New Hygrocins K–U and Streptophenylpropanamide A and Bioactive Compounds from the Marine-Associated Streptomyces sp. ZZ1956
Abstract
:1. Introduction
2. Results and Discussion
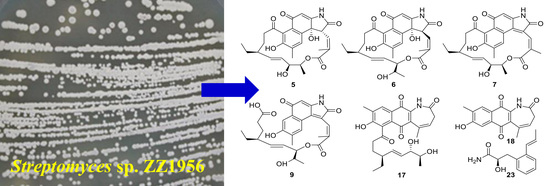
2.1. Structure Elucidation of the Isolated Compounds
2.2. Biological Activity Evaluation
3. Experimental Section
3.1. General Procedures
3.2. Isolation and Taxonomic Identity of Streptomyces sp. ZZ1956
3.3. Mass Culture of the Strain ZZ1956
3.4. Extraction and Isolation of Compounds 1–30
3.5. Compound Characterization Data
3.6. MTPA Esterification Hygrocin N (8)
3.7. ECD Calculations
3.8. Sulforhodamine B (SRB) Assay
3.9. Antibacterail Activity Determination
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kang, Q.J.; Shen, Y.M.; Bai, L.Q. Biosynthesis of 3,5-AHBA-derived natural products. Nat. Prod. Rep. 2012, 29, 243–263. [Google Scholar] [CrossRef]
- Skrzypczak, N.; Przybylski, P. Structural diversity and biological relevance of benzenoid and atypical ansamycins and their congeners. Nat. Prod. Rep. 2022, 39, 1678–1704. [Google Scholar] [CrossRef]
- Funayama, S.; Cordell, G.A. Ansamycin antibiotics, discovery, classification, biosynthesis and biological activities. Stud. Nat. Prod. Chem. 2000, 23, 51–106. [Google Scholar]
- Eckelmann, D.; Kusari, S.; Spiteller, M. Occurrence and spatial distribution of maytansinoids in Putterlickia pyracantha, an unexplored resource of anticancer compounds. Fitoterapia 2016, 113, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Kusari, S.; Lamshöft, M.; Kusari, P.; Gottfried, S.; Zühlke, S.; Louven, K.; Hentschel, U.; Kayser, O.; Spiteller, M. Endophytes are hidden producers of maytansine in Putterlickia roots. J. Nat. Prod. 2014, 77, 2577–2584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, F.F.; Wang, Z.S.; Lu, C.H.; Li, Y.Y.; Zhu, J.; Wang, H.X.; Shen, Y.M. Targeted discovery of pentaketide ansamycin aminoansamycins A–G. Org. Lett. 2019, 21, 7818–7822. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.Y.; Zhang, H.R.; Ju, J.H. On-PKS Baeyer-Villiger-type O-atom insertion catalyzed by luciferase-like monooxygenase ovmO during olimycin biosynthesis. Org. Lett. 2020, 22, 1780–1784. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.H.; Wei, Y.Y.; Xing, Y.N.; Chen, Y.; Wang, W.; Wang, K.B.; Liang, Y.; Jiao, R.H.; Zhang, B.; Ge, H.M. A BBE-like oxidase, asmF, dictates the formation of naphthalenic hydroxyl groups in ansaseomycin biosynthesis. Org. Lett. 2021, 23, 3724–3728. [Google Scholar] [CrossRef] [PubMed]
- Nong, X.H.; Tu, Z.C.; Qi, S.H. Ansamycin derivatives from the marine-derived Streptomyces sp. SCSGAA 0027 and their cytotoxic and antiviral activities. Bioorganic Med. Chem. Lett. 2020, 30, 7168. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.W.; Anjum, K.; Song, T.F.; Wang, W.L.; Liang, Y.; Chen, M.X.; Huang, H.C.; Lian, X.Y.; Zhang, Z.Z. Antiproliferative cyclodepsipeptides from the marine actinomycete Streptomyces sp. P11-23B downregulating the tumor metabolic enzymes of glycolysis, glutaminolysis, and lipogenesis. Phytochemistry 2017, 135, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Chai, W.Y.; Wang, W.L.; Song, T.F.; Lian, X.Y.; Zhang, Z.Z. Cytotoxic bagremycins from mangrove-derived Streptomyces sp. Q22. J. Nat. Prod. 2017, 80, 1450–1456. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.X.; Chai, W.Y.; Song, T.F.; Ma, M.Z.; Lian, X.Y.; Zhang, Z.Z. Anti-glioma natural products downregulating tumor glycolytic enzymes from marine actinomycetes Streptomyces sp. ZZ406. Sci. Rep. 2018, 8, 72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, T.F.; Chen, M.X.; Chai, W.Y.; Zhang, Z.Z.; Lian, X.Y. New bioactive pyrrospirones C-I from a marine-derived fungus Penicillium sp. ZZ380. Tetrahedron 2018, 74, 884–891. [Google Scholar] [CrossRef]
