The Role of Peptide Signals Hidden in the Structure of Functional Proteins in Plant Immune Responses
Abstract
:1. Introduction
2. Plant Cryptic Immune Peptides
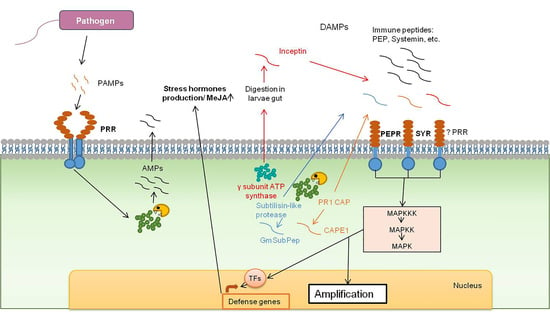
2.1. Inceptin
2.2. CAP-Derived Peptide 1
2.3. Glycine Max Subtilase Peptide
3. Peptide Signals Hidden in Phytopathogenic Proteins
3.1. Flagellin-Derived Peptides
3.2. Elf18
3.3. Sulfated Peptide RaxX
3.4. Pep-13
3.5. Cuscuta Factor
3.6. Csp22, Csp15
3.7. RxLR Avr Effectors
4. Functional Proteins as a Source of Antimicrobial Peptides
5. Perspectives
Funding
Conflicts of Interest
Abbreviations
| PRR | Pattern recognition receptor |
| RLK | Receptor-like kinases |
| PAMP | Pathogen-associated molecular pattern |
| DAMP | Damage-associated molecular pattern |
| PTI | Pattern-triggered immunity |
| ETI | Effector-triggered immunity |
| NB-LRR | Nucleotide binding leucine-rich repeat |
| PEP | Plant elicitor peptide |
| LRR-RLK | Leucine-rich repeat receptor-like kinases |
| PEPR | Plant elicitor peptide receptor |
| BAK1 | BRI1-associated kinase 1 |
| SA | Salicylic acid |
| MeJA | Methyl jasmonate |
| AMP | Antimicrobial peptides |
References
- Boller, T.; Felix, G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 2009, 60, 379–406. [Google Scholar] [CrossRef] [PubMed]
- Bigeard, J.; Colcombet, J.; Hirt, H. Signaling mechanisms in pattern-triggered immunity (PTI). Mol. Plant 2015, 8, 521–539. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhou, J.M. Plant immunity triggered by microbial molecular signatures. Mol. Plant 2010, 3, 783–793. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Gomez, L.; Boller, T. FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 2000, 5, 1003–1011. [Google Scholar] [CrossRef]
- Katagiri, F.; Tsuda, K. Understanding the plant immune system. Mol. Plant Microbe Interact. 2010, 23, 1531–1536. [Google Scholar] [CrossRef] [PubMed]
- Han, G.Z. Origin and evolution of the plant immune system. New Phytol. 2019, 222, 70–83. [Google Scholar] [CrossRef]
- Nawrot, R.; Barylski, J.; Nowicki, G.; Broniarczyk, J.; Buchwald, W.; Gozdzicka-Jozefiak, A. Plant antimicrobial peptides. Folia Microbiol. 2014, 59, 181–196. [Google Scholar] [CrossRef]
- Peng, Y.; van Wersch, R.; Zhang, Y. Convergent and Divergent Signaling in PAMP-Triggered Immunity and Effector-Triggered Immunity. Mol. Plant Microbe Interact. 2018, 31, 403–409. [Google Scholar] [CrossRef]
- Kanyuka, K.; Rudd, J.J. Cell surface immune receptors: The guardians of the plant’s extracellular spaces. Curr. Opin. Plant Biol. 2019, 50, 1–8. [Google Scholar] [CrossRef]
- Choi, H.W.; Klessig, D.F. DAMPs, MAMPs, and NAMPs in plant innate immunity. BMC Plant Biol. 2016, 16, 232. [Google Scholar] [CrossRef]
- Gust, A.A.; Pruitt, R.; Nurnberger, T. Sensing Danger: Key to Activating Plant Immunity. Trends Plant Sci. 2017, 22, 779–791. [Google Scholar] [CrossRef] [PubMed]
- Huffaker, A.; Pearce, G.; Ryan, C.A. An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc. Natl. Acad. Sci. USA 2006, 103, 10098–10103. [Google Scholar] [CrossRef] [PubMed]
- Hander, T.; Fernandez-Fernandez, A.D.; Kumpf, R.P.; Willems, P.; Schatowitz, H.; Rombaut, D.; Staes, A.; Nolf, J.; Pottie, R.; Yao, P.; et al. Damage on plants activates Ca(2+)-dependent metacaspases for release of immunomodulatory peptides. Science 2019, 363. [Google Scholar] [CrossRef]
