1. Introduction
Cross-linking, the formation of covalent bonds between protein molecules or parts, plays a crucial role in shaping protein structure and modulating their function. In the case of amyloidogenic proteins, which are associated with various neurodegenerative disorders, cross-linking and alterations in conformation can significantly influence the propensity for protein aggregation [
1,
2,
3,
4,
5].
Cross-linking occurs when reactive chemical groups, such as cysteine or lysine residues, form covalent linkages between protein regions or molecules. These inter- or intramolecular bonds restrict the conformational flexibility of proteins, resulting in a variety of structural consequences. Cross-linked proteins often adopt more compact and rigid structures, altering their overall shape and accessibility to other molecules. Consequently, the functional properties of proteins, including enzymatic activity, binding affinity, and cellular interactions, can be profoundly affected by cross-linking [
6,
7,
8,
9].
Amyloidogenic proteins are a class of proteins that can misfold and aggregate into insoluble amyloid fibrils, a hallmark of many neurodegenerative diseases, such as Alzheimer’s and Parkinson’s disease. The aggregation process is highly dependent on the conformational states of these proteins. Cross-linking of amyloidogenic proteins can occur through various mechanisms, including oxidative stress, post-translational modifications, or through the action of molecular chaperones [
1,
10,
11,
12,
13,
14]. The presence of cross-links within amyloidogenic proteins can influence the rate, extent, and stability of protein aggregation. Cross-linking often promotes the formation of larger protein aggregates, altering the kinetics of fibril formation. These cross-links can stabilize intermediate oligomeric species, which are believed to be the toxic entities responsible for cellular dysfunction and neuronal damage in neurodegenerative diseases. Additionally, cross-linking may hinder the clearance of aggregated proteins by the cellular proteostasis machinery, further exacerbating the pathological consequences [
3,
15]. Cross-linking could also be a potential approach for inhibition of aggregation processes. By creating spatial restraints within the protein’s folding landscape, cross-linking aims to restrict the conformational flexibility necessary for aggregation. The introduction of appropriate steric constraints could limit the formation of the β-sheet-rich conformations typically associated with aggregation-prone states. By specifically interacting with the regions prone to aggregation, the cross-linking agents prevent the formation of intermolecular contacts required for fibril assembly, effectively inhibiting the protein’s pathogenic behavior. The steric constraints imposed by intramolecular cross-linking not only can disrupt the aggregation-prone conformations but also confer enhanced stability to the protein, thereby reducing its susceptibility to proteolytic degradation. This added stability can further contribute to the inhibition of aggregation processes and provide potential therapeutic benefits [
3,
16].
In recent years, the cross-linking mass spectrometry (XL–MS) technique has emerged as a widely applied method for studying the structure of proteins that are difficult to investigate with conventional methods (e.g., NMR spectroscopy and X-ray crystallography), such as intrinsically disordered proteins or proteins with disordered regions. Cross-linking agents usually contain two reactive groups that are connected by a spacer with specific length. The spacer provides a distance information between amino acid residues of a protein or protein–protein partners which are hereby covalently cross-linked together. The cross-linked amino acids in the protein chains are then analyzed generally by bottom-up proteomic approaches using LC/ESI–MS/MS (liquid chromatography/electrospray ionization–tandem mass spectrometry).
