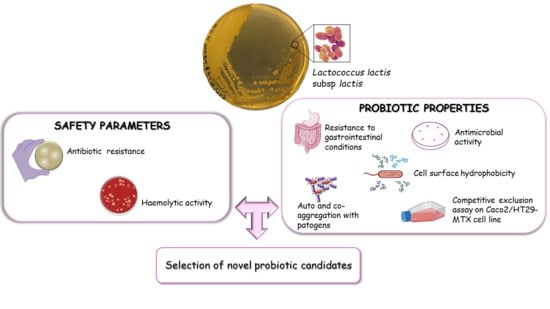
Probiotic Properties of Lactococcus lactis Strains Isolated from Natural Whey Starter Cultures
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Media, and Growth Conditions
2.2. Resistance to Simulated Gastrointestinal Conditions
2.3. Auto-Aggregation and Co-Aggregation Assays
2.4. Hydrophobicity Assay
2.5. Cell Cultures
2.6. Adhesion Assay
2.7. Production of Antimicrobial Substances
2.8. Safety Parameters
2.8.1. Antibiotic Susceptibility Test
2.8.2. Haemolytic Activity
2.9. Statistical Analysis
3. Results and Discussion
3.1. Survivability in Simulated Gastric Juice and Bile Salts
3.2. Hydrophobicity and Aggregative Potential
3.3. Activity of L. lactis Strains against S. Typhimurium and EIEC Adhesion to Epithelial Cells
3.4. Detection of Antimicrobial Activity
3.5. Antibiotic Susceptibility Test
3.6. Haemolytic Activity
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zielińska, D.; Kolożyn-Krajewska, D. Food-Origin Lactic Acid Bacteria May Exhibit Probiotic Properties: Review. Biomed. Res. Int. 2018, 2018, 5063185. [Google Scholar] [CrossRef]
- Mathur, H.; Beresford, T.P.; Cotter, P.D. Health Benefits of Lactic Acid Bacteria (LAB) Fermentates. Nutrients 2020, 12, 1679. [Google Scholar] [CrossRef]
- van de Wouw, M.; Walsh, A.M.; Crispie, F.; van Leuven, L.; Lyte, J.M.; Boehme, M.; Clarke, G.; Dinan, T.G.; Cotter, P.D.; Cryan, J.F. Distinct actions of the fermented beverage kefir on host behaviour, immunity and microbiome gut-brain modules in the mouse. Microbiome 2020, 8, 67. [Google Scholar] [CrossRef]
- Rai, R.; Tamang, J.P. In vitro and genetic screening of probiotic properties of lactic acid bacteria isolated from naturally fermented cow-milk and yak-milk products of Sikkim, India. World J. Microbiol. Biotechnol. 2022, 38, 25. [Google Scholar] [CrossRef] [PubMed]
- Aryana, K.J.; Olson, D.W. A 100-Year Review: Yogurt and other cultured dairy products. J. Dairy Sci. 2017, 100, 9987–10013. [Google Scholar] [CrossRef]
- Li, J.; Zhao, J.; Wang, X.; Qayum, A.; Hussain, M.A.; Liang, G.; Hou, J.; Jiang, Z.; Li, A. Novel Angiotensin-Converting Enzyme-Inhibitory Peptides From Fermented Bovine Milk Started by. Front. Microbiol. 2019, 10, 2643. [Google Scholar] [CrossRef]
- Akour, A. Probiotics and COVID-19: Is there any link? Lett. Appl. Microbiol. 2020, 71, 229–234. [Google Scholar] [CrossRef]
- Hu, J.; Zhang, L.; Lin, W.; Tang, W.; Chan, F.K.L.; Ng, S.C. Review article: Probiotics, prebiotics and dietary approaches during COVID-19 pandemic. Trends Food Sci. Technol. 2021, 108, 187–196. [Google Scholar] [CrossRef]
- Endres, K. Amyloidogenic Peptides in Human Neuro-Degenerative Diseases and in Microorganisms: A Sorrow Shared Is a Sorrow Halved? Molecules 2020, 25, 925. [Google Scholar] [CrossRef] [PubMed]
- Leblhuber, F.; Ehrlich, D.; Steiner, K.; Geisler, S.; Fuchs, D.; Lanser, L.; Kurz, K. The Immunopathogenesis of Alzheimer’s Disease Is Related to the Composition of Gut Microbiota. Nutrients 2021, 13, 361. [Google Scholar] [CrossRef] [PubMed]
- Gentile, M.T.; Camerino, I.; Ciarmiello, L.; Woodrow, P.; Muscariello, L.; De Chiara, I.; Pacifico, S. Neuro-Nutraceutical Polyphenols: How Far Are We? Antioxidants 2023, 12, 539. [Google Scholar] [CrossRef] [PubMed]