- Wrzaczek, M.; Vainonen, J.P.; Stael, S.; Tsiatsiani, L.; Help-Rinta-Rahko, H.; Gauthier, A.; Kaufholdt, D.; Bollhoner, B.; Lamminmaki, A.; Staes, A.; et al. GRIM REAPER peptide binds to receptor kinase PRK5 to trigger cell death in Arabidopsis. EMBO J. 2015, 34, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Ziemann, S.; van der Linde, K.; Lahrmann, U.; Acar, B.; Kaschani, F.; Colby, T.; Kaiser, M.; Ding, Y.; Schmelz, E.; Huffaker, A.; et al. An apoplastic peptide activates salicylic acid signalling in maize. Nat. Plants 2018, 4, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y.; Huffaker, A.; Bryan, A.C.; Tax, F.E.; Ryan, C.A. PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell 2010, 22, 508–522. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.L.; Du, L.; Huang, Y.; Gao, S.M.; Yu, M. Origin and diversification of leucine-rich repeat receptor-like protein kinase (LRR-RLK) genes in plants. BMC Evol. Biol. 2017, 17, 47. [Google Scholar] [CrossRef]
- McGurl, B.; Pearce, G.; Orozco-Cardenas, M.; Ryan, C.A. Structure, expression, and antisense inhibition of the systemin precursor gene. Science 1992, 255, 1570–1573. [Google Scholar] [CrossRef]
- Wang, L.; Einig, E.; Almeida-Trapp, M.; Albert, M.; Fliegmann, J.; Mithofer, A.; Kalbacher, H.; Felix, G. The systemin receptor SYR1 enhances resistance of tomato against herbivorous insects. Nat. Plants 2018, 4, 152–156. [Google Scholar] [CrossRef]
- Ryan, C.A. The systemin signaling pathway: Differential activation of plant defensive genes. Biochim. Biophys. Acta 2000, 1477, 112–121. [Google Scholar] [CrossRef]
- Beloshistov, R.E.; Dreizler, K.; Galiullina, R.A.; Tuzhikov, A.I.; Serebryakova, M.V.; Reichardt, S.; Shaw, J.; Taliansky, M.E.; Pfannstiel, J.; Chichkova, N.V.; et al. Phytaspase-mediated precursor processing and maturation of the wound hormone systemin. New Phytol. 2018, 218, 1167–1178. [Google Scholar] [CrossRef] [PubMed]
- Kastin, A.J. Handbook of Biologically Active Peptides, 2nd ed.; Academic Press: Cambridge, MA, USA, 2013. [Google Scholar]
- Arnison, P.G.; Bibb, M.J.; Bierbaum, G.; Bowers, A.A.; Bugni, T.S.; Bulaj, G.; Camarero, J.A.; Campopiano, D.J.; Challis, G.L.; Clardy, J.; et al. Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2013, 30, 108–160. [Google Scholar] [CrossRef] [PubMed]
- Filippova, A.; Lyapina, I.; Kirov, I.; Zgoda, V.; Belogurov, A.; Kudriaeva, A.; Ivanov, V.; Fesenko, I. Salicylic acid influences the protease activity and posttranslation modifications of the secreted peptides in the moss Physcomitrella patens. J. Pept. Sci. 2019, 25, e3138. [Google Scholar] [CrossRef] [PubMed]
- Samir, P.; Link, A.J. Analyzing the cryptome: Uncovering secret sequences. AAPS J. 2011, 13, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.L.; Lee, C.Y.; Cheng, K.T.; Chang, W.H.; Huang, R.N.; Nam, H.G.; Chen, Y.R. Quantitative peptidomics study reveals that a wound-induced peptide from PR-1 regulates immune signaling in tomato. Plant Cell 2014, 26, 4135–4148. [Google Scholar] [CrossRef] [PubMed]
- Pearce, G.; Munske, G.; Yamaguchi, Y.; Ryan, C.A. Structure-activity studies of GmSubPep, a soybean peptide defense signal derived from an extracellular protease. Peptides 2010, 31, 2159–2164. [Google Scholar] [CrossRef] [PubMed]