The most commonly used cross-linkers are amine-reactive
N-Hydroxysuccinimide (NHS) esters, which target primarily lysine side chains. Many different NHS ester-based cross-linkers with various solubilities, spacer arm lengths, and gas-phase cleavability are available commercially. The most frequently used NHS cross-linker is the water-soluble, non-gas-phase cleavable BS3 (bis(sulfosuccinimidyl)suberate) [
17] with a sulfonic acid group at both reactive ends and a spacer arm length of 11.4 Å. The sulfonic group is incorporated for increased water solubility. Additionally, cross-linking agents with longer (e.g., BS(PEG)5 (PEGylated bis(sulfosuccinimidyl)suberate) of 21.7 Å) or shorter (e.g., EDC (1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride) with zero length) distance constraints can foster the reconstruction of a protein 3D structure. Furthermore, targeting not only lysine residues, but for example the more acidic aspartic and glutamic acids like in the case of the EDC cross-linker, can provide complementary distance information to the lysine reactive cross-linkers [
18]. Other popular cross-linkers are DSP [
19] (dithiobis(succinimidylpropionate)) and its water-soluble sulfo-NHS ester variant, the DTSSP, (3,3′-dithiobis(sulfosuccinimidyl propionate)) with a spacer arm length of 12.0 Å, which are cleavable under reducing conditions in solution or in gas-phase during collision induced dissociation (CID). Gas-phase cleavability of a cross-linker may facilitate the unambiguous identification of the cross-linked peptide pairs through the production of characteristic fragmentation patterns in the MS2 spectra [
20].
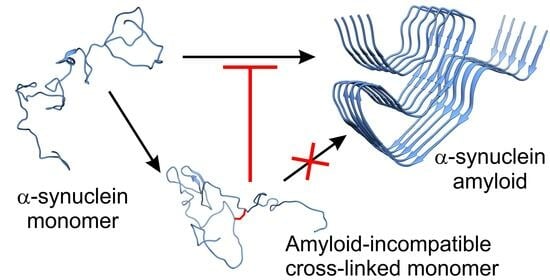
α-Synuclein (α-syn) is considered an intrinsically disordered protein (IDP) and its monomeric form is characterized by highly dynamic conformational ensembles. Aggregation and amyloid formation of α-syn is associated with Parkinson’s disease and other synucleinopathies [
21,
22]. In this work, we try to map the monomeric, natively disordered structure of α-syn by cross-linking and investigate the effect of steric constraints introduced by cross-linking on the aggregation and fibrillation of the protein. First, we carried out in silico structural analysis to predict the potential sites for cross-linking and their potential inhibitory effect on fibrillation. Then, we cross-linked monomeric α-syn intramolecularly with several cross-linkers and determined the location of the cross-links by mass spectrometry. Cross-linked α-syn samples were tested for their aggregation properties and for their inhibitory effect on the fibrillation of unmodified α-syn.
3. Discussion
The aggregation and fibrillation of amyloidogenic proteins are associated with numerous degenerative diseases in the human body [
10]. These proteins can be classified in different groups depending on the structural state of the native, monomeric protein. There are peptides which usually have hydrophobic properties and are produced by proteolytic cleavage in the body, such as amylin and the amyloid-β peptide. On the opposite end, there are globular, highly ordered proteins, which can be destabilized under certain environmental conditions, and their conformation undergoes a transition to a highly β-structured polymeric state of the amyloid fibrils (β
2-microglobulin, transthyretin, Ig light chain) [
10]. A special class of amyloidogenic proteins are natively disordered proteins, which are characterized by highly dynamic conformational ensembles. They have good solubility; however, they are still prone to form different types of aggregates and amyloid fibrils. α-Synuclein is an IDP which plays a role in synaptic vesicle trafficking in neurons [
26]. However, upon aggregation and amyloid formation, the protein is deposited in the brain and is responsible for Parkinson’s disease and other synucleinopathies [
21,
22]. Due to its disease relation, blocking α-syn aggregation is receiving high attention. Because of the high conformational multiplicity, it is difficult to target the monomeric state with peptide or small molecule binders [
27,
28,
29,
30,
31]. The amyloid formation is coupled with a large structural transformation to the highly non-native, cross-β-sheet structure.
There are few studies in the literature on α-syn using cross-linking. Selkoe and co-workers used cross-linking in vivo to study the oligomeric state of α-syn [
32]. Lucas and co-workers investigated the metal ion-induced structural alterations in α-syn by photo-chemical cross-linking and pointed out the different coordination and the related molecular mechanisms for copper and iron [
33]. Schmid et al. determined the effect of tissue transglutaminase catalyzed cross-linking on the oligomerization, fibrillization, and membrane binding of α-syn. Cross-linking at Gln79 and Gln109 had inhibitory effects on α-syn fibrillogenesis [
34].
