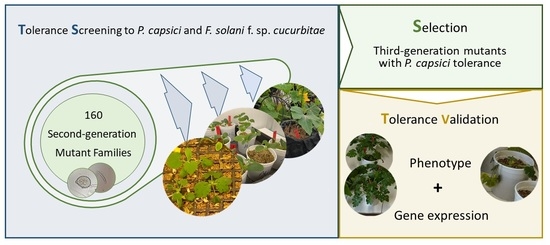
Tolerance Screening for Phytophthora capsici and Fusarium solani f. sp. cucurbitae in Cucurbita spp. and Gene Expression Responses in Mutant Families
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Pathogen Isolates
2.2. Inoculation
2.3. Test Conditions
2.4. Tolerance Evaluation
2.5. qPCR Sample Collection and Conditions
3. Results
3.1. Cucurbita Genotype Tolerance Screening to P. capsici and F. solani f. sp. cucurbitae
3.2. Mutant Tolerance Screening to P. capsici and Fusarium solani f. sp. cucurbitae
3.3. M3 Mutant Tolerance Analysis
3.4. M3 Gene Expression
4. Discussion
4.1. Tolerant Sources for Fusarium solani f. sp. cucurbitae and Phytophthora capsici in the Cucurbita Genus
4.2. Mutant Selection
4.3. qPCR
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A

References
- Babadoost, M. Oomycete Diseases of Cucurbits: History, Significance, and Management. In Horticultural Reviews; Wiley backwell: Hoboken, NJ, USA, 2016; Volume 44, pp. 279–314. [Google Scholar]
- Pérez-Hernández, A.; Rocha, L.O.; Porcel-Rodríguez, E.; Summerell, B.A.; Liew, E.C.Y.; Gómez-Vázquez, J.M. Pathogenic, Morphological, and Phylogenetic Characterization of Fusarium solani f. sp. cucurbitae Isolates From Cucurbits in Almería Province, Spain. Plant Dis. 2020, 104, 1465–1476. [Google Scholar] [CrossRef] [PubMed]
- Ayala-Doñas, A.; de Cara-García, M.; Talavera-Rubia, M.; Verdejo-Lucas, S. Management of Soil-Borne Fungi and Root-Knot Nematodes in Cucurbits through Breeding for Resistance and Grafting. Agronomy 2020, 10, 1641. [Google Scholar] [CrossRef]
- Krasnow, C.S.; Hammerschmidt, R.; Hausbeck, M.K. Characteristics of Resistance to Phytophthora Root and Crown Rot in Cucurbita pepo. Plant Dis. 2017, 101, 659–665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez-Hernández, A.; Porcel-Rodríguez, E.; Gómez-Vázquez, J. Survival of Fusarium solani f. sp. cucurbitae and Fungicide Application, Soil Solarization, and Biosolarization for Control of Crown and Foot Rot of Zucchini Squash. Plant Dis. 2017, 101, 1507–1514. [Google Scholar] [CrossRef] [Green Version]
- de Cara García, M.; Plaza, M.F.; Vázquez, J.M.G. Pathogenic and Biological Characterization of Phytophthora capsici Isolates from Zucchini and Pepper in Southeast Spain. Span. J. Agric. Res. 2018, 16, 22. [Google Scholar] [CrossRef]
- Sanogo, S.; Ji, P. Integrated Management of Phytophthora capsici on Solanaceous and Cucurbitaceous Crops: Current Status, Gaps in Knowledge and Research Needs. Can. J. Plant Pathol. 2012, 34, 479–492. [Google Scholar] [CrossRef]
- Davis, A.R.; Perkins-Veazie, P.; Sakata, Y.; López-Galarza, S.; Maroto, J.V.; Lee, S.-G.; Huh, Y.-C.; Sun, Z.; Miguel, A.; King, S.R.; et al. Cucurbit Grafting. Crit. Rev. Plant Sci. 2008, 27, 50–74. [Google Scholar] [CrossRef]
- Padley, L.D.; Kabelka, E.A.; Roberts, P.D. Evaluation of Cucurbita pepo Accessions for Crown Rot Resistance to Isolates of Phytophthora capsici. HortScience 2008, 43, 1996–1999. [Google Scholar] [CrossRef] [Green Version]