- Schmelz, E.A.; Carroll, M.J.; LeClere, S.; Phipps, S.M.; Meredith, J.; Chourey, P.S.; Alborn, H.T.; Teal, P.E. Fragments of ATP synthase mediate plant perception of insect attack. Proc. Natl. Acad. Sci. USA 2006, 103, 8894–8899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fesenko, I.; Azarkina, R.; Kirov, I.; Kniazev, A.; Filippova, A.; Grafskaia, E.; Lazarev, V.; Zgoda, V.; Butenko, I.; Bukato, O.; et al. Phytohormone treatment induces generation of cryptic peptides with antimicrobial activity in the Moss Physcomitrella patens. BMC Plant Biol. 2019, 19, 9. [Google Scholar] [CrossRef] [PubMed]
- Fesenko, I.A.; Arapidi, G.P.; Skripnikov, A.Y.; Alexeev, D.G.; Kostryukova, E.S.; Manolov, A.I.; Altukhov, I.A.; Khazigaleeva, R.A.; Seredina, A.V.; Kovalchuk, S.I.; et al. Specific pools of endogenous peptides are present in gametophore, protonema, and protoplast cells of the moss Physcomitrella patens. BMC Plant Biol. 2015, 15, 87. [Google Scholar] [CrossRef] [PubMed]
- Kmiec, B.; Branca, R.M.M.; Berkowitz, O.; Li, L.; Wang, Y.; Murcha, M.W.; Whelan, J.; Lehtio, J.; Glaser, E.; Teixeira, P.F. Accumulation of endogenous peptides triggers a pathogen stress response in Arabidopsis thaliana. Plant J. 2018, 96, 705–715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morimoto, K.; van der Hoorn, R.A.L. Plant Biology: Proteolytic Release of Damage Signals. Curr. Biol. CB 2019, 29, R378–R380. [Google Scholar] [CrossRef] [PubMed]
- Adams, E.H.G.; Spoel, S.H. The ubiquitin-proteasome system as a transcriptional regulator of plant immunity. J. Exp. Bot. 2018, 69, 4529–4537. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.Q.; Yan, S.; Saleh, A.; Wang, W.; Ruble, J.; Oka, N.; Mohan, R.; Spoel, S.H.; Tada, Y.; Zheng, N.; et al. NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 2012, 486, 228–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolf-Levy, H.; Javitt, A.; Eisenberg-Lerner, A.; Kacen, A.; Ulman, A.; Sheban, D.; Dassa, B.; Fishbain-Yoskovitz, V.; Carmona-Rivera, C.; Kramer, M.P.; et al. Revealing the cellular degradome by mass spectrometry analysis of proteasome-cleaved peptides. Nat. Biotechnol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Steinbrenner, A.D.; Muñoz-Amatriaín, M.; Aguilar Venegas, J.M.; Lo, S.; Shi, D.; Holton, N.; Zipfel, C.; Abagyan, R.; Huffaker, A.; Close, T.J.; et al. A receptor for herbivore-associated molecular patterns mediates plant immunity. Biorxiv 2019. [Google Scholar] [CrossRef]
- Felix, G.; Duran, J.D.; Volko, S.; Boller, T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 1999, 18, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Kunze, G.; Zipfel, C.; Robatzek, S.; Niehaus, K.; Boller, T.; Felix, G. The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 2004, 16, 3496–3507. [Google Scholar] [CrossRef]
- Zipfel, C.; Kunze, G.; Chinchilla, D.; Caniard, A.; Jones, J.D.; Boller, T.; Felix, G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 2006, 125, 749–760. [Google Scholar] [CrossRef]
- Song, W.Y.; Wang, G.L.; Chen, L.L.; Kim, H.S.; Pi, L.Y.; Holsten, T.; Gardner, J.; Wang, B.; Zhai, W.X.; Zhu, L.H.; et al. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 1995, 270, 1804–1806. [Google Scholar] [CrossRef]