- Leblhuber, F.; Steiner, K.; Schuetz, B.; Fuchs, D.; Gostner, J.M. Probiotic Supplementation in Patients with Alzheimer’s Dementia—An Explorative Intervention Study. Curr. Alzheimer Res. 2018, 15, 1106–1113. [Google Scholar] [CrossRef] [PubMed]
- Kesika, P.; Suganthy, N.; Sivamaruthi, B.S.; Chaiyasut, C. Role of gut-brain axis, gut microbial composition, and probiotic intervention in Alzheimer’s disease. Life Sci. 2021, 264, 118627. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Li, X.; Zhang, G.; Sadiq, F.A.; Simal-Gandara, J.; Xiao, J.; Sang, Y. Probiotics in the dairy industry-Advances and opportunities. Compr. Rev. Food Sci. Food Saf. 2021, 20, 3937–3982. [Google Scholar] [CrossRef] [PubMed]
- Nowak, A.; Zakłos-Szyda, M.; Rosicka-Kaczmarek, J.; Motyl, I. Anticancer Potential of Post-Fermentation Media and Cell Extracts of Probiotic Strains: An In Vitro Study. Cancers 2022, 14, 1853. [Google Scholar] [CrossRef] [PubMed]
- Şanlier, N.; Gökcen, B.B.; Sezgin, A.C. Health benefits of fermented foods. Crit. Rev. Food Sci. Nutr. 2019, 59, 506–527. [Google Scholar] [CrossRef]
- Ağagündüz, D.; Yılmaz, B.; Şahin, T.; Güneşliol, B.E.; Ayten, Ş.; Russo, P.; Spano, G.; Rocha, J.M.; Bartkiene, E.; Özogul, F. Dairy Lactic Acid Bacteria and Their Potential Function in Dietetics: The Food-Gut-Health Axis. Foods 2021, 10, 3099. [Google Scholar] [CrossRef]
- Monteagudo-Mera, A.; Rastall, R.A.; Gibson, G.R.; Charalampopoulos, D.; Chatzifragkou, A. Adhesion mechanisms mediated by probiotics and prebiotics and their potential impact on human health. Aug 2019, 103, 6463–6472. [Google Scholar] [CrossRef]
- Raheem, A.; Liang, L.; Zhang, G.; Cui, S. Modulatory Effects of Probiotics During Pathogenic Infections With Emphasis on Immune Regulation. Modul. Eff. Probiotics Dur. Pathog. Infect. Emphas. Immune Regul. 2021, 12, 616713. [Google Scholar] [CrossRef]
- Shokryazdan, P.; Faseleh Jahromi, M.; Liang, J.B.; Ho, Y.W. Probiotics: From Isolation to Application. J. Am. Coll. Nutr. 2017, 36, 666–676. [Google Scholar] [CrossRef]
- Ozdogan, D.K.; Akcelik, N.; Aslim, B.; Suludere, Z.; Akcelik, M. Probiotic and Antioxidative Properties of L. Lactis LL27 Isolated from Milk. Biotechnol. Biotechnol. Equip. 2012, 26, 2750–2758. [Google Scholar] [CrossRef]
- Yadav, K.; Bhardwaj, A.; Kaur, G.; Iyer, R.; De, S.; Malik, R.K. Potential of Lactococcus lactis as a probiotic and functional lactic acid bacteria in dairy industry. Int. J. Probiotics Prebiotics 2009, 4, 219–228. [Google Scholar]
- Mercier-Bonin, M.; Chapot-Chartier, M.P. Surface Proteins of Lactococcus lactis: Bacterial Resources for Muco-adhesion in the Gastrointestinal Tract. Front. Microbiol. 2017, 8, 2247. [Google Scholar] [CrossRef] [PubMed]
- Khemariya, P.; Singh, S.; Nath, G.; Gulati, A.K. Probiotic Lactococcus lactis: A Review. Turk. J. Agric. —Food Sci. Technol. 2017, 5, 556–562. [Google Scholar] [CrossRef]
- Carvalho, R.D.; Breyner, N.; Menezes-Garcia, Z.; Rodrigues, N.M.; Lemos, L.; Maioli, T.U.; da Gloria Souza, D.; Carmona, D.; de Faria, A.M.; Langella, P.; et al. Secretion of biologically active pancreatitis-associated protein I (PAP) by genetically modified dairy Lactococcus lactis NZ9000 in the prevention of intestinal mucositis. Microb. Cell Fact. 2017, 16, 27. [Google Scholar] [CrossRef] [PubMed]