- Padley, L.D.; Kabelka, E.A.; Roberts, P.D. Inheritance of Resistance to Crown Rot Caused by Phytophthora capsici in Cucurbita. HortScience 2009, 44, 211–213. [Google Scholar] [CrossRef] [Green Version]
- Chavez, D.J.; Kabelka, E.A.; Chaparro, J.X. Screening of Cucurbita moschata Duchesne Germplasm for Crown Rot Resistance to Floridian Isolates of Phytophthora capsici Leonian. HortScience 2011, 46, 536–540. [Google Scholar] [CrossRef] [Green Version]
- Ramos, A.; Fu, Y.; Michael, V.; Meru, G. QTL-Seq for Identification of Loci Associated with Resistance to Phytophthora Crown Rot in Squash. Sci. Rep. 2020, 10, 5326. [Google Scholar] [CrossRef] [PubMed]
- Vogel, G.; LaPlant, K.E.; Mazourek, M.; Gore, M.A.; Smart, C.D. A Combined BSA-Seq and Linkage Mapping Approach Identifies Genomic Regions Associated with Phytophthora Root and Crown Rot Resistance in Squash. Theor. Appl. Genet. 2020, 134, 1015–1031. [Google Scholar] [CrossRef]
- Enzenbacher, T.B.; Hausbeck, M.K. An Evaluation of Cucurbits for Susceptibility to Cucurbitaceous and Solanaceous Phytophthora capsici Isolates. Plant Dis. 2012, 96, 1404–1414. [Google Scholar] [CrossRef] [Green Version]
- Kabelka, E.; Les Padley, J.; Roberts, P.; Ramos, L.; Martinez, M.; Klassen, W. Resistance to Phytophthora capsici within Winter Squash (Cucurbita moschata) Derived from a Wild Cucurbita Species. In Proceedings of the HortScience; American Society for Horticultural Science: Alexandria, VA, USA, 2007; Volume 42, p. 1014. [Google Scholar]
- Vincent Michael, N.; Moon, P.; Fu, Y.; Meru, G. Genetic Diversity Among Accessions of Cucurbita pepo Resistant to Phytophthora Crown Rot. HortScience 2019, 54, 17–22. [Google Scholar] [CrossRef] [Green Version]
- Michael, V.N.; Fu, Y.; Meru, G. Inheritance of Resistance to Phytophthora Crown Rot in Cucurbita pepo. HortScience 2019, 54, 1156–1158. [Google Scholar] [CrossRef] [Green Version]
- Santos, J.S.D.; Dias, R.D.C.S.; Almeida, K.B.D.; Ribeiro Junior, P.M.; Nascimento, T.L.D. Resistance of Cucurbita spp. to Fusarium solani for Use as Rootstock. Rev. Caatinga 2020, 33, 384–394. [Google Scholar] [CrossRef]
- Ji, X.; Zhang, H.; Zhang, Y.; Wang, Y.; Gao, C. Establishing a CRISPR–Cas-like Immune System Conferring DNA Virus Resistance in Plants. Nat. Plants 2015, 1, 15144. [Google Scholar] [CrossRef]
- Pyott, D.E.; Sheehan, E.; Molnar, A. Engineering of CRISPR/Cas9-Mediated Potyvirus Resistance in Transgene-Free Arabidopsis Plants: CRISPR/Cas9-Induced Potyvirus Resistance. Mol. Plant Pathol. 2016, 17, 1276–1288. [Google Scholar] [CrossRef] [Green Version]
- Chandrasekaran, J.; Brumin, M.; Wolf, D.; Leibman, D.; Klap, C.; Pearlsman, M.; Sherman, A.; Arazi, T.; Gal-On, A. Development of Broad Virus Resistance in Non-Transgenic Cucumber Using CRISPR/Cas9 Technology: Virus Resistance in Cucumber Using CRISPR/Cas9. Mol. Plant Pathol. 2016, 17, 1140–1153. [Google Scholar] [CrossRef] [Green Version]