- Nurnberger, T.; Nennstiel, D.; Jabs, T.; Sacks, W.R.; Hahlbrock, K.; Scheel, D. High affinity binding of a fungal oligopeptide elicitor to parsley plasma membranes triggers multiple defense responses. Cell 1994, 78, 449–460. [Google Scholar] [CrossRef]
- Bressendorff, S.; Azevedo, R.; Kenchappa, C.S.; Ponce de Leon, I.; Olsen, J.V.; Rasmussen, M.W.; Erbs, G.; Newman, M.A.; Petersen, M.; Mundy, J. An Innate Immunity Pathway in the Moss Physcomitrella patens. Plant Cell 2016, 28, 1328–1342. [Google Scholar] [CrossRef] [PubMed]
- Felix, G.; Boller, T. Molecular sensing of bacteria in plants. The highly conserved RNA-binding motif RNP-1 of bacterial cold shock proteins is recognized as an elicitor signal in tobacco. J. Biol. Chem. 2003, 278, 6201–6208. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Weide, R.; Zhao, Z.; Msimuko, P.; Govers, F.; Bouwmeester, K. RXLR effector diversity in Phytophthora infestans isolates determines recognition by potato resistance proteins; the case study AVR1 and R1. Stud. Mycol. 2018, 89, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Schmelz, E.A.; LeClere, S.; Carroll, M.J.; Alborn, H.T.; Teal, P.E. Cowpea chloroplastic ATP synthase is the source of multiple plant defense elicitors during insect herbivory. Plant Physiol. 2007, 144, 793–805. [Google Scholar] [CrossRef] [PubMed]
- Schmelz, E.A.; Huffaker, A.; Carroll, M.J.; Alborn, H.T.; Ali, J.G.; Teal, P.E. An amino acid substitution inhibits specialist herbivore production of an antagonist effector and recovers insect-induced plant defenses. Plant Physiol. 2012, 160, 1468–1478. [Google Scholar] [CrossRef] [PubMed]
- Chien, P.S.; Nam, H.G.; Chen, Y.R. A salt-regulated peptide derived from the CAP superfamily protein negatively regulates salt-stress tolerance in Arabidopsis. J. Exp. Bot. 2015, 66, 5301–5313. [Google Scholar] [CrossRef] [PubMed]
- Pearce, G.; Yamaguchi, Y.; Barona, G.; Ryan, C.A. A subtilisin-like protein from soybean contains an embedded, cryptic signal that activates defense-related genes. Proc. Natl. Acad. Sci. USA 2010, 107, 14921–14925. [Google Scholar] [CrossRef] [Green Version]
- Launholt, D.; Merkle, T.; Houben, A.; Schulz, A.; Grasser, K.D. Arabidopsis chromatin-associated HMGA and HMGB use different nuclear targeting signals and display highly dynamic localization within the nucleus. Plant Cell 2006, 18, 2904–2918. [Google Scholar] [CrossRef]
- Merkle, T.; Grasser, K.D. Unexpected mobility of plant chromatin-associated HMGB proteins. Plant Signal. Behav. 2011, 6, 878–880. [Google Scholar] [CrossRef] [Green Version]
- Pedersen, D.S.; Grasser, K.D. The role of chromosomal HMGB proteins in plants. Biochim. Biophys. Acta 2010, 1799, 171–174. [Google Scholar] [CrossRef]
- Hann, D.R.; Rathjen, J.P. Early events in the pathogenicity of Pseudomonas syringae on Nicotiana benthamiana. Plant J. 2007, 49, 607–618. [Google Scholar] [CrossRef] [PubMed]
- Robatzek, S.; Bittel, P.; Chinchilla, D.; Kochner, P.; Felix, G.; Shiu, S.H.; Boller, T. Molecular identification and characterization of the tomato flagellin receptor LeFLS2, an orthologue of Arabidopsis FLS2 exhibiting characteristically different perception specificities. Plant Mol. Biol. 2007, 64, 539–547. [Google Scholar] [CrossRef] [PubMed]
- Takai, R.; Isogai, A.; Takayama, S.; Che, F.S. Analysis of flagellin perception mediated by flg22 receptor OsFLS2 in rice. Mol. Plant Microbe Interact. 2008, 21, 1635–1642. [Google Scholar] [CrossRef] [PubMed]