- Ballal, S.A.; Veiga, P.; Fenn, K.; Michaud, M.; Kim, J.H.; Gallini, C.A.; Glickman, J.N.; Quéré, G.; Garault, P.; Béal, C.; et al. Host lysozyme-mediated lysis of Lactococcus lactis facilitates delivery of colitis-attenuating superoxide dismutase to inflamed colons. Proc. Natl. Acad. Sci. USA 2015, 112, 7803–7808. [Google Scholar] [CrossRef] [PubMed]
- Aprea, G.; Del Matto, I.; Tucci, P.; Marino, L.; Scattolini, S.; Rossi, F. In Vivo Functional Properties of Dairy Bacteria. Microorganisms 2023, 11, 1787. [Google Scholar] [CrossRef] [PubMed]
- Yerlikaya, O. Probiotic potential and biochemical and technological properties of Lactococcus lactis ssp. lactis strains isolated from raw milk and kefir grains. J. Dairy. Sci. 2019, 102, 124–134. [Google Scholar] [CrossRef] [PubMed]
- Licitra, G.; Carpino, S. The Microfloras and Sensory Profiles of Selected Protected Designation of Origin Italian Cheeses. Microbiol. Spectr. 2014, 2, CM-0007-2012. [Google Scholar] [CrossRef]
- Blaya, J.; Barzideh, Z.; LaPointe, G. Symposium review: Interaction of starter cultures and nonstarter lactic acid bacteria in the cheese environment. J. Dairy Sci. 2018, 101, 3611–3629. [Google Scholar] [CrossRef]
- Choi, J.; In Lee, S.; Rackerby, B.; Frojen, R.; Goddik, L.; Ha, S.D.; Park, S.H. Assessment of overall microbial community shift during Cheddar cheese production from raw milk to aging. Appl. Microbiol. Biotechnol. 2020, 104, 6249–6260. [Google Scholar] [CrossRef] [PubMed]
- Mauriello, G.; Moio, L.; Genovese, A.; Ercolini, D. Relationships between flavoring capabilities, bacterial composition, and geographical origin of natural whey cultures used for traditional water-buffalo mozzarella cheese manufacture. J. Dairy Sci. 2003, 86, 486–497. [Google Scholar] [CrossRef]
- Gatti, M.; Bottari, B.; Lazzi, C.; Neviani, E.; Mucchetti, G. Invited review: Microbial evolution in raw-milk, long-ripened cheeses produced using undefined natural whey starters. J. Dairy Sci. 2014, 97, 573–591. [Google Scholar] [CrossRef] [PubMed]
- Losito, F.; Arienzo, A.; Bottini, G.; Priolisi, F.R.; Mari, A.; Antonini, G. Microbiological safety and quality of Mozzarella cheese assessed by the microbiological survey method. J. Dairy Sci. 2014, 97, 46–55. [Google Scholar] [CrossRef] [PubMed]
- Silva, L.F.; Casella, T.; Gomes, E.S.; Nogueira, M.C.; De Dea Lindner, J.; Penna, A.L. Diversity of lactic acid bacteria isolated from Brazilian water buffalo mozzarella cheese. J. Food Sci. 2015, 80, M411–M417. [Google Scholar] [CrossRef] [PubMed]
- Marasco, R.; Gazzillo, M.; Campolattano, N.; Sacco, M.; Muscariello, L. Isolation and Identification of Lactic Acid Bacteria from Natural Whey Cultures of Buffalo and Cow Milk. Foods 2022, 11, 233. [Google Scholar] [CrossRef] [PubMed]
- Talib, N.; Mohamad, N.E.; Yeap, S.K.; Hussin, Y.; Aziz, M.N.M.; Masarudin, M.J.; Sharifuddin, S.A.; Hui, Y.W.; Ho, C.L.; Alitheen, N.B. Isolation and Characterization of Lactobacillus spp. from Kefir Samples in Malaysia. Molecules 2019, 24, 2606. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Jeong, H.S.; Lee, H.Y.; Ahn, J. Assessment of cell surface properties and adhesion potential of selected probiotic strains. Lett. Appl. Microbiol. 2009, 49, 434–442. [Google Scholar] [CrossRef]
