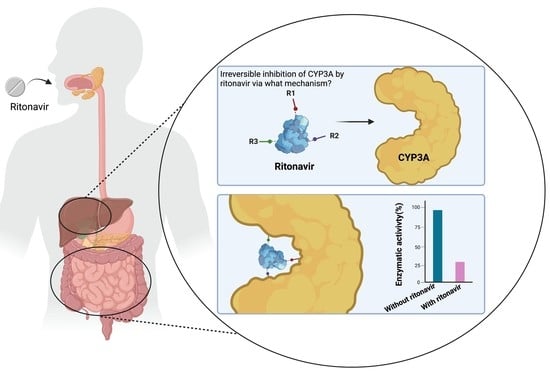
The Mechanism-Based Inactivation of CYP3A4 by Ritonavir: What Mechanism?
Abstract
:1. Introduction
1.1. Properties of Cytochrome P450 Enzymes including CYP3A4 and CYP3A5
1.2. Ritonavir as a Clinically Important CYP3A Inhibitor
1.3. The Ritonavir Analogue Cobicistat Has Very Similar CYP3A Inhibition Properties
1.4. Aim of This Review
2. Binding of Ritonavir to CYP3A4 and CYP3A5
3. Metabolism of Ritonavir by CYP3A4 and -3A5
4. Principal Mechanisms of Irreversible Inhibition of CYP Enzymes by Substrates
4.1. Inactivation of CYP3A by Formation of a Metabolic Intermediate Complex (MIC)
4.2. Inactivation of CYP3A through Tight Binding to the Heme Iron
4.3. Inactivation of CYP3A through Heme Modification
4.4. Inactivation of CYP3A by Putative Covalent Linkage of a Reactive Intermediate to the CYP3A4 Apoprotein
5. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Lolodi, O.; Wang, Y.-M.; Wright, W.C.; Chen, T. Differential Regulation of CYP3A4 and CYP3A5 and its Implication in Drug Discovery. Curr. Drug Metab. 2018, 18, 1095–1105. [Google Scholar] [CrossRef]
- Yadav, J.; Korzekwa, K.; Nagar, S. Improved Predictions of Drug–Drug Interactions Mediated by Time-Dependent Inhibition of CYP3A. Mol. Pharm. 2018, 15, 1979–1995. [Google Scholar] [CrossRef]
- Thelen, K.; Dressman, J.B. Cytochrome P450-mediated metabolism in the human gut wall. J. Pharm. Pharmacol. 2009, 61, 541–558. [Google Scholar] [CrossRef]
- Kim, J.H.; Sherman, M.E.; Curriero, F.C.; Guengerich, F.; Strickland, P.T.; Sutter, T.R. Expression of cytochromes P450 1A1 and 1B1 in human lung from smokers, non-smokers, and ex-smokers. Toxicol. Appl. Pharmacol. 2004, 199, 210–219. [Google Scholar] [CrossRef]
- Ghosh, C.; Marchi, N.; Desai, N.K.; Puvenna, V.; Hossain, M.; Gonzalez-Martinez, J.; Alexopoulos, A.V.; Janigro, D. Cellular localization and functional significance of CYP3A4 in the human epileptic brain. Epilepsia 2011, 52, 562–571. [Google Scholar] [CrossRef] [PubMed]
- Knops, N.; Heuvel, L.P.V.D.; Masereeuw, R.; Bongaers, I.; de Loor, H.; Levtchenko, E.; Kuypers, D. The Functional Implications of Common Genetic Variation in CYP3A5 and ABCB1 in Human Proximal Tubule Cells. Mol. Pharm. 2015, 12, 758–768. [Google Scholar] [CrossRef] [PubMed]
- Van Eijk, M.; Boosman, R.J.; Schinkel, A.H.; Huitema, A.D.R.; Beijnen, J.H. Cytochrome P450 3A4, 3A5, and 2C8 expression in breast, prostate, lung, endometrial, and ovarian tumors: Relevance for resistance to taxanes. Cancer Chemother. Pharmacol. 2019, 84, 487–499. [Google Scholar] [CrossRef]
- Zanger, U.M.; Schwab, M. Cytochrome P450 enzymes in drug metabolism: Regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 2013, 138, 103–141. [Google Scholar] [CrossRef]
- Woodland, C.; Huang, T.T.; Gryz, E.; Bendayan, R.; Fawcett, J.P. Expression, Activity and Regulation of CYP3A in Human and Rodent Brain. Drug Metab. Rev. 2008, 40, 149–168. [Google Scholar] [CrossRef]