Cross-linking of α-syn might occur naturally in vivo. Depending on the cross-linked structural form of the molecule (disordered vs. aggregated species), amyloid formation can be inhibited or the fibril structure can be promoted and stabilized. Nemes et al. studied the effect of the transglutaminase-mediated intramolecular cross-linking of α-syn, which was identified between Gln99 and Lys58 in Lewy bodies. In vitro cross-linking by transglutaminases produced a heterogeneously cross-linked solution, which inhibited amyloid formation [
35]. However, in the presence of phosphatidylserine-rich membranes and calcium, the formation of the Gln99–Lys58 cross-link was preferred, which triggered amyloid formation. In contrast, the cross-link between residues 99 and 10 had an inhibitory effect. In their recent work, Sahin et al. modeled the effect of oxidative stress on the α-syn structure and self-assembly. They found that mild oxidation led to intramolecular cross-linkages, compaction of the monomer molecule, and the inhibition of amyloid formation by steric hindrance, suggesting that mild oxidation has a role in preventing amyloid formation [
36].
In the present study, we introduced structural constraints to the monomeric α-syn molecule by cross-linking and studied its effect on the aggregation of the protein and investigated the possible inhibitory effect of cross-linked α-syn molecules on the native α-syn. First, we carried out an in silico structural analysis on native monomeric α-syn and on the available high-resolution amyloid structures to predict the potential cross-linking sites and their usability to block the aggregation of the protein. How to study the structure of the IDP α-syn monomer was an intriguing task. First, we tried to use the eight representative native α-syn structures published by Chen et al. [
24] and surveyed them for amines that lie within predefined cross-linking distances from each other. The observed amine pairs were compared to the potential cross-linking pairs in the structures of α-syn amyloid fibrils downloaded from the PDB (
Supplementary Table S1). The results revealed that most of the potential cross-links in the native monomeric structures are non-compatible with the amyloid forms; therefore, they might be suitable as constraints to block amyloid formation (
Figure 1,
Supplementary Figure S1). In the second experimental step of the study, α-syn was cross-linked with several cross-linkers (
Supplementary Figure S2) under conditions where the reactions occur intramolecularly (
Supplementary Figure S3) and mostly one cross-link is formed per molecule. The cross-linked samples were analyzed by LC–ESI–MS and LC–ESI–MS/MS techniques for native and bottom-up analyses, respectively (
Figure 2,
Supplementary Table S2 and
Supplementary Figures S5–S7) and proved to be heterogeneous in the cross-links (
Supplementary Figure S4). Taking advantage of the extra ion mobility separation opportunity with the Cyclic IMS instrument, it was possible to increase cross-link identifications of the heterogeneous chemically modified α-syn molecules. In previous studies, Cyclic IMS has been described to enhance overall protein coverage and peptide identifications in proteomics studies [
37]. Furthermore, the NHS ester cross-linkers we used tend to dissociate upon CID fragmentation at their cross-linker–lysine amide bond resulting in diagnostic, cross-linker-specific ions, which support the unambiguous identifications [
9]. Additionally, the DTSSP cross-linker is prone to dissociate at the linker’s S–S bond as well [
38] which further complements CSM detections. Fragmentation of the linker is potentiated when additional IMS was applied before CID fragmentation; thus, higher scoring and on average enhanced cross-link recognition can be obtained. For the above reasons, DTSSP was proved to be an efficient cross-linker, and it was possible to identify 60 different cross-linked amine pairs that were observed with different frequencies (
Supplementary Table S2). The experimental results overlapped only in half with the expected cross-linkable amine pairs from the eight structures (
Figure 1), revealing that those representative structures with a simple amine-distance analysis are insufficient to depict the entire conformational ensembles of α-syn. To provide a better description of the dynamic conformational ensembles, we carried out MD simulation with α-syn and analyzed the entire trajectory for potential amine cross-link distances and determined their frequencies (occurrence in % of time) (
Figure 4). All the possible 120 amine pairs showed distances within the cut-off for cross-linking at least for a few frames in the trajectory, which assume a significantly larger conformational space than that characterized by the eight structures. However, the calculated occurrences showed no correlation with the experimentally observed cross-linking frequencies (
Figure 4). We tried to carry out a more sophisticated analysis on the trajectory by examining if there is a steric hindrance between the two amines (see
Section 4). We found an altered frequency for the potential cross-links; however, there was still no real correlation with the experimental frequencies. These results reveal that the in silico analysis is unable to reliably describe the intrinsically disordered α-syn chain and to predict its complex behavior. Borchers and co-authors studied the structure of native α-syn using the ABAS cross-linker and identified 10 cross-links using MS techniques [
39]. These cross-links were incompatible with the α-syn NMR structure of Ulmer et al. [
40], and they concluded that the in-solution structure of α-syn may be significantly different from that of the micelle-bound structure of monomeric α-syn.
