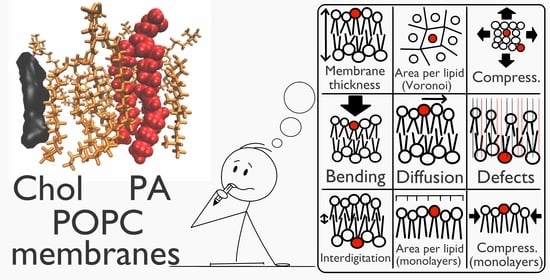
Simple Does Not Mean Trivial: Behavior of Phosphatidic Acid in Lipid Mono- and Bilayers
Abstract
:1. Introduction
2. Results and Discussion
2.1. Molecular Dynamics Characterization
2.2. Monolayers
2.3. Bending Rigidity Determination Using Flicker Noise Spectroscopy
2.4. Interactions of PA Lipids with PC and/or Cholesterol Wrapped Up in a Summary
3. Materials and Methods
3.1. Materials
3.2. Molecular Dynamics (MD) Simulations
3.3. MD System Characteristics
3.4. Surface Pressure–Area (π–A) Isotherm Measurement
3.5. Surface Pressure–Area (π–A) Isotherm Analysis
3.6. GUVs Electroformation
3.7. Flicker Noise Spectroscopy
3.8. Statistics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zegarlinska, J.; Piascik, M.; Sikorski, A.F.; Czogalla, A. Phosphatidic acid—A simple phospholipid with multiple faces. Acta Biochim. Pol. 2018, 65, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Zhukovsky, M.A.; Filograna, A.; Luini, A.; Corda, D.; Valente, C. Phosphatidic acid in membrane rearrangements. FEBS Lett. 2019, 593, 2428–2451. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.Y.; Frohman, M.A. Mitochondria: Signaling with phosphatidic acid. Int. J. Biochem. Cell Biol. 2012, 44, 1346–1350. [Google Scholar] [CrossRef] [Green Version]
- Stace, C.L.; Ktistakis, N.T. Phosphatidic acid- and phosphatidylserine-binding proteins. Biochim. Biophys. Acta 2006, 1761, 913–926. [Google Scholar] [CrossRef]
- Perez-Isidoro, R.; Ruiz-Suarez, J.C. Calcium and protons affect the interaction of neurotransmitters and anesthetics with anionic lipid membranes. Biochim. Biophys. Acta 2016, 1858, 2215–2222. [Google Scholar] [CrossRef]
- Putta, P.; Rankenberg, J.; Korver, R.A.; van Wijk, R.; Munnik, T.; Testerink, C.; Kooijman, E.E. Phosphatidic acid binding proteins display differential binding as a function of membrane curvature stress and chemical properties. Biochim. Biophys. Acta 2016, 1858, 2709–2716. [Google Scholar] [CrossRef]
- Testerink, C.; Munnik, T. Molecular, cellular, and physiological responses to phosphatidic acid formation in plants. J. Exp. Bot. 2011, 62, 2349–2361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kooijman, E.E.; Chupin, V.; Fuller, N.L.; Kozlov, M.M.; de Kruijff, B.; Burger, K.N.; Rand, P.R. Spontaneous curvature of phosphatidic acid and lysophosphatidic acid. Biochemistry 2005, 44, 2097–2102. [Google Scholar] [CrossRef] [PubMed]
- Kooijman, E.E.; Burger, K.N. Biophysics and function of phosphatidic acid: A molecular perspective. Biochim. Biophys. Acta 2009, 1791, 881–888. [Google Scholar] [CrossRef] [PubMed]
- Sezgin, E.; Gutmann, T.; Buhl, T.; Dirkx, R.; Grzybek, M.; Coskun, U.; Solimena, M.; Simons, K.; Levental, I.; Schwille, P. Adaptive lipid packing and bioactivity in membrane domains. PLoS ONE 2015, 10, e0123930. [Google Scholar] [CrossRef] [PubMed]
- Czogalla, A.; Grzybek, M.; Jones, W.; Coskun, U. Validity and applicability of membrane model systems for studying interactions of peripheral membrane proteins with lipids. Biochim. Biophys. Acta 2014, 1841, 1049–1059. [Google Scholar] [CrossRef]