- Krusekopf, S.; Roots, I.; Kleeberg, U. Differential drug-induced mRNA expression of human CYP3A4 compared to CYP3A5, CYP3A7 and CYP3A43. Eur. J. Pharmacol. 2003, 466, 7–12. [Google Scholar] [CrossRef]
- Zhou, S.-F. Drugs Behave as Substrates, Inhibitors and Inducers of Human Cytochrome P450 3A4. Curr. Drug Metab. 2008, 9, 310–322. [Google Scholar] [CrossRef] [PubMed]
- Ince, I.; Knibbe, C.A.J.; Danhof, M.; de Wildt, S.N. Developmental Changes in the Expression and Function of Cytochrome P450 3A Isoforms: Evidence from In Vitro and In Vivo Investigations. Clin. Pharmacokinet. 2013, 52, 333–345. [Google Scholar] [CrossRef]
- Hsu, M.-H.; Savas, U.; Johnson, E.F. The X-ray Crystal Structure of the Human Mono-Oxygenase Cytochrome P450 3A5-Ritonavir Complex Reveals Active Site Differences between P450s 3A4 and 3A5. Mol. Pharmacol. 2018, 93, 14–24. [Google Scholar] [CrossRef]
- Hsu, M.-H.; Johnson, E.F. Active-site differences between substrate-free and ritonavir-bound cytochrome P450 (CYP) 3A5 reveal plasticity differences between CYP3A5 and CYP3A4. J. Biol. Chem. 2019, 294, 8015–8022. [Google Scholar] [CrossRef]
- Kuehl, P.; Zhang, J.; Lin, Y.; Lamba, J.; Assem, M.; Schuetz, J.; Watkins, P.B.; Daly, A.; Wrighton, S.A.; Hall, S.D.; et al. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat. Genet. 2001, 27, 383–391. [Google Scholar] [CrossRef]
- Saito, Y.; Yamamoto, N.; Katori, N.; Maekawa, K.; Fukushima-Uesaka, H.; Sugimoto, D.; Kurose, K.; Sai, K.; Kaniwa, N.; Sawada, J.-I.; et al. Genetic Polymorphisms and Haplotypes of POR, Encoding Cytochrome P450 Oxidoreductase, in a Japanese Population. Drug Metab. Pharmacokinet. 2011, 26, 107–116. [Google Scholar] [CrossRef]
- Yoo, S.-E.; Yi, M.; Kim, W.-Y.; Cho, S.-A.; Lee, S.S.; Lee, S.-J.; Shin, J.-G. Influences of cytochrome b5 expression and its genetic variant on the activity of CYP2C9, CYP2C19 and CYP3A4. Drug Metab. Pharmacokinet. 2019, 34, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Gan, L.; von Moltke, L.L.; Trepanier, L.A.; Harmatz, J.S.; Greenblatt, D.J.; Court, M.H. Role of NADPH-cytochrome P450 reductase and cytochrome-b5/NADH-b5 reductase in variability of CYP3A activity in human liver microsomes. Drug Metab. Dispos. 2009, 37, 90–96. [Google Scholar]
- Rock, B.M.; Hengel, S.M.; Rock, D.A.; Wienkers, L.C.; Kunze, K.L. Characterization of Ritonavir-Mediated Inactivation of Cytochrome P450 3A4. Mol. Pharmacol. 2014, 86, 665–674. [Google Scholar] [CrossRef] [PubMed]
- Masubuchi, Y.; Horie, T. Toxicological Significance of Mechanism-Based Inactivation of Cytochrome P450 Enzymes by Drugs. Crit. Rev. Toxicol. 2007, 37, 389–412. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Zhou, X.; Mohutsky, M.A.; Desai, P.V. Structure–Property Relationships and Machine Learning Models for Addressing CYP3A4-Mediated Victim Drug–Drug Interaction Risk in Drug Discovery. Mol. Pharm. 2020, 17, 3600–3608. [Google Scholar] [CrossRef]