- Zuo, F.; Yu, R.; Feng, X.; Chen, L.; Zeng, Z.; Khaskheli, G.B.; Ma, H.; Chen, S. Characterization and in vitro properties of potential probiotic Bifidobacterium strains isolated from breast-fed infant feces. Ann. Microbiol. 2016, 66, 1027–1037. [Google Scholar] [CrossRef]
- Oliveira, L.C.; Saraiva, T.D.; Silva, W.M.; Pereira, U.P.; Campos, B.C.; Benevides, L.J.; Rocha, F.S.; Figueiredo, H.C.; Azevedo, V.; Soares, S.C. Analyses of the probiotic property and stress resistance-related genes of Lactococcus lactis subsp. lactis NCDO 2118 through comparative genomics and in vitro assays. PLoS ONE 2017, 12, e0175116. [Google Scholar] [CrossRef]
- Venegas-Ortega, M.G.; Flores-Gallegos, A.C.; Aguilar, C.N.; Rodríguez-Herrera, R.; Martínez-Hernández, J.L.; Nevárez-Moorillón, G.V. Multi-Functional Potential of Presumptive Lactic Acid Bacteria Isolated from Chihuahua Cheese. Foods 2020, 9, 276. [Google Scholar] [CrossRef]
- Prabhurajeshwar, C.; Chandrakanth, R.K. Probiotic potential of Lactobacilli with antagonistic activity against pathogenic strains: An in vitro validation for the production of inhibitory substances. Biomed. J. 2017, 40, 270–283. [Google Scholar] [CrossRef]
- Liasi, S.A.; Azmi, T.I.; Hassan, M.D.; Shuhaimi, M.; Rosfarizan, M.; Ariff, A.B. Antimicrobial activity and antibiotic sensitivity of three isolates of lactic acid bacteria from fermented fish product, Budu. Malays. J. Microbiol. 2009, 5, 33–37. [Google Scholar]
- Maragkoudakis, P.A.; Zoumpopoulou, G.; Miaris, C.; Kalantzopoulos, G.; Pot, B.; Tsakalidou, E. Probiotic potential of Lactobacillus strains isolated from dairy products. Int. Dairy J. 2006, 16, 189–199. [Google Scholar] [CrossRef]
- Sałański, P.; Kowalczyk, M.; Bardowski, J.K.; Szczepankowska, A.K. Health-Promoting Nature of Lactococcus lactis IBB109 and Lactococcus lactis IBB417 Strains Exhibiting Proliferation Inhibition and Stimulation of Interleukin-18 Expression in Colorectal Cancer Cells. Front. Microbiol. 2022, 13, 822912. [Google Scholar] [CrossRef]
- Islam, M.Z.; Uddin, M.E.; Rahman, M.T.; Islam, M.A.; Harun-ur-Rashid, M. Isolation and characterization of dominant lactic acid bacteria from raw goat milk: Assessment of probiotic potential and technological properties. Small Rumin. Res. 2021, 205, 106532. [Google Scholar] [CrossRef]
- Stasiak-Różańska, L.; Berthold-Pluta, A.; Pluta, A.S.; Dasiewicz, K.; Garbowska, M. Effect of Simulated Gastrointestinal Tract Conditions on Survivability of Probiotic Bacteria Present in Commercial Preparations. Int. J. Environ. Res. Public. Health 2021, 18, 1108. [Google Scholar] [CrossRef]
- Tsigkrimani, M.; Panagiotarea, K.; Paramithiotis, S.; Bosnea, L.; Pappa, E.; Drosinos, E.H.; Skandamis, P.N.; Mataragas, M. Microbial Ecology of Sheep Milk, Artisanal Feta, and Kefalograviera Cheeses. Part II: Technological, Safety, and Probiotic Attributes of Lactic Acid Bacteria Isolates. Foods 2022, 11, 459. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Moyano, S.; Gonçalves dos Santos, M.T.P.; Galván, A.I.; Merchán, A.V.; González, E.; Córdoba, M.d.G.; Benito, M.J. Screening of autochthonous lactic acid bacteria strains from artisanal soft cheese: Probiotic characteristics and prebiotic metabolism. LWT 2019, 114, 108388. [Google Scholar] [CrossRef]
