Bacterial Survey in the Guts of Domestic Silkworms, Bombyx mori L.
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
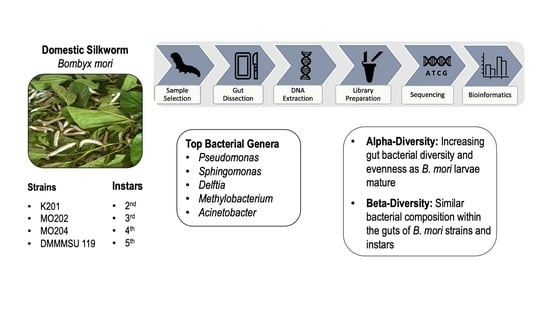
2.1. Sample Collection, Preservation, and Selection
2.2. Silkworm Gut Dissection and DNA Extraction
2.3. Library Preparation and Sequencing on MiSeq FGx
2.4. 16S rRNA Amplicon Sequence Analysis
3. Results
3.1. Analysis of Bacterial Abundance
3.2. Alpha-Diversity Analysis
3.3. Beta-Diversity Analysis
4. Discussion
4.1. Bacterial Abundance
4.2. Alpha-Diversity and Beta-Diversity
5. Summary and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mita, K.; Kasahara, M.; Sasaki, S.; Nagayasu, Y.; Yamada, T.; Kanamori, H.; Namiki, N.; Kitagawa, M.; Yamashita, H.; Yasukochi, Y.; et al. The genome sequence of silkworm, Bombyx mori. DNA Res. 2004, 11, 27–35. [Google Scholar] [CrossRef] [Green Version]
- Mohanta, M.K.; Saha, A.K.; Saleh, D.K.M.A.; Islam, M.S.; Mannan, K.S.B.; Fakruddin, M. Characterization of Klebsiella granulomatis pathogenic to silkworm, Bombyx mori L. Biotech 2010, 5, 577–583. [Google Scholar] [CrossRef] [Green Version]
- Chen, B.; Du, K.; Sun, C.; Vimalanathan, A.; Liang, X.; Li, Y.; Wang, B.; Lu, X.; Li, L.; Shao, Y. Gut bacterial and fungal communities of the domesticated silkworm (Bombyx mori) and wild mulberry-feeding relatives. ISME J. 2018, 12, 2252–2262. [Google Scholar] [CrossRef] [Green Version]
- Nagaraju, J.; Goldsmith, M.R. Silkworm genomics—Progress and prospects. Curr. Sci. 2002, 83, 415–425. [Google Scholar]
- Xia, Q.Y.; Zhou, Z.Y.; Lu, C.; Cheng, D.; Dai, F.-Y.; Liu, B.; Zhao, P.; Zha, X.; Cheng, T.; Biology Analysis Group; et al. A draft sequence for the genome of the domesticated silkworm (Bombyx mori). Science 2004, 306, 1937–1940. [Google Scholar] [PubMed] [Green Version]
- Pereira, N.C.; Munhoz, R.E.F.; Bignotto, T.S.; Bespalhuk, R.; Garay, L.B.; Saez, C.R.N.; Fernandez, M.A. Biological and molecular characterization of silkworm strains from the Brazilian germplasm bank of Bombyx mori. Genet. Mol. Res. 2013, 12, 2138–2147. [Google Scholar] [CrossRef] [PubMed]
- Zanatta, D.B.; Bravo, J.P.; Barbosa, J.F.; Munhoz, R.E.F.; Fernandez, M.A. Evaluation of economically important traits from sixteen parental strains of the silkworm Bombyx mori L. (Lepidoptera: Bombycidae). Neotrop. Entomol. 2009, 38, 327–331. [Google Scholar] [CrossRef] [Green Version]
- Matienzo, L.H.; Gatica, R.A. Sericulture: Silk Production in the Philippines. Available online: https://aboutphilippines.org/files/Silk-Production-in-the-Philippines.pdf (accessed on 6 December 2018).
- Alcudia-Catalma, M.N.; Conde, M.Y.E.D.; Dee Tan, I.Y.; Bautista, M.A.M. First report on the characterization of genetic diversity of Philippine-reared Bombyx mori strains based on COI and ITS2. Philipp. J. Sci. 2020, 150 (Suppl. S1), 503–517. [Google Scholar]
- Basaen, A.M.; Placido, A.; Bacuso, P.; Ladines, A.; Cabrito, F. Silkworm breeding for the development of Philippine purelines. NSTA Technol. J. 1982, 7, 72–85. [Google Scholar]
- Villanueva, R.C. Problems and Issues Affecting the Pace of Sericulture Development in the Philippines. 1999. Available online: https://agris.fao.org/agris-search/search.do?recordID=PH2007M00034 (accessed on 11 January 2022).
