24.4 | 7, 8 | 1.65, m | 24.3 | 6, 7, 8 | ||||
| 7 | 0.90, d (6.7) | 23.0 | 5, 6, 8 | 6 | 0.91, d (7.0) | 22.9 | 5, 6, 8 | 6 | ||
| 8 | 0.87, d (6.5) | 21.4 | 5, 6, 7 | 6 | 0.87, d (6.5) | 21.3 | 5, 6, 7 | 6 | ||
| NH | 8.58, d (8.4) | 9 | 4 | 8.56, d (8.5) | 9 | 4 | ||||
| 9 | 170.5 | 170.6 | ||||||||
| 10 | 4.83, br | c | 11 | 4.80, br | c | |||||
| 11 | 1.29, br d (7.2) | c | 9 | 10 | 1.28, br d (7.1) | c | 9 | 12 | ||
| 12 | 2.78, s | c | 13 | 2.78, s | c | 13 | 11 | |||
| 13 | 173.0 | 173.1 | ||||||||
| 14 | 130.2 | 130.4 | ||||||||
| 15 | 6.32, d (11.2) | 121.0 | 13, 17, 26 | 16 | 27 | 6.31, d (11.2) | 121.0 | 13, 26 | 16 | 27 |
| 16 | 6.09, d (11.2) | 121.1 | 14, 18, 27 | 15 | 18, 26, 28 | 6.10, d (11.2) | 121.0 | 18, 27 | 15 | 18, 26 |
| 17 | 142.6 | 142.2 | ||||||||
| 18 | 2.43, p (6.7) | 47.7 | 16, 17, 19, 20 | 19, 28 | 2.43, p (6.7) | 47.7 | 16, 17, 19, 28 | 19, 28 | 16 | |
| 19 | 3.95, t (8.0) | 78.2 | 18, 21, 28, 30 | 18, 20 | 27 | 3.91, t (8.0) | 78.0 | 18, 21, 28, 30 | 18, 20 | 21, 27, 28, 30 |
| 20 | 5.29, dd (15.7, 8.4) | 124.4 | 22 | 19, 21 | 29 | 5.26, dd (15.7, 8.4) | 124.6 | 22 | 19, 21 | 29 |
| 21 | 6.14, d (15.7) | 138.1 | 19, 22, 23, 29 | 20 | 6.11, d (15.7) | 138.0 | 19, 22, 23, 29 | 20 | 19 | |
| 22 | 131.8 | 131.8 | ||||||||
| 23 | 5.49, t (7.2) | 134.7 | 21, 24, 29 | 24 | 24 | 5.48, t (7.2) | 134.9 | 21, 24, 25, 29 | 24 | 24 |
| 24 | 2.08, m | 21.0 | 22, 23, 25 | 23, 25 | 2.07, m | 20.9 | 22, 23, 25 | 23, 25 | 23, 29 | |
| 25 | 0.93, t (7.5) | 14.0 | 23, 24 | 24 | 0.93, t (7.5) | 13.8 | 23, 24 | 24 | ||
| 26 | 1.83, s | 14.3 | 13, 14 | 1.82, s | 14.6 | 13, 14 | 16 | |||
| 27 | 1.65, s | 14.8 | 16, 17, 18 | 1.64, s | 14.3 | 16, 17, 18 | 15, 19 | |||
| 28 | 1.10, d (7.0) | 15.4 | 17, 18, 19 | 18 | 1.10, d (7.0) | 15.5 | 17, 18, 19 | 18 | ||
| 29 | 1.64, s | 12.1 | 21, 22 | 1.63, s | 12.1 | 21, 22, 23 | 20 | 24 | ||
| 30 | 4.69, d (1.8) | 93.0 | 19, 32, 34 | 31 | 31 | 4.55, br s | 96.2 | 19, 31, 32, 34 | 31 | 19, 31 |
| 31 | 3.47, m | 76.7 | 32, 33, 36 | 30 | 30 | 3.75, m | 66.6 | 30, 32 | 30, 32 | |
| 31-OH | - | 4.71, d (4.5) | 30, 31, 32 | 37 | ||||||
| 32 | 3.21, m | 81.0 | 31 | 34 | 3.12, dd (9.3, 3.2) | 80.7 | 33, 36 | 31, 33 | 31 | |
| 33 | 3.21, m | 70.9 | 3.28, m | 70.6 | 34, 35 | 32, 34 | ||||
| 33-OH | 4.97, br | 4.87, d (5.5) | 33, 34, 35 | |||||||
| 34 | 3.47, m | 69.0 | 32, 33 | 35 | 32, 36 | 3.48, dq (11.8, 6.0) | 68.9 | 33, 35 | 35 | |
| 35 | 1.12, d (6.2) | 18.0 | 33, 34 | 34 | 1.13, d (6.0) | 17.9 | 33, 34 | 34 | 34 | |
| 36 | 3.27, m | 58.3 | 31 | 34 | - | |||||
| 37 | 3.32, m | 56.8 | 32 | 3.31, m | 56.2 | 32 | ||||
| A549 | HeLa | HSAEC | Primary Cervical Epithelial Cells | |||
|---|---|---|---|---|---|---|
| IC50 | EC50 | IC50 | EC50 | IC50 | IC50 | |
| Iezoside (1) | 1.5 ± 0.5 | ~0.3 | 1.0 ± 0.4 | ~0.1 | >10 | 0.02 ± 0.01 |
| Iezoside B (2) | 3.0 ± 0.9 | ~0.6 | 2.4 ± 0.9 | ~0.5 | >10 | 0.05 ± 0.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kokkaliari, S.; Luo, D.; Paul, V.J.; Luesch, H. Isolation and Biological Activity of Iezoside and Iezoside B, SERCA Inhibitors from Floridian Marine Cyanobacteria. Mar. Drugs 2023, 21, 378. https://doi.org/10.3390/md21070378
Kokkaliari S, Luo D, Paul VJ, Luesch H. Isolation and Biological Activity of Iezoside and Iezoside B, SERCA Inhibitors from Floridian Marine Cyanobacteria. Marine Drugs. 2023; 21(7):378. https://doi.org/10.3390/md21070378
Chicago/Turabian StyleKokkaliari, Sofia, Danmeng Luo, Valerie J. Paul, and Hendrik Luesch. 2023. "Isolation and Biological Activity of Iezoside and Iezoside B, SERCA Inhibitors from Floridian Marine Cyanobacteria" Marine Drugs 21, no. 7: 378. https://doi.org/10.3390/md21070378