- Chaudhary, J.; Deshmukh, R.; Sonah, H. Mutagenesis Approaches and Their Role in Crop Improvement. Plants 2019, 8, 467. [Google Scholar] [CrossRef] [Green Version]
- Vicente-Dólera, N.; Troadec, C.; Moya, M.; del Río-Celestino, M.; Pomares-Viciana, T.; Bendahmane, A.; Picó, B.; Román, B.; Gómez, P. First TILLING Platform in Cucurbita pepo: A New Mutant Resource for Gene Function and Crop Improvement. PLoS ONE 2014, 9, e112743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García, A.; Aguado, E.; Parra, G.; Manzano, S.; Martínez, C.; Megías, Z.; Cebrián, G.; Romero, J.; Beltrán, S.; Garrido, D.; et al. Phenomic and Genomic Characterization of a Mutant Platform in Cucurbita pepo. Front. Plant Sci. 2018, 9, 1049. [Google Scholar] [CrossRef] [PubMed]
- García, A.; Manzano, S.; Megías, Z.; Aguado, E.; Martínez, C.; Garrido, D.; Rebolloso, M.M.; Valenzuela, J.L.; Jamilena, M. Use of Mutant Platforms to Discover Novel Postharvest Fruit-Quality Traits in Cucurbita pepo. Acta Hortic. 2018, 1194, 367–374. [Google Scholar] [CrossRef]
- Chen, X.; Guo, W.; Jiang, L.; Hayat, S.; Chen, B.; Yang, P.; Zhou, J.; Li, X.; Bai, Y. Screening of EMS-Induced NaCl-Tolerant Mutants in Cucurbita moschata Duchesne Ex Poir. Pak. J. Bot. 2018, 50, 1305–1312. [Google Scholar]
- Al-Kubati, A.M.S.; Kang, B.; Liu, L.; Abbas, A.; Gu, Q. Development of Bottle Gourd Lines Resistant to Zucchini Yellow Mosaic Virus Using Ethyl Methanesulfonate Mutagenesis. HortScience 2021, 56, 909–914. [Google Scholar] [CrossRef]
- Cebrián, G.; Iglesias-Moya, J.; García, A.; Martínez, J.; Romero, J.; Regalado, J.J.; Martínez, C.; Valenzuela, J.L.; Jamilena, M. Involvement of Ethylene Receptors in the Salt Tolerance Response of Cucurbita pepo. Hortic. Res. 2021, 8, 73. [Google Scholar] [CrossRef]
- Liu, Z.; Shi, L.; Yang, S.; Lin, Y.; Weng, Y.; Li, X.; Hussain, A.; Noman, A.; He, S. Functional and Promoter Analysis of ChiIV3, a Chitinase of Pepper Plant, in Response to Phytophthora capsici Infection. Int. J. Mol. Sci. 2017, 18, 1661. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Liu, X.; Guo, J.; Liu, C.; Fu, N.; Shen, H. Identification and Expression Analysis of Candidate Genes Associated with Defense Responses to Phytophthora capsici in Pepper Line “PI 201234”. Int. J. Mol. Sci. 2015, 16, 11417–11438. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Wu, H.; Zhao, G.; He, Y.; Kong, W.; Zhang, J.; Liu, S.; Liu, M.; Hu, K.; Liu, L.; et al. Transcriptome Analysis Clarified Genes Involved in Resistance to Phytophthora capsici in Melon. PLoS ONE 2020, 15, e0227284. [Google Scholar] [CrossRef]
- Zhang, M.; Xu, J.H.; Liu, G.; Yao, X.F.; Li, P.F.; Yang, X.P. Characterization of the Watermelon Seedling Infection Process by Fusarium oxysporum f. sp. niveum. Plant Pathol. 2015, 64, 1076–1084. [Google Scholar] [CrossRef]