- Naito, K.; Taguchi, F.; Suzuki, T.; Inagaki, Y.; Toyoda, K.; Shiraishi, T.; Ichinose, Y. Amino acid sequence of bacterial microbe-associated molecular pattern flg22 is required for virulence. Mol. Plant Microbe Interact. 2008, 21, 1165–1174. [Google Scholar] [CrossRef]
- Hind, S.R.; Strickler, S.R.; Boyle, P.C.; Dunham, D.M.; Bao, Z.; O’Doherty, I.M.; Baccile, J.A.; Hoki, J.S.; Viox, E.G.; Clarke, C.R.; et al. Tomato receptor FLAGELLIN-SENSING 3 binds flgII-28 and activates the plant immune system. Nat. Plants 2016, 2, 16128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buscaill, P.; Chandrasekar, B.; Sanguankiattichai, N.; Kourelis, J.; Kaschani, F.; Thomas, E.L.; Morimoto, K.; Kaiser, M.; Preston, G.M.; Ichinose, Y.; et al. Glycosidase and glycan polymorphism control hydrolytic release of immunogenic flagellin peptides. Science 2019, 364. [Google Scholar] [CrossRef]
- Furukawa, T.; Inagaki, H.; Takai, R.; Hirai, H.; Che, F.S. Two distinct EF-Tu epitopes induce immune responses in rice and Arabidopsis. Mol. Plant Microbe Interact. 2014, 27, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Chinchilla, D.; Zipfel, C.; Robatzek, S.; Kemmerling, B.; Nurnberger, T.; Jones, J.D.; Felix, G.; Boller, T. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 2007, 448, 497–500. [Google Scholar] [CrossRef]
- Heese, A.; Hann, D.R.; Gimenez-Ibanez, S.; Jones, A.M.; He, K.; Li, J.; Schroeder, J.I.; Peck, S.C.; Rathjen, J.P. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. USA 2007, 104, 12217–12222. [Google Scholar] [CrossRef] [Green Version]
- Luu, D.D.; Joe, A.; Chen, Y.; Parys, K.; Bahar, O.; Pruitt, R.; Chen, L.J.G.; Petzold, C.J.; Long, K.; Adamchak, C.; et al. Sulfated RaxX, which represents an unclassified group of ribosomally synthesized post-translationally modified peptides, binds a host immune receptor. Biorxiv 2018. [Google Scholar] [CrossRef]
- Pruitt, R.N.; Joe, A.; Zhang, W.; Feng, W.; Stewart, V.; Schwessinger, B.; Dinneny, J.R.; Ronald, P.C. A microbially derived tyrosine-sulfated peptide mimics a plant peptide hormone. New Phytol. 2017, 215, 725–736. [Google Scholar] [CrossRef] [PubMed]
- Brunner, F.; Rosahl, S.; Lee, J.; Rudd, J.J.; Geiler, C.; Kauppinen, S.; Rasmussen, G.; Scheel, D.; Nurnberger, T. Pep-13, a plant defense-inducing pathogen-associated pattern from Phytophthora transglutaminases. EMBO J. 2002, 21, 6681–6688. [Google Scholar] [CrossRef] [PubMed]
- Furst, U.; Hegenauer, V.; Kaiser, B.; Korner, M.; Welz, M.; Albert, M. Parasitic Cuscuta factor(s) and the detection by tomato initiates plant defense. Commun. Integr. Biol. 2016, 9, e1244590. [Google Scholar] [CrossRef] [PubMed]
- Hegenauer, V.; Furst, U.; Kaiser, B.; Smoker, M.; Zipfel, C.; Felix, G.; Stahl, M.; Albert, M. Detection of the plant parasite Cuscuta reflexa by a tomato cell surface receptor. Science 2016, 353, 478–481. [Google Scholar] [CrossRef] [PubMed]
- Salas, C.E.; Badillo-Corona, J.A.; Ramirez-Sotelo, G.; Oliver-Salvador, C. Biologically active and antimicrobial peptides from plants. Biomed. Res. Int. 2015, 2015, 102129. [Google Scholar] [CrossRef]