Analyzing further the experimental results, we found that the sequential environment (neighboring residues) has a large effect on the cross-linking frequencies. Not surprisingly, the amino terminal with the lowest
pKa value participated at the highest frequency in the crosslinks. Among the Lys side chains, the one in the middle of the KXKXG motif showed the highest frequency, whereas the negative charge was not preferable at the end of the motif (
Figure 5,
Supplementary Tables S2 and S3). These results revealed that cross-linking efficiency in the natively disordered chain of α-syn is rather determined by the physico-chemical properties/local environment, affecting the chemical reaction rate and is difficult to predict simply based on distances in the structure.
Compared to the amyloid structures (
Supplementary Table S1), the experimentally identified cross-links and their frequencies in the samples assumed that although the samples are heterogeneous, most of the cross-links are incompatible with the amyloid structures and can block the fibrillation of the protein. Indeed, none of the cross-linked samples showed aggregation in the ThT fluorescence assays (
Figure 6).
An important observation is that the DTSSP-cross-linked α-syn samples displayed a significant inhibitory potency at sub-stoichiometric concentrations on the aggregation of the unmodified α-synuclein even at a 0.01:1 cross-linked/native ratio (
Figure 6C). TEM images also proved the inhibitory effect of DTSSP-cross-linked α-syn (
Figure 7). In order to inhibit aggregation at sub-stoichiometric ratios, the cross-linked molecule has to bind with high affinity to α-syn aggregates and block the further growth by not being able to transform into the amyloid conformation, which is assured by the cross-linking.
Here, we used cross-linked samples containing a heterogeneous mixture of intramolecularly cross-linked molecules (see
Supplementary Table S2 for components and their frequencies). In the future, by selective purification, cross-links with the most beneficial effects could be sorted out.
In this work, we studied the effect of cross-linking on aggregation and fibrillation on α-syn as model protein. It is a question whether our results have therapeutic or in vivo relevance. The inhibitory effect at sub-stoichiometric concentrations is promising in this regard, and moreover, the cross-linked α-syn is probably non-immunogenic. However, we would rather think that such cross-linked α-syn, instead of a highly dynamic native molecule, might be a good target for screening of small molecules, peptides, or proteins that will bind and stabilize the “non-amyloid” conformation of α-syn. Moreover, such a conformation might have an inhibitory effect on the aggregation of α-syn.
4. Materials and Methods
4.1. Expression and Purification of α-Synuclein
E. coli BL21 Star (DE3) pLysS cells were transformed with pT7-7 plasmid containing the WT α-synuclein (
H. sapiens) cDNA sequence, grown in LB medium containing 100 µM/mL ampicillin and induced with 1 mM IPTG at OD600 = 0.6 for 3 h at 37 °C. Following cell harvesting by centrifugation at 7000×
g for 20 min at 4 °C, monomeric α-syn was purified as described earlier [
41,
42]. After lyophilization, the protein was kept at −20 °C until usage.