- Sarkar, K. Management of nutritional and climatic factors for silkworm rearing in West Bengal: A review. Int. J. Agric. Environ. Biotechnol. 2018, 11, 769–780. [Google Scholar] [CrossRef]
- Barretto, D.A.; Gadwala, M.; Vootla, S.K. Chapter 1—The silkworm gut microbiota: A potential source for biotechnological applications. Methods Microbiol. 2021, 49, 1–26. [Google Scholar]
- Sun, Z.; Kumar, D.; Cao, G.; Zhu, L.; Liu, B.; Zhu, M.; Liang, Z.; Kuang, S.; Chen, F.; Feng, Y.; et al. Effects of transient high temperature treatment on the intestinal flora of the silkworm Bombyx mori. Sci. Rep. 2017, 7, 3349. [Google Scholar] [CrossRef] [Green Version]
- National Research Council (US). Committee on Metagenomics: Challenges and Functional Applications. In The New Science of Metagenomics: Revealing Secrets of Our Microbial Planet; National Academies Press (US): Washington, DC, USA, 2007. Available online: https://www.ncbi.nlm.nih.gov/books/NBK54011/ (accessed on 7 May 2019).
- Thomas, T.; Gilbert, J.; Meyer, F. Metagenomics—A guide from sampling to data analysis. Microb. Inform. Exp. 2021, 2, 3. [Google Scholar] [CrossRef] [Green Version]
- Sun, Z.; Lu, Y.; Zhang, H.; Kumar, D.; Liu, B.; Gong, Y.; Zhu, M.; Zhu, L.; Liang, Z.; Kuang, S.; et al. Effects of BmCPV infection on silkworm Bombyx mori intestinal bacteria. PLoS ONE 2016, 11, e0146313. [Google Scholar] [CrossRef]
- Hammer, T.; Dickerson, J.; Fierer, N. Evidence-based recommendations on storing and handling specimens for analyses of insect microbiota. PeerJ 2015, 3, e1190. [Google Scholar] [CrossRef] [Green Version]
- Herlemann, D.P.R.; Labrenz, M.; Jürgens, K.; Bertilsson, S.; Waniek, J.J.; Andersson, A.F. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 2011, 5, 1571–1579. [Google Scholar] [CrossRef] [Green Version]
- Suenami, S.; Nobu, M.K.; Miyazaki, R. Community analysis of gut microbiota in hornets, the largest eusocial wasps, Vespa mandarinia and V. simillima. Sci. Rep. 2019, 9, 9830. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Garrity, G.; Tiedje, J.; Cole, J. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [Green Version]
- Schoch, C.L.; Ciufo, S.; Domrachev, M.; Hotton, C.L.; Kannan, S.; Khovanskaya, R.; Leipe, D.; McVeigh, R.; O’Neill, K.; Robbertse, B.; et al. NCBI Taxonomy: A comprehensive update on curation, resources and tools. Database 2020, 2020, 1–21. [Google Scholar] [CrossRef]
- Ou, T.; Xu, W.-F.; Wang, F.; Strobel, G.; Zhou, Z.-Y.; Xiang, Z.-H.; Liu, J.; Xie, J. A microbiome study reveals seasonal variation in endophytic bacteria among different mulberry cultivars. Comput. Struct. Biotechnol. J. 2019, 17, 1091–1100. [Google Scholar] [CrossRef]
- Lukša, J.; Servienė, E. White mulberry (Morus alba L.) fruit-associated bacterial and fungal microbiota. J. Environ. Eng. Landsc. Manag. 2020, 28, 183–191. [Google Scholar] [CrossRef]
- Dong, H.L.; Zhang, S.X.; Chen, Z.H.; Tao, H.; Li, X.; Qiu, J.F.; Xu, S.Q. Differences in gut microbiota between silkworms (Bombyx mori) reared on fresh mulberry (Morus alba var. multicaulis) leaves or an artificial diet. RSC Adv. 2018, 8, 26188–26200. [Google Scholar]
- Flury, P.; Aellen, N.; Ruffner, B.; Péchy-Tarr, M.; Fataar, S.; Metla, Z.; Maurhofer, M. Insect pathogenicity in plant-beneficial pseudomonads: Phylogenetic distribution and comparative genomics. ISME J. 2016, 10, 2527–2542. [Google Scholar] [CrossRef] [PubMed]
