Using All-Atom Potentials to Refine RNA Structure Predictions of SARS-CoV-2 Stem Loops
Abstract
:1. Introduction
2. Results
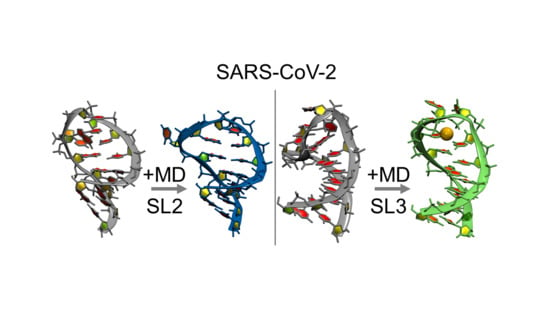
2.1. MD Refinement of SL2
2.1.1. Refinement of SL2
2.1.2. Validation of SL2 Prediction
2.2. MD Refinement of SL3
2.2.1. Predicted SL3 Structures
2.2.2. The Role of the Solvent and Ions in SL3
3. Discussion
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| SARS | Severe acute respiratory syndrome |
| CoV | Coronavirus |
| MD | Molecular Dynamics |
| NMR | Nuclear Magnetic Resonance |
| RNA | Ribonucleic Acid |
| UTR | Untranslated Region |
| TRS | Transcription Regulating Sequences |
| SL | Stem Loop |
References
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef] [Green Version]
- Hermann, T. Small molecules targeting viral RNA. Wiley Interdiscip. Rev. RNA 2016, 7, 726–743. [Google Scholar] [CrossRef]
- Cleveland, M.; Vallone, P.; Romsos, E.; Steffen, B.; Valiant, W. RGTM 10169-The SARS-CoV-2 Research Grade Test Material; National Institute of Standards and Technology: Gaithersburg, MD, USA, 2020. [Google Scholar]
- Coronavirus Standards Working Group—The Joint Initiative for Metrology in Biology. Available online: https://jimb.stanford.edu/covid-19-standards (accessed on 30 July 2020).
- Madhugiri, R.; Karl, N.; Petersen, D.; Lamkiewicz, K.; Fricke, M.; Wend, U.; Scheuer, R.; Marz, M.; Ziebuhr, J. Structural and functional conservation of cis-acting RNA elements in coronavirus 5′-terminal genome regions. Virology 2018, 517, 44–55. [Google Scholar] [CrossRef]
- Yang, D.; Leibowitz, J.L. The structure and functions of coronavirus genomic 3′ and 5′ ends. Virus Res. 2015, 206, 120–133. [Google Scholar] [CrossRef]
- Sawicki, S.G.; Sawicki, D.L.; Siddell, S.G. A contemporary view of coronavirus transcription. J. Virol. 2007, 81, 20–29. [Google Scholar] [CrossRef] [Green Version]
- Liu, P.; Li, L.; Millership, J.J.; Kang, H.; Leibowitz, J.L.; Giedroc, D.P. A U-turn motif-containing stem-loop in the coronavirus 5′ untranslated region plays a functional role in replication. RNA 2007, 13, 763–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sola, I.; Almazán, F.; Zúñiga, S.; Enjuanes, L. Continuous and Discontinuous RNA Synthesis in Coronaviruses. Annu. Rev. Virol. 2015, 2, 265–288. [Google Scholar] [CrossRef] [Green Version]
- Dufour, D.; Mateos-Gomez, P.A.; Enjuanes, L.; Gallego, J.; Sola, I. Structure and Functional Relevance of a Transcription-Regulating Sequence Involved in Coronavirus Discontinuous RNA Synthesis. J. Virol. 2011, 85, 4963–4973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van den Born, E.; Posthuma, C.C.; Gultyaev, A.P.; Snijder, E.J. Discontinuous subgenomic RNA synthesis in arteriviruses is guided by an RNA hairpin structure located in the genomic leader region. J. Virol. 2005, 79, 6312–6324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rangan, R.; Zheludev, I.N.; Das, R. RNA genome conservation and secondary structure in SARS-CoV-2 and SARS-related viruses: A first look. RNA 2020. [Google Scholar] [CrossRef]