4.2. Cross-Linking Reactions of α-Synuclein by Chosen Agents and Optimization
Reagents were products of Sigma-Aldrich (Merck Life Science Kft., Budapest, Hungary or Merck KGaA, Darmstadt, Germany). First attempts were carried out by general protocols from the manufacturers for DTSSP (Sigma-Aldrich, 803200 (3,3′-dithiobis(sulfosuccinimidyl propionate))), BS3 (Sigma-Aldrich, S5799 Suberic acid bis(3-sulfo-
N-hydroxysuccinimide ester) sodium salt), EDC (Sigma-Aldrich, 8.00907
N-(3-Dimethylaminopropyl)-
N′-ethylcarbodiimide hydrochloride) and Sulfo-NHS (Sigma-Aldrich, 56485
N-Hydroxysulfosuccinimide sodium salt), BS(PEG)5 (Sigma-Aldrich, 803537 PEGylated bis(sulfosuccinimidyl)suberate). Reactions were optimized for intramolecular cross-linking, using an adapted protocol from Pál-Gábor et al. [
43]. Samples were incubated with cross-linkers at a protein to cross-linker ratio of 2:1 (
w/
w) for 1 h at 37 °C. The reactions were quenched by addition of 100 mM TRIS, pH 7.4, at a 50-fold molar excess to the cross-linker. The quenching reaction was allowed to proceed for 30 min. After quenching, free cross-linkers were removed on a PD-10 column (Sephadex G-25M, GE17-0851-01). Cross-linked α-syn solutions were concentrated and transferred into MQ by centrifugal filtration (Amicon Ultra–3K, Merck Millipore Ltd., Carrigtwohill, Ireland). Protein concentration was measured at 214 nm [
44] and 205 nm [
45]. The monomeric state of the proteins was verified using SDS PAGE under non-reducing conditions.
4.3. Protein Measurement with LC–MS and Data Analysis
For all MS analyses chemicals were from Sigma-Aldrich (Munich, Germany) except where indicated otherwise.
Approximately 2 nmol of cross-linked α-syn dissolved in MQ water was injected into a Waters Acquity I-Class UPLC system connected to a Waters Select Series Cyclic IMS (Waters Corporation, Milford, UK), hybrid quadrupole–TOF mass spectrometer. Liquid chromatographic separation of the protein and its modified variants was performed on an Acquity UPLC BEH300 C4 column, 300 Å, 1.7 μm, 1 mm × 150 mm. Mobile phase (A) was composed of 0.1% trifluoroacetic acid (TFA) in water; mobile phase (B) was composed of 0.1% TFA in acetonitrile. The elution method at a flow rate 400 µL/min included the following gradient: 1 min: 5% B, 12 min: 60% B, 12.5 min: 90% B at 80 oC. MS data acquisition was performed with the following parameters: m/z 350–2000, V-mode, scan time: 0.3 s, single Lock Mass: leucine–enkephalin.
Data were analyzed using Waters MassLynx 4.2 and spectra were inspected manually.
4.4. Sample Preparation for LC–MS/MS Analysis, Measurement, and Data Evaluation
Protein samples of 20–20 µg were denatured with 0.01% Rapigest (Waters Corporation, Milford, UK). Then (except for the DTSSP-cross-linked samples), they were reduced with 10 mM of TCEP at 37 °C for 30 min and alkylated with 55 mM of iodoacetamide at room temperature for 30 min in the dark. Samples were treated with trypsin (Promega Corporation, Madison, WI, USA) at an enzyme-to-protein ratio of 1:100 (wt/wt). Digestion was performed at 37 °C for 4 h. Enzymatic digestion was quenched by adding formic acid to reach a final concentration of ~1%. Peptides were purified and enriched using Pierce C18 Spin Columns according to the manufacturer’s description. Eluted samples were dried using SpeedVac and kept at −20 °C until LC–MS/MS analysis.