- Jayakanthan, M.; Chandrasekar, S.; Muthukumaran, J.; Mathur, P.P. Analysis of CYP3A4-HIV-1 protease drugs interactions by computational methods for Highly Active Antiretroviral Therapy in HIV/AIDS. J. Mol. Graph. Model. 2010, 28, 455–463. [Google Scholar] [CrossRef]
- Von Hentig, N.; Haberl, A. Safety of pharmacoenhancers for HIV therapy. Expert Rev. Clin. Pharmacol. 2012, 5, 557–568. [Google Scholar] [CrossRef]
- Kempf, D.J.; Marsh, K.C.; Denissen, J.F.; McDonald, E.; Vasavanonda, S.; Flentge, C.A.; Green, B.E.; Fino, L.; Park, C.H.; Kong, X.P. ABT-538 is a potent inhibitor of human immunodeficiency virus protease and has high oral bioavailability in humans. Proc. Natl. Acad. Sci. USA 1995, 92, 2484–2488. [Google Scholar] [CrossRef]
- Koudriakova, T.; Iatsimirskaia, E.; Utkin, I.; Gangl, E.; Vouros, P.; Storozhuk, E.; Orza, D.; Marinina, J.; Gerber, N. Metabolism of the human immunodeficiency virus protease inhibitors indinavir and ritonavir by human intestinal microsomes and expressed cytochrome P4503A4/3A5: Mechanism-based inactivation of cytochrome P4503A by ritonavir. Drug Metab. Dispos. 1998, 26, 552–561. [Google Scholar]
- Kumar, G.N.; Rodrigues, A.D.; Buko, A.M.; Denissen, J.F. Cytochrome P450-mediated metabolism of the HIV-1 protease inhibitor ritonavir (ABT-538) in human liver microsomes. J. Pharmacol. Exp. Ther. 1996, 277, 423–431. [Google Scholar]
- Margolis, A.M.; Heverling, H.; Pham, P.A.; Stolbach, A. A review of the toxicity of HIV medications. J. Med. Toxicol. 2014, 10, 26–39. [Google Scholar] [CrossRef]
- Kageyama, M.; Namiki, H.; Fukushima, H.; Terasaka, S.; Togawa, T.; Tanaka, A.; Ito, Y.; Shibata, N.; Takada, K. Effect of Chronic Administration of Ritonavir on Function of Cytochrome P450 3A and P-Glycoprotein in Rats. Biol. Pharm. Bull. 2005, 28, 130–137. [Google Scholar] [CrossRef]
- Fukushima, K.; Kobuchi, S.; Mizuhara, K.; Aoyama, H.; Takada, K.; Sugioka, N. Time-Dependent Interaction of Ritonavir in Chronic Use: The Power Balance between Inhibition and Induction of P-Glycoprotein and Cytochrome P450 3A. J. Pharm. Sci. 2013, 102, 2044–2055. [Google Scholar] [CrossRef]
- Kirby, B.J.; Collier, A.C.; Kharasch, E.D.; Dixit, V.; Desai, P.; Whittington, D.; Thummel, K.E.; Unadkat, J.D. Complex Drug Interactions of HIV Protease Inhibitors 2: In Vivo Induction and In Vitro to In Vivo Correlation of Induction of Cytochrome P450 1A2, 2B6, and 2C9 by Ritonavir or Nelfinavir. Drug Metab. Dispos. 2011, 39, 2329–2337. [Google Scholar] [CrossRef]