- Vanni, S.; Hirose, H.; Barelli, H.; Antonny, B.; Gautier, R. A sub-nanometre view of how membrane curvature and composition modulate lipid packing and protein recruitment. Nat. Commun. 2014, 5, 4916. [Google Scholar] [CrossRef] [Green Version]
- Lingwood, D.; Binnington, B.; Rog, T.; Vattulainen, I.; Grzybek, M.; Coskun, U.; Lingwood, C.A.; Simons, K. Cholesterol modulates glycolipid conformation and receptor activity. Nat. Chem. Biol. 2011, 7, 260–262. [Google Scholar] [CrossRef]
- Sarkis, J.; Vie, V. Biomimetic Models to Investigate Membrane Biophysics Affecting Lipid-Protein Interaction. Front. Bioeng. Biotechnol. 2020, 8, 270. [Google Scholar] [CrossRef]
- Ouberai, M.M.; Wang, J.; Swann, M.J.; Galvagnion, C.; Guilliams, T.; Dobson, C.M.; Welland, M.E. alpha-Synuclein senses lipid packing defects and induces lateral expansion of lipids leading to membrane remodeling. J. Biol. Chem. 2013, 288, 20883–20895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kassas, N.; Tanguy, E.; Thahouly, T.; Fouillen, L.; Heintz, D.; Chasserot-Golaz, S.; Bader, M.F.; Grant, N.J.; Vitale, N. Comparative Characterization of Phosphatidic Acid Sensors and Their Localization during Frustrated Phagocytosis. J. Biol. Chem. 2017, 292, 4266–4279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ekroos, K.; Ejsing, C.S.; Bahr, U.; Karas, M.; Simons, K.; Shevchenko, A. Charting molecular composition of phosphatidylcholines by fatty acid scanning and ion trap MS3 fragmentation. J. Lipid Res. 2003, 44, 2181–2192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maxfield, F.R.; van Meer, G. Cholesterol, the central lipid of mammalian cells. Curr. Opin. Cell Biol. 2010, 22, 422–429. [Google Scholar] [CrossRef] [Green Version]
- Boyd, K.J.; Alder, N.N.; May, E.R. Molecular Dynamics Analysis of Cardiolipin and Monolysocardiolipin on Bilayer Properties. Biophys. J. 2018, 114, 2116–2127. [Google Scholar] [CrossRef] [Green Version]
- Vanni, S.; Vamparys, L.; Gautier, R.; Drin, G.; Etchebest, C.; Fuchs, P.F.; Antonny, B. Amphipathic lipid packing sensor motifs: Probing bilayer defects with hydrophobic residues. Biophys. J. 2013, 104, 575–584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hristova, K.; Wimley, W.C.; Mishra, V.K.; Anantharamiah, G.M.; Segrest, J.P.; White, S.H. An amphipathic alpha-helix at a membrane interface: A structural study using a novel X-ray diffraction method. J. Mol. Biol. 1999, 290, 99–117. [Google Scholar] [CrossRef] [Green Version]
- Villasuso, A.L.; Wilke, N.; Maggio, B.; Machado, E. Zn2+-dependent surface behavior of diacylglycerol pyrophosphate and its mixtures with phosphatidic acid at different pHs. Front. Plant Sci. 2014, 5, 371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Demel, R.A.; Yin, C.C.; Lin, B.Z.; Hauser, H. Monolayer characterstics and thermal behaviour of phosphatidic acids. Chem. Phys. Lipids 1992, 60, 209–223. [Google Scholar] [CrossRef]