Samples were reconstituted in a 25 µL 2% ACN, 0.1% formic acid solution before injection into a Waters Acquity I-Class UPLC system connected to a Waters Select Series Cyclic IMS (Waters Corporation, Milford, UK), hybrid quadrupole–TOF mass spectrometer with high resolution and high mass accuracy, equipped with a cyclic ion mobility separation device. For peptide separation, samples were loaded on Waters Acquity Peptide CSH column (1.7 µm, 1 mm × 150 mm) or multi-step gradient elution. Mobile phase (A) was composed of 0.1% formic acid in water; mobile phase (B) was composed of 0.1% formic acid in acetonitrile. The elution method at a flow rate 20 µL/min included the following: 1 min: 5% B, 45 min: 35% B, 46 min: 85% B at 45 °C. MS data acquisition was performed with the following parameters: m/z 50–2000, V-mode, scan time: 0.5 s, single Lock Mass: leucine–enkephalin. HDMSE fragmentation was performed in the transfer cell: low energy: 6 V, high energy: ramping 19–45 V, after one ion mobility separation cycle. MSE fragmentation was performed in the transfer cell: low energy: 6 V, high energy: ramping 19–45 V.
Raw data were converted to mzML file format and searched for contaminants using Waters ProteinLynx Global Server (PLGS) v3.0.3 software in the SwissProt database. Parent and fragment ion tolerance was set to 20 ppm and 30 ppm, respectively. Digestion enzyme was trypsin and missed cleavages were set to 2. Carbamidomethyl modification on cysteines was set as fixed modification and for variable modifications oxidation on M. The false discovery rate was 2%, and only proteins with a minimal probability of 95% were counted. Processing parameters were the following: low energy threshold: 200 counts; elevated energy threshold: 20 counts; minimum fragment ion matches per peptide: 3; minimum fragment ion matches per protein: 7; minimum peptide matches per protein: 2. The cross-link analysis was performed using Protein Prospector. The digestion enzyme was set to trypsin with 2 missed cleavages; constant modification was carbamidomethyl (C) and variable modifications were: acetyl (Protein N-term), Gln to pyro-Glu (N-term Q), Met-loss (Protein N-term M), Met-loss + Acetyl (Protein N-term M), oxidation (M). Cross-linkers were set to EDC, DSP for DTSSP, DSS (disuccinimidyl suberate) for BS3, and BS(PEG)5 was set as user-defined cross-link, bridge components: C14O7H22, linking at lysine residues and protein N-terminus. The parent ion tolerance was set to 20 ppm; the fragment ion tolerance was 30 ppm; and the instrument was set to ESI-Q-TOF. The database was created with proteins previously found in the SwissProt search, random concatenated; the FDR cut off was 1%. The mass spectrometry-based proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE [
46] partner repository with the dataset identifier PXD042501.
4.5. MD Simulations and Trajectory Analysis for Potential Cross-Linking Pairs
The monomer structure of α-syn was subjected to MD simulations as implemented in GROMACS [
47], using the AMBER-ff99SB*-ILDNP force field [
48]. As initial monomeric conformation, we used the third model of the ensemble structure of the α-syn monomer determined using FRET data and discrete MD simulations [
24] (PDBDEV_00000082, Model #3). The system was solvated by ~10,032 water molecules with TIP4P parametrized water molecules [
49]. The total charge of the system was neutralized, and the physiological salt concentration was set by placing Na
+ and Cl
− ions. Energy minimization of starting structures was followed by sequential relaxation of constraints on protein atoms in three steps and an additional NVT step (all of 200 ps) to stabilize pressure. One µs trajectory of NPT simulations was carried out at 283 K and at 1 bar by collecting snapshots at every 20 ps.
The possibility of DTSSP cross-linking was investigated in silico based on the distance between the N-terminal and lysine side chains. The distance was calculated using the amino N atom of the N-terminal and of the lysine side chains, using in-house Matlab scripts. To calculate the distances in the monomeric form, first we used the 8 structural models reported by Chen et al. [
24], and for the amyloid form, we used structures from the PDB (
Supplementary Table S1). For intramolecular cross-linking with the DTSSP cross-linker, we used a cut-off of 15 Å in all cases. These distances were also calculated for the MD simulations, and we determined for each N–N pair the frequency (percentage) in the entire 1 µs trajectory (5001 frames), when the pair is within the cut-off distance.