- Li, Q.; Chen, Y.; Wang, J.; Zou, F.; Jia, Y.; Shen, D.; Zhang, Q.; Jing, M.; Dou, D.; Zhang, M. A Phytophthora capsici Virulence Effector Associates with NPR1 and Suppresses Plant Immune Responses. Phytopathol. Res. 2019, 1, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Ayala-Doñas, A.; Gómez, P.; de Cara-García, M. Gene Expression in Cucurbita spp. Root and Crown during Phytophthora capsici Infection. Plants 2021, 10, 2718. [Google Scholar] [CrossRef] [PubMed]
- Gomez, J.; Guerra-Sanz, J.M.; Sanchez-Guerrero, M.C.; Serrano, Y.; Melero-Vara, M. Crown Rot of Zucchini Squash Caused by Fusarium solani f. sp. cucurbitae in Almería Province, Spain. Plant Dis. 2008, 92, 1137. [Google Scholar] [CrossRef]
- Meyer, M.D.; Hausbeck, M.K. Age-Related Resistance to Phytophthora Fruit Rot in ‘Dickenson Field’ Processing Pumpkin and ‘Golden Delicious’ Winter Squash Fruit. Plant Dis. 2013, 97, 446–552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Obrero, Á.; Die, J.V.; Román, B.; Gómez, P.; Nadal, S.; González-Verdejo, C.I. Selection of Reference Genes for Gene Expression Studies in Zucchini (Cucurbita pepo) Using QPCR. J. Agric. Food Chem. 2011, 59, 5402–5411. [Google Scholar] [CrossRef] [Green Version]
- Hellemans, J.; Mortier, G.; Paepe, A.D.; Speleman, F.; Vandesompele, J. QBase Relative Quantification Framework and Software for Management and Automated Analysis of Real-Time Quantitative PCR Data. Genome Biol. 2007, 8, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pomares-Viciana, T.; Die, J.; Del Río-Celestino, M.; Román, B.; Gómez, P. Auxin Signalling Regulation during Induced and Parthenocarpic Fruit Set in Zucchini. Mol. Breed. 2017, 37, 56. [Google Scholar] [CrossRef]
- Krasnow, C.S.; Naegele, R.P.; Hausbeck, M.K. Evaluation of Fruit Rot Resistance in Cucurbita Germplasm Resistant to Phytophthora capsici Crown Rot. HortScience 2014, 49, 285–288. [Google Scholar] [CrossRef] [Green Version]
- Boughalleb, N.; Mhamdi, M.; El Assadi, B.; El Bourgi, Z.; Tarchoun, N.; Romdhani, M.S. Resistance Evaluation of Grafted Watermelon (Citrullus lanatus L.) Against Fusarium Wilt and Fusarium Crown and Root Rot. Asian J. Plant Pathol. 2008, 2, 24–29. [Google Scholar] [CrossRef]
- Kousik, C.S.; Donahoo, R.S.; Hassell, R. Resistance in Watermelon Rootstocks to Crown Rot Caused by Phytophthora capsici. Crop Prot. 2012, 39, 18–25. [Google Scholar] [CrossRef]
- Tezuka, T.; Waki, K.; Kuzuya, M.; Ishikawa, T.; Takatsu, Y.; Miyagi, M. Development of New DNA Markers Linked to the Fusarium Wilt Resistance Locus Fom-1 in Melon: New DNA Markers Linked to Fom-1 in Melon. Plant Breed. 2011, 130, 261–267. [Google Scholar] [CrossRef]
- Win, K.T.; Zhang, C.; Lee, S. Genome-Wide Identification and Description of MLO Family Genes in Pumpkin (Cucurbita maxima Duch.). Hortic. Environ. Biotechnol. 2018, 59, 397–410. [Google Scholar] [CrossRef]