- Yeh, R.F.; Gaver, V.E.; Patterson, K.B.; Rezk, N.L.; Baxter-Meheux, F.; Blake, M.J.; Eron, J.J.; Klein, C.E.; Rublein, J.C.; Kashuba, A.D. Lopinavir/Ritonavir Induces the Hepatic Activity of Cytochrome P450 Enzymes CYP2C9, CYP2C19, and CYP1A2 But Inhibits the Hepatic and Intestinal Activity of CYP3A as Measured by a Phenotyping Drug Cocktail in Healthy Volunteers. JAIDS J. Acquir. Immune Defic. Syndr. 2006, 42, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Foisy, M.M.; Yakiwchuk, E.M.; Hughes, C.A. Induction Effects of Ritonavir: Implications for Drug Interactions. Ann. Pharmacother. 2008, 42, 1048–1059. [Google Scholar] [CrossRef]
- Gupta, A.; Mugundu, G.M.; Desai, P.B.; Thummel, K.E.; Unadkat, J.D. Intestinal Human Colon Adenocarcinoma Cell Line LS180 Is an Excellent Model to Study Pregnane X Receptor, but Not Constitutive Androstane Receptor, Mediated CYP3A4 and Multidrug Resistance Transporter 1 Induction: Studies with Anti-Human Immunodeficiency Virus Protease Inhibitors. Drug Metab. Dispos. 2008, 36, 1172–1180. [Google Scholar] [CrossRef] [PubMed]
- Fahmi, O.A.; Maurer, T.S.; Kish, M.; Cardenas, E.; Boldt, S.; Nettleton, D. A Combined Model for Predicting CYP3A4 Clinical Net Drug-Drug Interaction Based on CYP3A4 Inhibition, Inactivation, and Induction Determined In Vitro. Drug Metab. Dispos. 2008, 36, 1698–1708. [Google Scholar] [CrossRef] [PubMed]
- Eagling, V.A.; Back, D.J.; Barry, M.G. Differential inhibition of cytochrome P450 isoforms by the protease inhibitors, ritonavir, saquinavir and indinavir. Br. J. Clin. Pharmacol. 1997, 44, 190–194. [Google Scholar] [CrossRef] [PubMed]
- Von Moltke, L.L.; Greenblatt, D.J.; Grassi, J.M.; Granda, B.W.; Duan, S.X.; Fogelman, S.M.; Daily, J.P.; Harmatz, J.S.; Shader, R.I. Protease inhibitors as inhibitors of human cytochromes P450: High risk associated with ritonavir. J. Clin. Pharmacol. 1998, 38, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Zeldin, R.K.; Petruschke, R.A. Pharmacological and therapeutic properties of ritonavir-boosted protease inhibitor therapy in HIV-infected patients. J. Antimicrob. Chemother. 2004, 53, 4–9. [Google Scholar] [CrossRef]
- Pasternak, A.O.; Vroom, J.; Kootstra, N.A.; Wit, F.W.; de Bruin, M.; De Francesco, D.; Bakker, M.; Sabin, C.A.; Winston, A.; Prins, J.M.; et al. Non-nucleoside reverse transcriptase inhibitor-based combination antiretroviral therapy is associated with lower cell-associated HIV RNA and DNA levels compared to protease inhibitor-based therapy. eLife 2021, 10, e68174. [Google Scholar] [CrossRef]
- Carrillo, A.; Stewart, K.D.; Sham, H.L.; Norbeck, D.W.; Kohlbrenner, W.E.; Leonard, J.M.; Kempf, D.J.; Molla, A. In vitro selection and characterization of human immunodeficiency virus type 1 variants with increased resistance to ABT-378, a novel protease inhibitor. J. Virol. 1998, 72, 7532–7541. [Google Scholar] [CrossRef]
- Highleyman, L. ABT-378: A second generation protease inhibitor. BETA 1998, 8, 55. [Google Scholar]