- Breen, S.; Solomon, P.S.; Bedon, F.; Vincent, D. Surveying the potential of secreted antimicrobial peptides to enhance plant disease resistance. Front. Plant Sci. 2015, 6, 900. [Google Scholar] [CrossRef] [Green Version]
- Khazigaleeva, R.A.; Vinogradova, S.V.; Petrova, V.L.; Fesenko, I.A.; Arapidi, G.P.; Kamionskaya, A.M.; Govorun, V.M.; Ivanov, V.T. Antimicrobial activity of endogenous peptides of the moss Physcomitrella patens. Russ. J. Bioorg. Chem. 2017, 43, 248–254. [Google Scholar] [CrossRef]
- Ramada, M.H.S.; Brand, G.D.; Abrao, F.Y.; Oliveira, M.; Filho, J.L.C.; Galbieri, R.; Gramacho, K.P.; Prates, M.V.; Bloch, C., Jr. Encrypted Antimicrobial Peptides from Plant Proteins. Sci. Rep. 2017, 7, 13263. [Google Scholar] [CrossRef]
- Brand, G.D.; Magalhaes, M.T.; Tinoco, M.L.; Aragao, F.J.; Nicoli, J.; Kelly, S.M.; Cooper, A.; Bloch, C., Jr. Probing protein sequences as sources for encrypted antimicrobial peptides. PLoS ONE 2012, 7, e45848. [Google Scholar] [CrossRef]
- Candido-Ferreira, I.L.; Kronenberger, T.; Sayegh, R.S.; Batista, I.F.; da Silva Junior, P.I. Evidence of an Antimicrobial Peptide Signature Encrypted in HECT E3 Ubiquitin Ligases. Front. Immunol. 2016, 7, 664. [Google Scholar] [CrossRef]
- Vanhoye, D.; Bruston, F.; Nicolas, P.; Amiche, M. Antimicrobial peptides from hylid and ranin frogs originated from a 150-million-year-old ancestral precursor with a conserved signal peptide but a hypermutable antimicrobial domain. Eur. J. Biochem. 2003, 270, 2068–2081. [Google Scholar] [CrossRef] [PubMed]
- De Coninck, B.; De Smet, I. Plant peptides—Taking them to the next level. J. Exp. Bot. 2016, 67, 4791–4795. [Google Scholar] [CrossRef] [PubMed]


| Name | Precursor | Receptor | Reference | Peptide Sequence |
|---|---|---|---|---|
| Plant cryptic peptides | ||||
| Inceptin | ATP synthase γ-subunit | Inceptin receptor (INR) | [28,36] | ICDINGVCVDA |
| CAP-derived peptide 1 (CAPE1) | PR1b preproprotein of pathogenesis-related 1 proteins (CAP) superfamily | ? | [26] | PVGNWIGQRPY |
| Glycine max Subtilase Peptide (GmSubPep) | Subtilisin-like protease | ? | [27] | NYYDKHQLTRGH |
| PAMPs | ||||
| flg22 | Flagellin | FLAGELLIN SENSING 2 (FLS22) | [4,37] | QRLSTGSRINSAKDDAAGLQIA |
| elf18 | Elongation factor Tu | EF-Tu Receptor (EFR) | [38,39] | SKEKFERTKPHVNVGTIG |
| RaxX | preRaxX | XA21 | [11,40] | DYPPPGANPKHDPPPR |
| Pep-13 | Glycoprotein GP42 | R protein | [41] | VWNQPVRGFKVYE |
| Cuscuta factor (CuF) | ? | Cuscuta receptor 1 (CuRe1) | [42] | ? |
| Csp22, csp15 | Cold shock protein | ? | [43] | (AVGT)VKWFNAEKGFGFITP(DDG) |
| RxLR Avr effector | Avr1 etc. | R protein | [44] | ? |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lyapina, I.; Filippova, A.; Fesenko, I. The Role of Peptide Signals Hidden in the Structure of Functional Proteins in Plant Immune Responses. Int. J. Mol. Sci. 2019, 20, 4343. https://doi.org/10.3390/ijms20184343
Lyapina I, Filippova A, Fesenko I. The Role of Peptide Signals Hidden in the Structure of Functional Proteins in Plant Immune Responses. International Journal of Molecular Sciences. 2019; 20(18):4343. https://doi.org/10.3390/ijms20184343
Chicago/Turabian StyleLyapina, Irina, Anna Filippova, and Igor Fesenko. 2019. "The Role of Peptide Signals Hidden in the Structure of Functional Proteins in Plant Immune Responses" International Journal of Molecular Sciences 20, no. 18: 4343. https://doi.org/10.3390/ijms20184343