- Cheng, M.H.; Liu, L.T.; Saladino, A.C.; Xu, Y.; Tang, P. Molecular dynamics simulations of ternary membrane mixture: Phosphatidylcholine, phosphatidic acid, and cholesterol. J. Phys. Chem. B 2007, 111, 14186–14192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Meijere, K.; Brezesinski, G.; Mohwald, H. Polyelectrolyte coupling to a charged lipid monolayer. Macromolecules 1997, 30, 2337–2342. [Google Scholar] [CrossRef]
- Estrela-Lopis, I.; Brezesinski, G.; Mohwald, H. Miscibility of DPPC and DPPA in monolayers at the air/water interface. Chem. Phys. Lipids 2004, 131, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Minones, J.; Patino, J.M.R.; Minones, J.; Dynarowicz-Latka, P.; Carrera, C. Structural and topographical characteristics of dipalmitoyl phosphatidic acid in Langmuir monolayers. J. Colloid Interf. Sci. 2002, 249, 388–397. [Google Scholar] [CrossRef] [PubMed]
- Kulig, W.; Korolainen, H.; Zatorska, M.; Kwolek, U.; Wydro, P.; Kepczynski, M.; Rog, T. Complex Behavior of Phosphatidylcholine-Phosphatidic Acid Bilayers and Monolayers: Effect of Acyl Chain Unsaturation. Langmuir ACS J. Surf. Colloids 2019, 35, 5944–5956. [Google Scholar] [CrossRef]
- Lee, Y.L.; Lin, J.Y.; Chang, C.H. Thermodynamic characteristics and Langmuir-Blodgett deposition behavior of mixed DPPA/DPPC monolayers at air/liquid interfaces. J. Colloid Interf. Sci. 2006, 296, 647–654. [Google Scholar] [CrossRef] [PubMed]
- Mildner, J.; Wnetrzak, A.; Dynarowicz-Latka, P. Cholesterol and Cardiolipin Importance in Local Anesthetics-Membrane Interactions: The Langmuir Monolayer Study. J. Membr. Biol. 2019, 252, 31–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huynh, L.; Perrot, N.; Beswick, V.; Rosilio, V.; Curmi, P.A.; Sanson, A.; Jamin, N. Structural properties of POPC monolayers under lateral compression: Computer simulations analysis. Langmuir ACS J. Surf. Colloids 2014, 30, 564–573. [Google Scholar] [CrossRef] [PubMed]
- Jurak, M. Thermodynamic aspects of cholesterol effect on properties of phospholipid monolayers: Langmuir and Langmuir-Blodgett monolayer study. J. Phys. Chem. B 2013, 117, 3496–3502. [Google Scholar] [CrossRef]
- Marsh, D. Lateral pressure in membranes. Biochim. Biophys. Acta 1996, 1286, 183–223. [Google Scholar] [CrossRef]
- Marsh, D. Elastic curvature constants of lipid monolayers and bilayers. Chem Phys Lipids 2006, 144, 146–159. [Google Scholar] [CrossRef] [Green Version]
- Brockman, J.M.; Wang, Z.; Notter, R.H.; Dluhy, R.A. Effect of hydrophobic surfactant proteins SP-B and SP-C on binary phospholipid monolayers: II. Infrared external reflectance-absorption spectroscopy. Biophys. J. 2003, 84, 326–340. [Google Scholar] [CrossRef] [Green Version]
- Brown, R.E.; Brockman, H.L. Using monomolecular films to characterize lipid lateral interactions. Methods Mol. Biol. 2007, 398, 41–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwolek, U.; Kulig, W.; Wydro, P.; Nowakowska, M.; Rog, T.; Kepczynski, M. Effect of Phosphatidic Acid on Biomembrane: Experimental and Molecular Dynamics Simulations Study. J. Phys. Chem. B 2015, 119, 10042–10051. [Google Scholar] [CrossRef]