- Reuben, R.C.; Roy, P.C.; Sarkar, S.L.; Rubayet Ul Alam, A.S.M.; Jahid, I.K. Characterization and evaluation of lactic acid bacteria from indigenous raw milk for potential probiotic properties. J. Dairy Sci. 2020, 103, 1223–1237. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Castilho, N.P.A.; Todorov, S.D.; Nero, L.A. Beneficial properties of lactic acid bacteria naturally present in dairy production. BMC Microbiol. 2018, 18, 219. [Google Scholar] [CrossRef] [PubMed]
- Krausova, G.; Hyrslova, I.; Hynstova, I. In Vitro Evaluation of Adhesion Capacity, Hydrophobicity, and Auto-Aggregation of Newly Isolated Potential Probiotic Strains. Fermentation 2019, 5, 100. [Google Scholar] [CrossRef]
- Zareie, Z.; Moayedi, A.; Garavand, F.; Tabar-Heydar, K.; Khomeiri, M.; Maghsoudlou, Y. Probiotic Properties, Safety Assessment, and Aroma-Generating Attributes of Some Lactic Acid Bacteria Isolated from Iranian Traditional Cheese. Fermentation 2023, 9, 338. [Google Scholar] [CrossRef]
- Taheri, H.R.; Moravej, H.; Tabandeh, F.; Zaghari, M.; Shivazad, M. Screening of lactic acid bacteria toward their selection as a source of chicken probiotic. Poult. Sci. 2009, 88, 1586–1593. [Google Scholar] [CrossRef]
- Mohanty, D.; Panda, S.; Kumar, S.; Ray, P. In vitro evaluation of adherence and anti-infective property of probiotic Lactobacillus plantarum DM 69 against Salmonella enterica. Microb. Pathog. 2019, 126, 212–217. [Google Scholar] [CrossRef]
- de Souza, B.M.S.; Borgonovi, T.F.; Casarotti, S.N.; Todorov, S.D.; Penna, A.L.B. Lactobacillus casei and Lactobacillus fermentum Strains Isolated from Mozzarella Cheese: Probiotic Potential, Safety, Acidifying Kinetic Parameters and Viability under Gastrointestinal Tract Conditions. Probiotics Antimicrob. Proteins 2019, 11, 382–396. [Google Scholar] [CrossRef] [PubMed]
- van Zyl, W.F.; Deane, S.M.; Dicks, L.M.T. Molecular insights into probiotic mechanisms of action employed against intestinal pathogenic bacteria. Gut Microbes 2020, 12, 1831339. [Google Scholar] [CrossRef]
- Gorreja, F.; Walker, W.A. The potential role of adherence factors in probiotic function in the gastrointestinal tract of adults and pediatrics: A narrative review of experimental and human studies. Gut Microbes 2022, 14, 2149214. [Google Scholar] [CrossRef]
- Chang, Y.H.; Jeong, C.H.; Cheng, W.N.; Choi, Y.; Shin, D.M.; Lee, S.; Han, S.G. Quality characteristics of yogurts fermented with short-chain fatty acid-producing probiotics and their effects on mucin production and probiotic adhesion onto human colon epithelial cells. J. Dairy Sci. 2021, 104, 7415–7425. [Google Scholar] [CrossRef]
- Singh, T.P.; Kaur, G.; Kapila, S.; Malik, R.K. Antagonistic Activity of Lactobacillus reuteri Strains on the Adhesion Characteristics of Selected Pathogens. Front. Microbiol. 2017, 8, 486. [Google Scholar] [CrossRef]
- Kiepś, J.; Juzwa, W.; Olejnik, A.; Sip, A.; Tomaszewska-Gras, J.; Dembczyński, R. The Effects of Cellular Membrane Damage on the Long-Term Storage and Adhesion of Probiotic Bacteria in Caco-2 Cell Line. Nutrients 2023, 15, 3484. [Google Scholar] [CrossRef]