- Sham, H.L.; Kempf, D.J.; Molla, A.; Marsh, K.C.; Kumar, G.N.; Chen, C.-M.; Kati, W.; Stewart, K.; Lal, R.; Hsu, A.; et al. ABT-378, a Highly Potent Inhibitor of the Human Immunodeficiency Virus Protease. Antimicrob. Agents Chemother. 1998, 42, 3218–3224. [Google Scholar] [CrossRef] [PubMed]
- Hsu, A.; Granneman, G.R.; Cao, G.; Carothers, L.; El-Shourbagy, T.; Baroldi, P.; Erdman, K.; Brown, F.; Sun, E.; Leonard, J.M. Pharmacokinetic interactions between two human immunodeficiency virus protease inhibitors, ritonavir and saquinavir. Clin. Pharmacol. Ther. 1998, 63, 453–464. [Google Scholar] [CrossRef]
- Ahmad, B.; Batool, M.; Ain, Q.U.; Kim, M.S.; Choi, S. Exploring the Binding Mechanism of PF-07321332 SARS-CoV-2 Protease Inhibitor through Molecular Dynamics and Binding Free Energy Simulations. Int. J. Mol. Sci. 2021, 22, 9124. [Google Scholar] [CrossRef]
- Macchiagodena, M.; Pagliai, M.; Procacci, P. Characterization of the non-covalent interaction between the PF-07321332 inhibitor and the SARS-CoV-2 main protease. J. Mol. Graph. Model. 2021, 110, 108042. [Google Scholar] [CrossRef]
- Hammond, J.; Leister-Tebbe, H.; Gardner, A.; Abreu, P.; Bao, W.; Wisemandle, W.; Baniecki, M.; Hendrick, V.M.; Damle, B.; Simón-Campos, A.; et al. Oral Nirmatrelvir for High-Risk, Nonhospitalized Adults with COVID-19. N. Engl. J. Med. 2022, 386, 1397–1408. [Google Scholar] [CrossRef]
- Yu, H.; Janssen, J.M.; Sawicki, E.; Van Hasselt, J.G.C.; De Weger, V.A.; Nuijen, B.; Schellens, J.H.M.; Beijnen, J.H.; Huitema, A.D.R. A Population Pharmacokinetic Model of Oral Docetaxel Coadministered with Ritonavir to Support Early Clinical Development. J. Clin. Pharmacol. 2020, 60, 340–350. [Google Scholar] [CrossRef]
- De Weger, V.A.; Stuurman, F.E.; Koolen, S.L.W.; Moes, J.J.; Hendrikx, J.J.M.A.; Sawicki, E.; Thijssen, B.; Keessen, M.; Rosing, H.; Mergui-Roelvink, M.; et al. A Phase I Dose Escalation Study of Once-Weekly Oral Administration of Docetaxel as ModraDoc001 Capsule or ModraDoc006 Tablet in Combination with Ritonavir. Clin. Cancer Res. 2019, 25, 5466–5474. [Google Scholar] [CrossRef]
- Greenblatt, D.J.; Harmatz, J.S. Ritonavir is the best alternative to ketoconazole as an index inhibitor of cytochrome P450-3A in drug-drug interaction studies. Br. J. Clin. Pharmacol. 2015, 80, 342–350. [Google Scholar] [CrossRef]
- Darwich, A.S.; Aslam, U.; Ashcroft, D.M.; Rostami-Hodjegan, A. Meta-Analysis of the Turnover of Intestinal Epithelia in Preclinical Animal Species and Humans. Drug Metab. Dispos. 2014, 42, 2016–2022. [Google Scholar] [CrossRef]
- Culmmerdek, K.; Vonmoltke, L.; Gan, L.; Horan, K.A.; Reynolds, R.; Harmatz, J.S.; Court, M.H.; Greenblatt, D.J.; Moltke, L.L. Effect of extended exposure to grapefruit juice on cytochrome P450 3A activity in humans: Comparison with ritonavir. Clin. Pharmacol. Ther. 2006, 79, 243–254. [Google Scholar] [CrossRef]