- Yu, Y.; Kramer, A.; Venable, R.M.; Brooks, B.R.; Klauda, J.B.; Pastor, R.W. CHARMM36 Lipid Force Field with Explicit Treatment of Long-Range Dispersion: Parametrization and Validation for Phosphatidylethanolamine, Phosphatidylglycerol, and Ether Lipids. J. Chem. Theory Comput. 2021, 17, 1581–1595. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Kramer, A.; Venable, R.M.; Simmonett, A.C.; MacKerell, A.D., Jr.; Klauda, J.B.; Pastor, R.W.; Brooks, B.R. Semi-automated Optimization of the CHARMM36 Lipid Force Field to Include Explicit Treatment of Long-Range Dispersion. J. Chem. Theory Comput. 2021, 17, 1562–1580. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Ghosh, S.K.; DiLena, D.A.; Bera, S.; Lurio, L.B.; Parikh, A.N.; Sinha, S.K. Cholesterol Partition and Condensing Effect in Phase-Separated Ternary Mixture Lipid Multilayers. Biophys. J. 2016, 110, 1355–1366. [Google Scholar] [CrossRef] [Green Version]
- Akabori, K.; Nagle, J.F. Structure of the DMPC lipid bilayer ripple phase. Soft. Matter 2015, 11, 918–926. [Google Scholar] [CrossRef] [Green Version]
- Faizi, H.A.; Reeves, C.J.; Georgiev, V.N.; Vlahovska, P.M.; Dimova, R. Fluctuation spectroscopy of giant unilamellar vesicles using confocal and phase contrast microscopy. Soft. Matter 2020. [Google Scholar] [CrossRef] [PubMed]
- Drabik, D.; Chodaczek, G.; Kraszewski, S.; Langner, M. Mechanical Properties Determination of DMPC, DPPC, DSPC, and HSPC Solid-Ordered Bilayers. Langmuir ACS J. Surf. Colloids 2020, 36, 3826–3835. [Google Scholar] [CrossRef] [PubMed]
- Nagle, J.F.; Evans, E.A.; Bassereau, P.; Baumgart, T.; Tristram-Nagle, S.; Dimova, R. A needless but interesting controversy. Proc. Natl. Acad. Sci. USA 2021, 118, e2025011118. [Google Scholar] [CrossRef] [PubMed]
- Drabik, D.; Gavutis, M.; Valiokas, R.N.; Ulcinas, A.R. Determination of the Mechanical Properties of Model Lipid Bilayers Using Atomic Force Microscopy Indentation. Langmuir ACS J. Surf. Colloids 2020, 36, 13251–13262. [Google Scholar] [CrossRef] [PubMed]
- Philips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorsid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klauda, J.B.; Venable, R.M.; Freites, J.A.; O’Connor, J.W.; Tobias, D.J.; Mondragon-Ramirez, C.; Vorobyov, I.; MacKerell, A.D., Jr.; Pastor, R.W. Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types. J. Phys. Chem. B 2010, 114, 7830–7843. [Google Scholar] [CrossRef] [Green Version]
- Doktorova, M.; Harries, D.; Khelashvili, G. Determination of bending rigidity and tilt modulus of lipid membranes from real-space fluctuation analysis of molecular dynamics simulations. Phys. Chem. Chem. Phys. PCCP 2017, 19, 16806–16818. [Google Scholar] [CrossRef]
- Doktorova, M.; LeVine, M.V.; Khelashvili, G.; Weinstein, H. A New Computational Method for Membrane Compressibility: Bilayer Mechanical Thickness Revisited. Biophys. J. 2019, 116, 487–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagle, J.F. Area Compressibility Moduli of the Monolayer Leaflets of Asymmetric Bilayers from Simulations. Biophys. J. 2019, 117, 1051–1056. [Google Scholar] [CrossRef] [PubMed]