- Rangan, R.; Watkins, A.M.; Kladwang, W.; Das, R. De novo 3D models of SARS-CoV-2 RNA elements and small-molecule-binding RNAs to guide drug discovery. bioRxiv 2020, 1–31. [Google Scholar] [CrossRef]
- Cheng, C.Y.; Chou, F.C.; Das, R. Modeling complex RNA tertiary folds with Rosetta. Methods Enzymol. 2015, 553, 35–64. [Google Scholar] [CrossRef] [PubMed]
- Sripakdeevong, P.; Kladwang, W.; Das, R. An enumerative stepwise ansatz enables atomic-accuracy RNA loop modeling. Proc. Natl. Acad. Sci. USA 2011, 108, 20573–20578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergonzo, C.; Grishaev, A. Maximizing accuracy of RNA structure in refinement against residual dipolar couplings. J. Biomol. NMR 2019, 73, 117–139. [Google Scholar] [CrossRef] [PubMed]
- Bergonzo, C.; Hall, K.B.; Cheatham, T.E. Stem-Loop v of Varkud Satellite RNA Exhibits Characteristics of the Mg2+ Bound Structure in the Presence of Monovalent Ions. J. Phys. Chem. B 2015. [Google Scholar] [CrossRef]
- Giambaşu, G.M.; York, D.M.; Case, D.A. Structural fidelity and NMR relaxation analysis in a prototype RNA hairpin. RNA 2015, 21, 963–974. [Google Scholar] [CrossRef] [Green Version]
- Bergonzo, C.; Galindo-Murillo, R.; Cheatham, T.E., 3rd. Molecular Modeling of Nucleic Acid Structure: Electrostatics and Solvation. Curr. Protoc. Nucleic Acid Chem. 2013, 55, 7.9.1–7.9.27. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.W.; Li, L.; Giedroc, D.P. The solution structure of coronaviral stem-loop 2 (SL2) reveals a canonical CUYG tetraloop fold. FEBS Lett. 2011, 585, 1049–1053. [Google Scholar] [CrossRef] [Green Version]
- Kührová, P.; Banáš, P.; Best, R.B.; Šponer, J.; Otyepka, M. Computer folding of RNA tetraloops? Are we there yet? J. Chem. Theory Comput. 2013, 9, 2115–2125. [Google Scholar] [CrossRef]
- Woese, C.R.; Winkers, S.; Gutell, R.R. Architecture of ribosomal RNA: Constraints on the sequence of “tetra-loops”. Proc. Natl. Acad. Sci. USA 1990, 87, 8467–8471. [Google Scholar] [CrossRef] [Green Version]
- Draper, D.E. A guide to ions and RNA structure. RNA 2004, 10, 335–343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mancinelli, R.; Botti, A.; Bruni, F.; Ricci, M.A.; Soper, A.K. Hydration of sodium, potassium, and chloride ions in solution and the concept of structure maker/breaker. J. Phys. Chem. B 2007, 111, 13570–13577. [Google Scholar] [CrossRef]
- Lan, T.C.T.; Allan, M.F.; Malsick, L.E.; Khandwala, S.; Nyeo, S.S.Y.; Bathe, M.; Griffiths, A.; Rouskin, S. Structure of the full SARS-CoV-2 RNA genome in infected cells. bioRxiv 2020, 1–22. [Google Scholar] [CrossRef]
- Case, D.A.; Brozell, S.R.; Cerutti, D.S.; Cheatham III, T.E.; Cruzeiro, V.W.D.; Darden, T.A.; Duke, R.E.; Ghoreishi, D.; Gohlke, H.; Goetz, A.W.; et al. AMBER 2018; University of California: San Francisco, CA, USA, 2018. [Google Scholar]
- Bergonzo, C.; Cheatham, T.E., 3rd. Improved Force Field Parameters Lead to a Better Description of RNA Structure. J. Chem. Theory Comput. 2015, 11, 3969–3972. [Google Scholar] [CrossRef] [PubMed]