We have further sorted the amine pairs that lie within cut-off distance for steric availability for cross-linking, i.e., whether there are molecule parts between them hindering a possible reaction. To check this in a simple and rapid way with low computational power needs, we placed three spheres with a radius of 2.5 Å on the line connecting the two nitrogen atoms (
Supplementary Figure S8). These spheres may overlap if the space between the two N atoms is less than 15 Å, and we use only one sphere if the distance between the two nitrogen atoms is less than 5 Å. The other atoms of the molecule were represented by spheres of their van der Waals radii. If such a sphere has intersection with one of the spheres between the two N atoms, the case was treated as blocked for cross-linking. Note that the atoms in the space “behind” the nitrogen atoms were not treated as blockers (
Supplementary Figure S8).
4.6. α-Synuclein Aggregation and Thioflavin T Fluorescence Assay
Purified α-syn was dissolved in PBS or 20 mM Na-phosphate, 100 mM NaCl (pH 7.4), at a concentration of 400 µM. Cross-linked α-syn samples in MQ water were adjusted to the required buffer conditions. After monomeric WT and cross-linked α-syn solutions were diluted to the required working concentrations (100–200 µM) in the used ratios, the samples were split into at least three replicates in Eppendorf tubes containing each a Teflon polyball (1/800 diameter) and placed into a shaker (Eppendorf ThermoMixer C, Eppendorf A.G., Hamburg, Germany). Aggregation processes were carried out at 37 °C, 500 rpm for 48–100 h. For the thioflavin T (ThT) fluorescence assay, reaction mixtures were mixed with 20 μM ThT. The fluorescence intensity was measured on a SPEX FluoroMax fluorometer (SPEX Industries, Edison, NJ, USA) with excitation and emission wavelengths of 445 and 485 nm, respectively, at 25 °C.
For seeded aggregation measurements, a final concentration of 200 μM DTSSP-cross-linked and WT α-syn samples were used in 20 mM NaH2PO4, 100 mM NaCl (pH 7.4) containing 20 µM ThT and 0.05% sodium azide. Seeds were prepared with 3 min ultrasonication using a Branson Ultrasonic B200 Equipment of a 200 μM WT α-syn amyloid solution (grown in a shaking incubator for 2 days at 500 rpm, 37 °C), then added to the freshly prepared monomer solutions at 1–10% final concentration in a 96-well plate (polystyrene non-binding microplate, Greiner, sealed). The ThT fluorescence intensities were measured using a plate reader (Synergy H4 Hybrid Reader, BioTek Instruments, Winooski, VT, USA). Instrument settings: xenon lamp, 37 °C, 48 h, measurements every 30 min, rapid shaking constantly, excitation 445 nm, emission 485 nm, gain 80.
For dose-response measurements, the aggregation of 200 μM α-syn WT monomer solutions were monitored in the presence of different DTSSP-cross-linked α-syn fractions (between 1:1–1:0.00005, equal to 200 μM–10 nM cross-linked protein concentrations). Samples were aggregated in a shaker incubator (Eppendorf ThermoMixer C, Eppendorf A.G., Hamburg, Germany) at 500 rpm at 37 °C for 48 h. Fluorescence intensities of 20 µM ThT were measured on a FluoroMax fluorometer (SPEX Industries, Edison, NJ, USA) with excitation and emission wavelengths of 445 and 485 nm, respectively, at 25 °C.
4.7. Transmission Electron Microscopy (TEM)
Aggregated WT, cross-linked, and mixed α-syn samples from final point reactions were collected and diluted to a protein concentration of 10–12 µM. Samples of 5 µL were placed on carbon-coated formvar copper grids and incubated for 2 min. Then, samples were carefully removed using the edge of a filter paper, and 5 μL 1% (w/v) of uranyl acetate was applied for negative staining. After 1 min, the excess staining solution was removed, and the grids were air-dried. The samples were imaged using a Jeol J1100 transmission electron microscope (JEOL, Tokyo, Japan) operating at an accelerating voltage of 80 kV. At least 30 fields were screened from parallel samples to obtain representative images.