- Tomé, A.R.; Carvalho, F.M.; Teixeira-Santos, R.; Burmølle, M.; Mergulhão, F.J.M.; Gomes, L.C. Use of Probiotics to Control Biofilm Formation in Food Industries. Antibiotics 2023, 12, 754. [Google Scholar] [CrossRef]
- Guan, C.; Chen, X.; Jiang, X.; Zhao, R.; Yuan, Y.; Chen, D.; Zhang, C.; Lu, M.; Lu, Z.; Gu, R. Studies of adhesion properties of six lactic acid bacteria isolated from the longevous population of China. RSC Adv. 2020, 10, 24234–24240. [Google Scholar] [CrossRef]
- Woo, J.; Ahn, J. Probiotic-mediated competition, exclusion and displacement in biofilm formation by food-borne pathogens. Lett. Appl. Microbiol. 2013, 56, 307–313. [Google Scholar] [CrossRef] [PubMed]
- Collado, M.C.; Meriluoto, J.; Salminen, S. Role of commercial probiotic strains against human pathogen adhesion to intestinal mucus. Lett. Appl. Microbiol. 2007, 45, 454–460. [Google Scholar] [CrossRef]
- Viana de Souza, J.; Silva Dias, F. Protective, technological, and functional properties of select autochthonous lactic acid bacteria from goat dairy products. Curr. Opin. Food Sci. 2017, 13, 1–9. [Google Scholar] [CrossRef]
- Mgomi, F.C.; Yang, Y.R.; Cheng, G.; Yang, Z.Q. Lactic acid bacteria biofilms and their antimicrobial potential against pathogenic microorganisms. Biofilm 2023, 5, 100118. [Google Scholar] [CrossRef]
- Ibrahim, S.A.; Ayivi, R.D.; Zimmerman, T.; Siddiqui, S.A.; Altemimi, A.B.; Fidan, H.; Esatbeyoglu, T.; Bakhshayesh, R.V. Lactic Acid Bacteria as Antimicrobial Agents: Food Safety and Microbial Food Spoilage Prevention. Foods 2021, 10, 3131. [Google Scholar] [CrossRef] [PubMed]
- Fadiji, T.; Rashvand, M.; Daramola, M.O.; Iwarere, S.A. A Review on Antimicrobial Packaging for Extending the Shelf Life of Food. Processes 2023, 11, 590. [Google Scholar] [CrossRef]
- Mínguez, E.; Arqués, J.; Rodríguez, R.; Peirotén, A.; Iranzo, J.M.; Medina, M. Antimicrobial properties of probiotic strains isolated from breast-fed infants. J. Funct. Foods 2012, 4, 542–551. [Google Scholar] [CrossRef]
- Reuben, R.C.; Roy, P.C.; Sarkar, S.L.; Alam, R.U.; Jahid, I.K. Isolation, characterization, and assessment of lactic acid bacteria toward their selection as poultry probiotics. BMC Microbiol. 2019, 19, 253. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.C.; Zhang, L.W.; Ma, W.; Yi, H.X.; Yang, X.; Du, M.; Shan, Y.J.; Han, X.; Zhang, L.L. Screening of probiotic lactobacilli for inhibition of Shigella sonnei and the macromolecules involved in inhibition. Anaerobe 2012, 18, 498–503. [Google Scholar] [CrossRef] [PubMed]
- Jawan, R.; Abbasiliasi, S.; Tan, J.S.; Mustafa, S.; Halim, M.; Ariff, A.B. Influence of Culture Conditions and Medium Compositions on the Production of Bacteriocin-Like Inhibitory Substances by. Microorganisms 2020, 8, 1454. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Zhou, Q.; Li, P.; Gu, Q. Purification, characterization, and mode of action of Paracin 54, a novel bacteriocin against Staphylococci. Appl. Microbiol. Biotechnol. 2021, 105, 6735–6748. [Google Scholar] [CrossRef]
- Foudjing, G.G.D.; Sarmast, E.; Allahdad, Z.; Salmieri, S.; Lacroix, M. Influence of growth parameters on bacteriocin-like inhibitory substances (BLIS) production by lactic acid bacteria. Lett. Appl. Microbiol. 2023, 76, ovac013. [Google Scholar] [CrossRef]