- Katzenmaier, S.; Markert, C.; Riedel, K.-D.; Burhenne, J.; Haefeli, W.E.; Mikus, G. Determining the Time Course of CYP3A Inhibition by Potent Reversible and Irreversible CYP3A Inhibitors Using a Limited Sampling Strategy. Clin. Pharmacol. Ther. 2011, 90, 666–673. [Google Scholar] [CrossRef] [PubMed]
- Knox, T.A.; Oleson, L.; von Moltke, L.L.; Kaufman, R.C.; Wanke, C.A.; Greenblatt, D.J. Ritonavir Greatly Impairs CYP3A Activity in HIV Infection with Chronic Viral Hepatitis. J. Acquir. Immune Defic. Syndr. 2008, 49, 358–368. [Google Scholar] [CrossRef] [PubMed]
- Ernest, C.S., 2nd; Hall, S.D.; Jones, D.R. Mechanism-based inactivation of CYP3A by HIV protease inhibitors. J. Pharmacol. Exp. Ther. 2005, 312, 583–591. [Google Scholar] [CrossRef]
- Von Moltke, L.L.; Durol, A.L.B.; Duan, S.X.; Greenblatt, D.J. Potent mechanism-based inhibition of human CYP3A in vitro by amprenavir and ritonavir: Comparison with ketoconazole. Eur. J. Clin. Pharmacol. 2000, 56, 259–261. [Google Scholar] [CrossRef]
- Hossain, M.A.; Tran, T.; Chen, T.; Mikus, G.; Greenblatt, D.J. Inhibition of human cytochromes P450 in vitro by ritonavir and cobicistat. J. Pharm. Pharmacol. 2017, 69, 1786–1793. [Google Scholar] [CrossRef]
- Sevrioukova, I.F.; Poulos, T.L. Ritonavir analogues as a probe for deciphering the cytochrome P450 3A4 inhibitory mechanism. Curr. Top. Med. Chem. 2014, 14, 1348–1355. [Google Scholar] [CrossRef]
- Samuels, E.R.; Sevrioukova, I. Inhibition of Human CYP3A4 by Rationally Designed Ritonavir-like Compounds: Impact and Interplay of the Side Group Functionalities. Mol. Pharm. 2018, 15, 279–288. [Google Scholar] [CrossRef]
- Mathias, A.A.; German, P.; Murray, B.P.; Wei, L.; Jain, A.; West, S.; Warren, D.; Hui, J.; Kearney, B.P. Pharmacokinetics and Pharmacodynamics of GS-9350: A Novel Pharmacokinetic Enhancer without Anti-HIV Activity. Clin. Pharmacol. Ther. 2010, 87, 322–329. [Google Scholar] [CrossRef]
- Greenblatt, D.J. Antiretroviral boosting by cobicistat, a structural analog of ritonavir. Clin. Pharmacol. Drug Dev. 2014, 3, 335–337. [Google Scholar] [CrossRef]
- Marzolini, C.; Gibbons, S.; Khoo, S.; Back, D. Cobicistat versus ritonavir boosting and differences in the drug–drug interaction profiles with co-medications. J. Antimicrob. Chemother. 2016, 71, 1755–1758. [Google Scholar] [CrossRef]
- Sevrioukova, I.F.; Poulos, T.L. Structure and mechanism of the complex between cytochrome P4503A4 and ritonavir. Proc. Natl. Acad. Sci. USA 2010, 107, 18422–18427. [Google Scholar] [CrossRef] [PubMed]
- Rendic, S. Summary of information on human CYP enzymes: Human P450 metabolism data. Drug Metab. Rev. 2002, 34, 83–448. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.-L.; D’Agostino, J.; Kenaan, C.; Calinski, D.; Hollenberg, P.F. The Effect of Ritonavir on Human CYP2B6 Catalytic Activity: Heme Modification Contributes to the Mechanism-Based Inactivation of CYP2B6 and CYP3A4 by Ritonavir. Drug Metab. Dispos. 2013, 41, 1813–1824. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Lu, J.; Ma, X. Metabolomic Screening and Identification of the Bioactivation Pathways of Ritonavir. Chem. Res. Toxicol. 2011, 24, 2109–2114. [Google Scholar] [CrossRef] [Green Version]