- Stam, R.; Jupe, J.; Howden, A.J.M.; Morris, J.A.; Boevink, P.C.; Hedley, P.E.; Huitema, E. Identification and Characterisation CRN Effectors in Phytophthora capsici Shows Modularity and Functional Diversity. PLoS ONE 2013, 8, e59517. [Google Scholar] [CrossRef]
- Li, Q.; Wang, J.; Bai, T.; Zhang, M.; Jia, Y.; Shen, D.; Zhang, M.; Dou, D. A Phytophthora capsici Effector Suppresses Plant Immunity via Interaction with EDS1. Mol. Plant Pathol. 2020, 21, 502–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.-R.; Zhang, Y.; Li, H.-Y.; Zhang, Z.-H.; Sheng, G.-L.; Li, Y.-P.; Xing, Y.-P.; Huang, S.-X.; Tao, H.; Kuan, T.; et al. The RXLR Effector PcAvh1 Is Required for Full Virulence of Phytophthora capsici. Mol. Plant-Microbe Interact. 2019, 32, 986–1000. [Google Scholar] [CrossRef]
- Bellini, A.; Pugliese, M.; Guarnaccia, V.; Meloni, G.R.; Gullino, L.M. Calcium Oxide, Potassium Phosphite and a Trichoderma Enriched Compost Water Suspension Protect Capsicum annuum against Phytophthora capsici by Priming the Immune System. Pest. Manag. Sci. 2021, 77, 3484–3490. [Google Scholar] [CrossRef] [PubMed]




| Test | Description | Inoculation Method | Inoculation Plant Stage | Pathogen | Cucurbita spp. Material | Duration (Days) 1 | N 2 (rep. 3) |
|---|---|---|---|---|---|---|---|
| 1 4 | Cucurbita spp. screening | Root dipping | Seedlings | Pc5 | Commercial and Conserved genotypes | 37 | 18 (16) |
| Fsc6 | 18 (16) | ||||||
| 2 | Mutant first screening | Root dipping | Seedlings | Pc | M2 mutant populations | 27 | 160 (3) |
| Fsc | 160 (3) | ||||||
| 3 | Mutant second screening | Substrate infestation | 2–3 true leaves | Pc | M2 selected mutants | 27 | 50 (3) |
| Fsc | 26 (3) | ||||||
| 4 | Mutant third screening. Final selection | Substrate infestation | 2–3 true leaves | Pc | M2 selected mutants | 34 (97 8) | 6 (10) |
| AMA | 1 (4) | ||||||
| M64 | 1 (4) | ||||||
| Fsc | M2 selected mutants | 6 (10) | |||||
| AMA 7 | 1 (4) | ||||||
| M64 | 1 (4) | ||||||
| 5 4 | M3 tolerance and RNA expression | Substrate infestation | 2–3 true leaves | Pc | Cp106 (M3) | 14 | 1 (32) |
| Cp117 (M3) | 1 (32) | ||||||
| MUCU-16 | 1 (32) | ||||||
| M63 | 1 (32) |
| Genotype | cv. | Fusarium solani f. sp. cucurbitae | Phytophthora capsici | ||||
|---|---|---|---|---|---|---|---|
| RAUDPC 1 | Phenotype 2 | RAUDPC | Phenotype | ||||
| Cucurbita moschata | M63 | 0.29 | ab | LS | 0.09 | c | T |
| M64 | 0.18 | ab | LS | 0.08 | c | T | |
| TZ3 | 0.04 | b | T | 0.27 | bc | LS | |
| TER | 0.23 | ab | LS | 0.25 | bc | LS | |
| C. maxima × C. moschata | SHI | 0.67 | ab | IS | 0.62 | abc | IS |
| ROU | 0.61 | ab | IS | 0.81 | ab | HS | |
| AZM | 0.79 | a | HS | 0.50 | abc | IS | |
| CAM | 0.74 | a | HS | 0.64 | abc | IS | |
| HER | 0.84 | a | HS | 0.88 | ab | HS | |
| C. pepo subsp. ovifera | SCA | 0.86 | ab | HS | 1.00 | a | HS |
| C. pepo subsp. pepo (marrow) | PIC | 0.63 | ab | IS | 0.74 | abc | HS |
| ZEB | 0.67 | ab | IS | 0.29 | bc | LS | |
| C. pepo subsp. pepo (zucchini) | AMA | 0.72 | ab | HS | 0.72 | abc | HS |