- Demir, E.; Başbülbül, G. Screening of Bacteriocin Production in Lactic Acid Bacteria Isolated From Fermented Dairy Products. Biotechnol. J. Int. 2017, 18, 1–9. [Google Scholar] [CrossRef]
- Poelarends, G.J.; Mazurkiewicz, P.; Konings, W.N. Multidrug transporters and antibiotic resistance in Lactococcus lactis. Biochim. Biophys. Acta 2002, 1555, 1–7. [Google Scholar] [CrossRef]
- FAO/WHO. Guidelines for the Evaluation of Probiotics in Food. In Proceedings of the Joint FAO/WHO Working Group Report on Drafting Guidelines for the Evaluation of Probiotics in Food, London, ON, Canada, 30 April–1 May 2002. [Google Scholar]


| Survival to GI Conditions * (% logCFU/mL) | Cell Surface Properties * (%) | ||||
|---|---|---|---|---|---|
| STRAINS | Acid Tolerance (pH 2.00) | Acid Tolerance (pH 3.00) | Bile Tolerance (pH 8.00) | Hydrophobicity | Auto-Aggregation |
| A3 | ND | 100 ± 0.01 a | 85.0 ± 0.04 bd | 68.7 ± 0.01 A | 50.0 ± 0.4 G |
| A5 | ND | 94.0 ± 0.06 a | 52.0 ± 0.02 c | 78.8 ± 0.03 B | 66.0 ± 0.4 H |
| B1 | ND | 98.0 ± 0.03 a | 81.0 ± 0.02 db | 33.9 ± 0.01 C | 46.4 ± 0.1 I |
| D1 | ND | 98.0 ± 0.02 a | 71.0 ± 0.02 ed | 25.5 ± 0.02 D | 43.3 ± 0.2 J |
| D3 | ND | 98.0 ± 0.02 a | 67.0 ± 0.01 f | 66.5 ± 0.02 EA | 41.0 ± 0.1 K |
| I1 | ND | 100 ± 0.01 a | 100 ± 0.01 g | 35.9 ± 0.01 C | 53.0 ± 0.5 G |
| I4 | ND | 95.0 ± 0.01 a | 77.0 ± 0.01 de | 51.6 ± 0.01 F | 60.0 ± 0.3 L |
| I7 | ND | 99.0 ± 0.02 a | 89.0 ± 0.01 bd | 64.0 ± 0.02 E | 33.0 ± 0.1 M |
| Inhibition | Competition | Displacement | |
|---|---|---|---|
| S. Typhimurium + A3 | 2.1 × 108 | 5.7 × 107 | 6.7 × 107 |
| S. Typhimurium + A5 | 2.4 × 108 | 6.0 × 107 | 1.2 × 108 |
| S. Typhimurium + I4 | 1.9 × 108 | 4.7 × 107 | 1.3 × 108 |
| S. Typhimurium + I7 | 1.0 × 108 | 3.7 × 107 | 5.7 × 107 |
| EIEC + A3 | 5.7 × 106 | 1.5 × 106 | 5.4 × 106 |
| EIEC + A5 | 2.7 × 106 | 8.3 × 104 | 4.7 × 106 |
| EIEC + I4 | 4.7 × 106 | 5.4 × 106 | 3.0 × 106 |
| EIEC + I7 | 5.4 × 106 | 5.5 × 106 | 5.0 × 105 |
| Strains | AM 10 µg | P 10 µg | CN 10 µg | TE 30 µg | VA 30 µg |
|---|---|---|---|---|---|
| A3 | S | S | S | S | S |
| A5 | S | S | R | S | S |
| B1 | S | S | S | S | S |
| D1 | S | S | S | S | S |
| D3 | S | S | S | S | S |
| I1 | S | S | R | S | S |
| I4 | S | S | R | S | S |
| I7 | S | S | S | S | S |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Chiara, I.; Marasco, R.; Della Gala, M.; Fusco, A.; Donnarumma, G.; Muscariello, L. Probiotic Properties of Lactococcus lactis Strains Isolated from Natural Whey Starter Cultures. Foods 2024, 13, 957. https://doi.org/10.3390/foods13060957
De Chiara I, Marasco R, Della Gala M, Fusco A, Donnarumma G, Muscariello L. Probiotic Properties of Lactococcus lactis Strains Isolated from Natural Whey Starter Cultures. Foods. 2024; 13(6):957. https://doi.org/10.3390/foods13060957
Chicago/Turabian StyleDe Chiara, Ida, Rosangela Marasco, Milena Della Gala, Alessandra Fusco, Giovanna Donnarumma, and Lidia Muscariello. 2024. "Probiotic Properties of Lactococcus lactis Strains Isolated from Natural Whey Starter Cultures" Foods 13, no. 6: 957. https://doi.org/10.3390/foods13060957