- Flury, P.; Vesga, P.; Péchy-Tarr, M.; Aellen, N.; Dennert, F.; Hofer, N.; Kupferschmied, K.P.; Kupferschmied, P.; Metla, Z.; Ma, Z.; et al. Antimicrobial and insecticidal: Cyclic lipopeptides and hydrogen cyanide produced by plant-beneficial Pseudomonas strains CHA0, CMR12a, and PCL1391 contribute to insect killing. Front. Microbiol. 2017, 8, 100. [Google Scholar] [CrossRef]
- Prem Anand, A.A.; Vennison, S.J.; Sankar, S.G.; Gilwax Prabhu, D.I.; Vasan, P.T.; Raghuraman, T.; Vendan, S.E. Isolation and characterization of bacteria from the gut of Bombyx mori that degrade cellulose, xylan, pectin and starch and their impact on digestion. J. Insect. Sci. 2010, 10, 107. [Google Scholar]
- Briones-Roblero, C.I.; Rodríguez-Díaz, R.; Santiago-Cruz, J.A.; Zúñiga, G.; Rivera-Orduña, F.N. Degradation capacities of bacteria and yeasts isolated from the gut of Dendroctonus rhizophagus (Curculionidae: Scolytinae). Folia Microbiol. (Praha) 2017, 62, 1–9. [Google Scholar] [CrossRef]
- Delalibera, I., Jr.; Handelsman, J.; Raffa, K.F. Contrasts in cellulolytic activities of gut microorganisms between the wood borer, Saperda vestita (Coleoptera: Cerambycidae), and the bark beetles, Ips pini and Dendroctonus frontalis (Coleoptera: Curculionidae). Environ. Entomol. 2005, 34, 541–547. [Google Scholar] [CrossRef]
- Gong, B.; Wu, P.; Huang, Z.; Li, Y.; Dang, Z.; Ruan, B.; Kang, C.; Zhu, N. Enhanced degradation of phenol by Sphingomonas sp. GY2B with resistance towards suboptimal environment through adsorption on kaolinite. Chemosphere 2016, 148, 388–394. [Google Scholar] [CrossRef]
- Silva, C.; Cabral, J.M.S.; van Keulen, F. Isolation of a beta-carotene over-producing soil bacterium, Sphingomonas sp. Biotechnol. Lett. 2004, 26, 257–262. [Google Scholar] [CrossRef]
- Wang, X.; Xu, P.; Yuan, Y.; Liu, C.; Zhang, D.; Yang, Z.; Yang, C.; Ma, C. Modeling for gellan gum production by Sphingomonas paucimobilis ATCC 31461 in a simplified medium. Appl. Environ. Microbiol. 2006, 72, 3367–3374. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Li, Y.; Guo, S.; Yin, H.; Lei, C.; Wang, X. Association between gut microbiota and diapause preparation in the cabbage beetle: A new perspective for studying insect diapause. Sci. Rep. 2016, 6, 38900. [Google Scholar] [CrossRef]
- Hu, X.; Yu, J.; Wang, C.; Chen, H. Cellulolytic bacteria associated with the gut of Dendroctonus armandi larvae (Coleoptera: Curculionidae: Scolytinae). Forests 2014, 5, 455–465. [Google Scholar] [CrossRef]
- Xu, W.; Wang, F.; Zhang, M.; Ou, T.; Wang, R.; Strobel, G.; Xiang, Z.; Zhou, Z.; Xie, J. Diversity of cultivable endophytic bacteria in mulberry and their potential for antimicrobial and plant growth-promoting activities. Microbiol. Res. 2019, 229, 126328. [Google Scholar] [CrossRef] [PubMed]
- Amer, A.; Hamdy, B.; Mahmoud, D.; Elanany, M.; Rady, M.; Alahmadi, T.; Alharbi, S.; AlAshaal, S. Antagonistic activity of bacteria isolated from the Periplaneta americana L. gut against some multidrug-resistant human pathogens. Antibiotics 2021, 10, 294. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dee Tan, I.Y.; Bautista, M.A.M. Bacterial Survey in the Guts of Domestic Silkworms, Bombyx mori L. Insects 2022, 13, 100. https://doi.org/10.3390/insects13010100
Dee Tan IY, Bautista MAM. Bacterial Survey in the Guts of Domestic Silkworms, Bombyx mori L. Insects. 2022; 13(1):100. https://doi.org/10.3390/insects13010100
Chicago/Turabian StyleDee Tan, Ivan Y., and Ma. Anita M. Bautista. 2022. "Bacterial Survey in the Guts of Domestic Silkworms, Bombyx mori L." Insects 13, no. 1: 100. https://doi.org/10.3390/insects13010100