- Anandakrishnan, R.; Baker, C.; Izadi, S.; Onufriev, A.V. Point Charges Optimally Placed to Represent the Multipole Expansion of Charge Distributions. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [Green Version]
- Joung, I.S.; Cheatham, T.E., 3rd. Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J. Phys. Chem. B 2008, 112, 9020–9041. [Google Scholar] [CrossRef] [Green Version]
- Bergonzo, C.; Hall, K.B.; Cheatham, T.E., 3rd. Divalent Ion Dependent Conformational Changes in an RNA Stem-Loop Observed by Molecular Dynamics. J. Chem. Theory Comput. 2016, 12, 3382–3389. [Google Scholar] [CrossRef]
- Hopkins, C.W.; Le Grand, S.; Walker, R.C.; Roitberg, A.E. Long time step molecular dynamics through hydrogen mass repartitioning. J. Chem. Theory Comput. 2015, 11, 1864–1874. [Google Scholar] [CrossRef]
- Ryckaert, J.-P.; Ciccotti, G.; Berendsen, H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef] [Green Version]
- Götz, A.W.; Williamson, M.J.; Xu, D.; Poole, D.; Le Grand, S.; Walker, R.C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. Generalized born. J. Chem. Theory Comput. 2012, 8, 1542–1555. [Google Scholar] [CrossRef]
- Salomon-Ferrer, R.; Götz, A.W.; Poole, D.; Le Grand, S.; Walker, R.C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh Ewald. J. Chem. Theory Comput. 2013, 9, 3878–3888. [Google Scholar] [CrossRef] [PubMed]
- Loncharich, R.J.; Brooks, B.R.; Pastor, R.W. Langevin dynamics of peptides: The frictional dependence of isomerization rates of N-acetylalanyl-N′-methylamide. Biopolymers 1992, 32, 523–535. [Google Scholar] [CrossRef] [PubMed]
- Sindhikara, D.J.; Kim, S.; Voter, A.F.; Roitberg, A.E. Bad seeds sprout perilous dynamics: Stochastic thermostat induced trajectory synchronization in biomolecules. J. Chem. Theory Comput. 2009, 5, 1624–1631. [Google Scholar] [CrossRef] [PubMed]
- Roe, D.R.; Cheatham, T.E., 3rd. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef] [PubMed]
- Roe, D.R.; Cheatham, T.E. Parallelization of CPPTRAJ enables large scale analysis of molecular dynamics trajectory data. J. Comput. Chem. 2018, 39, 2110–2117. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Xu, X.; Moon, S.; Case, D.A. SHIFTS v 5.1. Rutgers; The State University of New Jersey: New Brunswick, NJ, USA, 2001. [Google Scholar]




| Starting Ensemble | NMR_MD Ensemble | FAR_CG Ensemble | FAR_CU Ensemble | |
|---|---|---|---|---|
| χ2 | 0.11 | 3.26 | 8.59 | 59.84 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bergonzo, C.; Szakal, A.L. Using All-Atom Potentials to Refine RNA Structure Predictions of SARS-CoV-2 Stem Loops. Int. J. Mol. Sci. 2020, 21, 6188. https://doi.org/10.3390/ijms21176188
Bergonzo C, Szakal AL. Using All-Atom Potentials to Refine RNA Structure Predictions of SARS-CoV-2 Stem Loops. International Journal of Molecular Sciences. 2020; 21(17):6188. https://doi.org/10.3390/ijms21176188
Chicago/Turabian StyleBergonzo, Christina, and Andrea L. Szakal. 2020. "Using All-Atom Potentials to Refine RNA Structure Predictions of SARS-CoV-2 Stem Loops" International Journal of Molecular Sciences 21, no. 17: 6188. https://doi.org/10.3390/ijms21176188