| JED | 0.46 | ab | IS | 0.71 | abc | HS | |
| NAT | 1.00 | a | HS | 1.00 | a | HS | |
| MUC | 0.91 | a | HS | 0.97 | a | HS | |
| MIL | 0.97 | a | HS | 0.99 | a | HS | |
| VIC | 0.72 | ab | HS | 0.97 | a | HS | |
| Pathogen | Genotype | Days Post-Inoculation | RAUDPC 4 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 7 | 15 | 27 | 34 | |||||||
| DSI 1 | %s.p.2 | DSI | %s.p. | DSI | %s.p. | DSI | %s.p. | |||
| Fusarium solani f. sp. cucurbitae | Cp005 3 | 0.80 | 0.80 | 3.70 | 1 | 3.90 | 1 | 4.00 | 1 | 0.99 |
| Cp059 | 1.40 | 1 | 3.70 | 1 | 4.00 | 1 | 4.00 | 1 | 1.00 | |
| Cp060 | 1.11 | 0.89 | 3.67 | 1 | 4.00 | 1 | 4.00 | 1 | 0.99 | |
| Cp109 | 0.75 | 0.75 | 2.50 | 0.88 | 3.00 | 1 | 4.00 | 1 | 0.91 | |
| Cp138 | 2.00 | 1 | 3.80 | 1 | 4.00 | 1 | 4.00 | 1 | 1.00 | |
| Cp157 | 1.13 | 0.75 | 3.13 | 1 | 4.00 | 1 | 4.00 | 1 | 0.99 | |
| AMA | 1 | 0.75 | 3.25 | 1 | 4.00 | 1 | 4.00 | 1 | 0.99 | |
| M64 | 0 | 0 | 0 | 0 | 0.30 | 0.25 | 0.50 | 0.25 | 0.24 | |
| Phytophthora capsici | Cp045 | 0.22 | 0.22 | 0.44 | 0.33 | 1.44 | 0.56 | 1.89 | 0.78 | 0.95 |
| Cp107 | 0 | 0 | 0 | 0 | 2.00 | 0.50 | 2.00 | 0.50 | 0.63 | |
| Cp116 | 0 | 0 | 0 | 0 | 0.25 | 0.13 | 0.50 | 0.13 | 0.10 | |
| Cp136 | 0.17 | 0.17 | 1.17 | 0.33 | 1.67 | 0.67 | 1.67 | 0.67 | 0.90 | |
| Cp139 | 0 | 0 | 0 | 0 | 0.17 | 0.17 | 0.83 | 0.33 | 0.18 | |
| Cp144 | 0 | 0 | 0.14 | 0.14 | 0.57 | 0.57 | 1 | 0.57 | 0.55 | |
| AMA | 0 | 0 | 0.50 | 0.25 | 2.50 | 0.75 | 4.00 | 1 | 1.00 | |
| M64 | 0 | 0 | 0 | 0 | 0.25 | 0.25 | 1 | 0.50 | 0.26 | |
| Genotypes | Cucurbita Species | Dpi 1 3 (n 2 ≥ 32) | dpi 10 (n ≥ 24) | dpi 14 (n ≥ 16) | RAUDPC 5 | |||
|---|---|---|---|---|---|---|---|---|
| DSI 3 | %s.p.4 | DSI | %s.p. | DSI | %s.p. | |||
| M63 | Cucurbita moschata | 0 | 0 | 0.08 | 0.08 | 0.50 | 0.21 | 0.34b |
| Cp116 | Cucurbita pepo | 0 | 0 | 0.36 | 0.18 | 0.80 | 0.20 | 0.56b |
| Cp107 | Cucurbita pepo | 0 | 0 | 0.21 | 0.13 | 1.12 | 0.29 | 0.48b |
| MUCU-16 | Cucurbita pepo | 0.06 | 0.03 | 0.35 | 0.19 | 1.28 | 0.44 | 1.00a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ayala-Doñas, A.; Gómez, P.; de Cara-García, M. Tolerance Screening for Phytophthora capsici and Fusarium solani f. sp. cucurbitae in Cucurbita spp. and Gene Expression Responses in Mutant Families. Horticulturae 2022, 8, 191. https://doi.org/10.3390/horticulturae8030191
Ayala-Doñas A, Gómez P, de Cara-García M. Tolerance Screening for Phytophthora capsici and Fusarium solani f. sp. cucurbitae in Cucurbita spp. and Gene Expression Responses in Mutant Families. Horticulturae. 2022; 8(3):191. https://doi.org/10.3390/horticulturae8030191
Chicago/Turabian StyleAyala-Doñas, Alejandro, Pedro Gómez, and Miguel de Cara-García. 2022. "Tolerance Screening for Phytophthora capsici and Fusarium solani f. sp. cucurbitae in Cucurbita spp. and Gene Expression Responses in Mutant Families" Horticulturae 8, no. 3: 191. https://doi.org/10.3390/horticulturae8030191