- Hsu, A.; Granneman, G.R.; Bertz, R.J. Erratum to Ritonavir: Clinical pharmacokinetics and interactions with other anti-HIV agents. Clin. Pharmacokinet. 1998, 35, 473. [Google Scholar] [CrossRef]
- Kaspera, R.; Kirby, B.J.; Sahele, T.; Collier, A.C.; Kharasch, E.D.; Unadkat, J.D.; Totah, R.A. Investigating the contribution of CYP2J2 to ritonavir metabolism in vitro and in vivo. Biochem. Pharmacol. 2014, 91, 109–118. [Google Scholar] [CrossRef]
- Lin, H.L.; Hollenberg, P.F. The inactivation of cytochrome P450 3A5 by 17alpha-ethynylestradiol is cytochrome b5-dependent: Metabolic activation of the ethynyl moiety leads to the formation of glutathione conjugates, a heme adduct, and covalent binding to the apoprotein. J. Pharmacol. Exp. Ther. 2007, 321, 276–287. [Google Scholar] [CrossRef]
- Hollenberg, P.F.; Kent, U.M.; Bumpus, N.N. Mechanism-Based Inactivation of Human Cytochromes P450s: Experimental Characterization, Reactive Intermediates, and Clinical Implications. Chem. Res. Toxicol. 2008, 21, 189–205. [Google Scholar] [CrossRef]
- Ho, H.K.; Chan, J.C.Y.; Hardy, K.D.; Chan, E.C.Y. Mechanism-based inactivation of CYP450 enzymes: A case study of lapatinib. Drug Metab. Rev. 2015, 47, 21–28. [Google Scholar] [CrossRef]
- Sevrioukova, I.F.; Poulos, T.L. Interaction of human cytochrome P4503A4 with ritonavir analogs. Arch. Biochem. Biophys. 2012, 520, 108–116. [Google Scholar] [CrossRef]
- Henderson, C.J.; McLaughlin, L.A.; Scheer, N.; Stanley, L.A.; Wolf, C.R. Cytochrome b5 is a major determinant of human cytochrome P450 CYP2D6 and CYP3A4 activity in vivo. Mol. Pharmacol. 2015, 87, 733–739. [Google Scholar] [CrossRef]
- Lee, S.-J.; Goldstein, J.A. Comparison of CYP3A4 and CYP3A5: The Effects of Cytochrome b5 and NADPH-cytochrome P450 Reductase on Testosterone Hydroxylation Activities. Drug Metab. Pharmacokinet. 2012, 27, 663–667. [Google Scholar] [CrossRef]
- Samuels, E.R.; Sevrioukova, I.F. Rational Design of CYP3A4 Inhibitors: A One-Atom Linker Elongation in Ritonavir-like Compounds Leads to a Marked Improvement in the Binding Strength. Int. J. Mol. Sci. 2021, 22, 852. [Google Scholar] [CrossRef]







| Reference | Suggested Primary Mechanism of Inactivation | Used Enzyme Preparations | Use of Added Cytb5 or CPR in the Experiments * | Incubation Time | Assay(s) | Ritonavir Concentration |
|---|---|---|---|---|---|---|
| Koudriakova et al. (1998) [25] | Reactive intermediate formation | Enterocyte microsomes and HLMs expressing CYP3A4, -3A5, and -2D6 | - No Cytb5 - No CPR | 1 h | Time-course assay using HPLC to examine the rate of ritonavir metabolism | 2 or 5 µM |