- Giorgino, T. Computing diffusion coefficients in macromolecular simulations: The Diffusion Coefficient Tool for VMD. J. Open Source Softw. 2019, 4, 1698. [Google Scholar] [CrossRef] [Green Version]
- Guixa-Gonzalez, R.; Rodriguez-Espigares, I.; Ramirez-Anguita, J.M.; Carrio-Gaspar, P.; Martinez-Seara, H.; Giorgino, T.; Selent, J. MEMBPLUGIN: Studying membrane complexity in VMD. Bioinformatics 2014, 30, 1478–1480. [Google Scholar] [CrossRef] [Green Version]
- Rouser, G.; Fleischer, S.; Yamamoto, A. Two dimensional thin layer chromatographic separation of polar lipids and determination of phospholipids by phosphorus analysis of spots. Lipids 1970, 5, 494–496. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, T.; Kato, S. Detailed Analysis of the Surface Area and Elasticity in the Saturated 1,2-Diacylphosphatidylcholine/Cholesterol Binary Monolayer System. Langmuir ACS J. Surf. Colloids 2015, 31, 9086–9096. [Google Scholar] [CrossRef]
- Grzybek, M.; Kubiak, J.; Lach, A.; Przybylo, M.; Sikorski, A.F. A raft-associated species of phosphatidylethanolamine interacts with cholesterol comparably to sphingomyelin. A Langmuir-Blodgett monolayer study. PLoS ONE 2009, 4, e5053. [Google Scholar] [CrossRef] [PubMed]
- Savva, M.; Acheampong, S. The interaction energies of cholesterol and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine in spread mixed monolayers at the air-water interface. J. Phys. Chem. B 2009, 113, 9811–9820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jurak, M.; Szafran, K.; Cea, P.; Martin, S. Analysis of Molecular Interactions between Components in Phospholipid-Immunosuppressant-Antioxidant Mixed Langmuir Films. Langmuir ACS J. Surf. Colloids 2021, 37, 5601–5616. [Google Scholar] [CrossRef]
- Drabik, D.; Doskocz, J.; Przybyło, M. Effects of electroformation protocol parameters on quality of homogeneous GUV populations. Chem. Phys. Lipids 2018, 212, 88–95. [Google Scholar] [CrossRef]
- Drabik, D.; Przybylo, M.; Chodaczek, G.; Iglic, A.; Langner, M. The modified fluorescence based vesicle fluctuation spectroscopy technique for determination of lipid bilayer bending properties. Biochim. Biophys. Acta 2016, 1858, 244–252. [Google Scholar] [CrossRef]






|
Lipid Bilayer Composition (Molar Ratios) | MTP-P | APL | κ | κtilt | KA | 2D Diffusion | Interdigitation |
|---|---|---|---|---|---|---|---|
| nm | Å2 | J | J | mN/m | µm2/s | Å | |
| POPC | 3.78 ± 0.04 | 60.7 ± 1.4 | (1.12 ± 0.03) × 10−19 | (2.86 ± 0.05) × 10−20 | 235 ± 23 | 7.7 ± 0.6 | 5.85 ± 0.43 |
| POPA | 4.10 ± 0.04 | 56.1 ± 1.4 | (1.27 ± 0.03) × 10−19 | (3.82 ± 0.07) × 10−20 | 846 ± 73 | 12.35 ± 0.12 | 5.09 ± 0.43 |
| DPPA | 4.64 ± 0.02 | 42.9 ± 1.2 | (4.55 ± 0.13) × 10−19 | (17.86 ± 0.54) × 10−20 | 4969 ± 208 | 1.57 ± 0.03 | 4.7 ± 0.3 |
| SAPA | 4.99 ± 0.03 | 61.5 ± 1.5 | (1.23 ± 0.04) × 10−19 | (3.33 ± 0.05) × 10−20 | 201.5 ± 23 | 9.65 ± 0.15 | 6.3 ± 0.5 |
| Chol | 3.18 ± 0.08 | 37.19 ± 0.35 | (4.34 ± 0.01) × 10−19 | (7.78 ± 0.06) × 10−20 | 16,526 ± 453 | 1.12 ± 0.01 | 3.37 ± 0.21 |