| 20 min | Inactivation of CYP enzymes assay using HPLC | 0.075 µM | ||||
| Ernest et al. (2005) [53] | MIC formation | HLMs expressing CYP3A4 and -3A5 | - Recombinant CYP3A4: with Cytb5 - Recombinant CYP3A5: no Cytb5 - All with CPR | Maximally 60 min | CYP3A4/5 inactivation and high-affinity binding assay with testosterone substrate to quantify time- and concentration-dependent loss of CYP3A activity | 0.05, 0.10, 0.20, 0.50, and 1 µM |
| Sevrioukova et al. (2010) [61] | Strong ligation of ritonavir to heme iron | Isolated CYP3A4Δ3-24 | - No Cytb5 - No CPR | - | Kinetic assay of CYP3A4-ritonavir binding using stopped-flow spectrophotometry to measure the kinetics of ritonavir binding to ferric and ferrous P450 by monitoring absorbance changes at 426 and 442 nm | 0.5–30 μM |
| Crystallization and structure determination with bound ritonavir | Ritonavir-bound CYP3A4 protein (50–60 mg/mL) | |||||
| Lin et al. (2013) [63] | Heme destruction and linkage of heme to apoprotein | Purified CYP3A4 and CYP2B6, and HLMs | - No Cytb5 - With CPR | 30 min | Enzyme and inactivation assay of CYP3A4 and CYP2B6 to determine catalytic activity using a fluorescence plate reader | 0.5–20 µM |
| 10 min | HPLC analysis of heme iron to study the loss of native heme and formation of heme adducts | 10 µM for CYP2B6; 2 µM for CYP3A4 | ||||
| 10 min | ESI–LC/MS analysis of the apoprotein to study the mass spectra | 10 µM | ||||
| 20 min | LC-MS/MS analysis of ritonavir metabolites and the GSH conjugate formed | 40 µM | ||||
| Rock et al. (2014) [19] | Reactive intermediate formation with covalent adduct binding to apoprotein (Lys257) | CYP3A4 supersomes or HLMs | - With Cytb5 - With CPR | 30 min, after 3 min pre-incubation | CYP3A4 activity and inactivation assay using midazolam with a UPLC system and LC-MS/MS for the inactivation assay | 0–10 µM of ritonavir or N-ritonavir |
| MIC formation assay using spectrophotometric repetitive scanning from 430–495 nm over 30 min | 10 µM | |||||
| With and without NADPH | 10 min | Mass spectral analysis of CYP3A4 peptides using a liquid chromatography - radioisotope counting system | 10 µM |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Loos, N.H.C.; Beijnen, J.H.; Schinkel, A.H. The Mechanism-Based Inactivation of CYP3A4 by Ritonavir: What Mechanism? Int. J. Mol. Sci. 2022, 23, 9866. https://doi.org/10.3390/ijms23179866
Loos NHC, Beijnen JH, Schinkel AH. The Mechanism-Based Inactivation of CYP3A4 by Ritonavir: What Mechanism? International Journal of Molecular Sciences. 2022; 23(17):9866. https://doi.org/10.3390/ijms23179866
Chicago/Turabian StyleLoos, Nancy H. C., Jos H. Beijnen, and Alfred H. Schinkel. 2022. "The Mechanism-Based Inactivation of CYP3A4 by Ritonavir: What Mechanism?" International Journal of Molecular Sciences 23, no. 17: 9866. https://doi.org/10.3390/ijms23179866