| POPC/POPA 8:2 | 3.92 ± 0.03 | 61.1 ± 1.4 | (1.19 ± 0.03) × 10−19 | (3.47 ± 0.06) × 10−20 | 455 ± 24 | 10.3 ± 0.2 | 6.0 ± 0.5 |
| POPC/DPPA 8:2 | 3.99 ± 0.03 | 59.4 ± 1.4 | (1.34 ± 0.03) × 10−19 | (3.52 ± 0.06) × 10−20 | 502 ± 33 | 13.16 ± 0.13 | 5.57 ± 0.44 |
| POPC/SAPA 8:2 | 3.95 ± 0.03 | 62.0 ± 1.3 | (1.22 ± 0.03) × 10−19 | (3.39 ± 0.05) × 10−20 | 156 ± 38 | 10.7 ± 0.1 | 6.1 ± 0.5 |
| Chol/POPA 1:1 | 3.97 ± 0.01 | 40.1 ± 0.9 | (1.60 ± 0.04) × 10−19 | (7.27 ± 0.01) × 10−20 | 8320 ± 162 | 3.85 ± 0.04 | 3.3 ± 0.3 |
| Chol/DPPA 1:1 | 4.04 ± 0.01 | 38.0 ± 0.8 | (1.59 ± 0.04) × 10−19 | (10.1 ± 0.1) × 10−20 | 13,898 ± 270 | 2.83 ± 0.03 | 2.9 ± 0.3 |
| Chol/SAPA 1:1 | 4.13 ± 0.01 | 40.0 ± 1.1 | (1.52 ± 0.04) × 10−19 | (6.48 ± 0.01) × 10−20 | 8304 ± 221 | 2.40 ± 0.06 | 3.7 ± 0.3 |
| POPC/Chol/POPA 5:3:2 | 4.08 ± 0.02 | 41.8 ± 2.0 | (1.51 ± 0.03) × 10−19 | (5.9 ± 0.1) × 10−20 | 3005 ± 110 | 6.44 ± 0.08 | 3.8 ± 0.3 |
| POPC/Chol/DPPA 5:3:2 | 4.17 ± 0.02 | 42.0 ± 1.5 | (1.66 ± 0.03) × 10−19 | (7.4 ± 0.1) × 10−20 | 7521 ± 257 | 5.49 ± 0.07 | 3.69 ± 0.42 |
| POPC/Chol/SAPA 5:3:2 | 4.12 ± 0.02 | 43.9 ± 1.8 | (1.43 ± 0.02) × 10−19 | (5.6 ± 0.1) × 10−20 | 3908 ± 116 | 5.46 ± 0.07 | 4.22 ± 0.41 |
| POPC/Chol 7:3 | 4.14 ± 0.02 | 45.3 ± 2.3 | (1.54 ± 0.03) × 10−19 | (5.7 ± 0.1) × 10−20 | 1948 ± 95 | 2.85 ± 0.02 | 3.75 ± 0.33 |
|
Lipid Bilayer Composition (Molar Ratios) | APL | Cs | ΔG | Κ |
|---|---|---|---|---|
| Å2 | mN/m | J | ||
| POPC | 59 ± 1 | 59 ± 10 | - | (1.9 ± 0.8) × 10−19 |
| POPA | 50.3 ± 2.2 | 53 ± 7 | - | - |
| DPPA | 39.3 ± 1.3 | 648 ± 58 | - | - |
| SAPA | 47 ± 2 | 53.4 ± 2.5 | - | - |
| Chol | 39.5 ± 1.2 | 227 ± 84 | - | - |
| POPC/POPA 8:2 | 38.6 ± 2.3 | 35.1 ± 7.2 | −666 ± 10 | (1.8 ± 0.8) × 10−19 |
| POPC/DPPA 8:2 | 54.1 ± 1.4 | 68 ± 13 | 50 ± 5 | (1.1 ± 0.5) × 10−19 |
| POPC/SAPA 8:2 | 44.6 ± 2.1 | 42.8 ± 4.3 | −302 ± 4 | (1.9 ± 0.6) × 10−19 |
| Chol/POPA 1:1 | 42.1 ± 0.7 | 102 ± 34 | −230 ± 6 | - |
| Chol/DPPA 1:1 | 44.8 ± 1.4 | 227 ± 53 | 560 ± 2 | - |
| Chol/SAPA 1:1 | 39.8 ± 1.3 | 68 ± 14 | −63 ± 4 | - |
| POPC/Chol/POPA 5:3:2 | 48.1 ± 2.3 | 58.5 ± 12.2 | −167 ± 8 | (10.0 ± 2.6) × 10−19 |
| POPC/Chol/DPPA 5:3:2 | 36.4 ± 1 | 45 ± 7 | −239 ± 4 | (10 ± 3) × 10−19 |
| POPC/Chol/SAPA 5:3:2 | 46.7 ± 1.5 | 57.1 ± 6.3 | −133 ± 5 | (7.8 ± 3.1) × 10−19 |
| POPC/Chol 7:3 | 44.3 ± 1.1 | 51 ± 11 | −18 ± 4 | (2.6 ± 0.9) × 10−19 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Drabik, D.; Czogalla, A. Simple Does Not Mean Trivial: Behavior of Phosphatidic Acid in Lipid Mono- and Bilayers. Int. J. Mol. Sci. 2021, 22, 11523. https://doi.org/10.3390/ijms222111523
Drabik D, Czogalla A. Simple Does Not Mean Trivial: Behavior of Phosphatidic Acid in Lipid Mono- and Bilayers. International Journal of Molecular Sciences. 2021; 22(21):11523. https://doi.org/10.3390/ijms222111523
Chicago/Turabian StyleDrabik, Dominik, and Aleksander Czogalla. 2021. "Simple Does Not Mean Trivial: Behavior of Phosphatidic Acid in Lipid Mono- and Bilayers" International Journal of Molecular Sciences 22, no. 21: 11523. https://doi.org/10.3390/ijms222111523