- Song, T.F.; Chen, M.X.; Ge, Z.W.; Chai, W.Y.; Li, X.C.; Zhang, Z.Z.; Lian, X.Y. Bioactive penicipyrrodiether A, an adduct of GKK1032 analogue and phenol A derivative, from a marine-sourced fungus Penicillium sp. ZZ380. J. Org. Chem. 2018, 83, 13395–13401. [Google Scholar] [CrossRef] [PubMed]
- Song, T.F.; Tang, M.M.; Ge, H.J.; Chen, M.X.; Lian, X.Y.; Zhang, Z.Z. Novel bioactive penicipyrroether A and pyrrospirone J from the marine-derived Penicillium sp. ZZ380. Mar. Drugs 2019, 17, 292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.; Yi, W.W.; Ge, H.J.; Zhang, Z.Z.; Wu, B. Bioactive streptoglutarimides A–J from the marine-derived Streptomyces sp. ZZ741. J. Nat. Prod. 2019, 82, 2800–2808. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Yi, W.W.; Lian, X.Y.; Zhang, Z.Z. Bioactive alkaloids from the actinomycete Actinoalloteichus sp. ZZ1866. J. Nat. Prod. 2020, 83, 2686–2695. [Google Scholar] [CrossRef] [PubMed]
- Ge, H.J.; Zhang, D.; Shi, M.R.; Lian, X.Y.; Zhang, Z.Z. Antiproliferative activity and potential mechanism of marine-sourced streptoglutarimide H against lung cancer cells. Mar. Drugs 2021, 19, 79. [Google Scholar] [CrossRef] [PubMed]
- Yong, K.; Kaleem, S.; Wu, B.; Zhang, Z.Z. New antiproliferative compounds against glioma cells from the marine-sourced fungus Penicillium sp. ZZ1750. Mar. Drugs 2021, 19, 483. [Google Scholar] [CrossRef]
- Yi, W.W.; Lian, X.Y.; Zhang, Z.Z. Cytotoxic metabolites from the marine-associated Streptomyces sp. ZZ1944. Phytochemistry 2022, 201, 113292. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.H.; Li, Y.Y.; Deng, J.J.; Li, S.R.; Shen, Y.; Wang, H.X.; Shen, Y.M. Hygrocins C–G, cytotoxic naphthoquinone ansamycins from gdmAI-disrupted Streptomyces sp. LZ35. J. Nat. Prod. 2013, 76, 2175–2179. [Google Scholar] [CrossRef] [PubMed]
- Cai, P.; Kong, F.M.; Ruppen, M.E.; Glasier, G.; Carter, G.T. Hygrocins A and B, naphthoquinone macrolides from Streptomyces hygroscopicus. J. Nat. Prod. 2005, 68, 1736–1742. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Ferrando, F. Assignment of quaternary carbons via two-bond heteronuclear selective NOE difference, a useful structure elucidation aid. Magn. Reson. Chem. 1985, 23, 185–191. [Google Scholar] [CrossRef]
- Nagao, T.; Otsuka, H.; Kohda, H.; Sato, T.; Yamasaki, K. Benzoxazinones from Coix lachryma-jobi var. ma-yuen. Phytochemistry 1985, 24, 2959–2962. [Google Scholar] [CrossRef]
- Supong, K.; Sripreechasak, P.; Tanasupawat, S.; Danwisetkanjana, K.; Rachtawee, P.; Pittayakhajonwut, P. Investigation on antimicrobial agents of the terrestrial Streptomyces sp. BCC71188. Appl. Microbiol. Biotechnol. 2017, 101, 533–543. [Google Scholar] [CrossRef]
- Hager, A.; Kuttruff, C.A.; Herrero-Gómez, E.; Trauner, D. Toward the total synthesis of ansalactam A. Tetrahedron Lett. 2014, 55, 59–62. [Google Scholar] [CrossRef]
- Spiessens, L.I.; Anteunis, M.J.O. NMR studies on imidines. III. 1H and 13C nuclear magnetic resonance study on the tautomerism and geometrical isomerism of 3-iminoisoindolinone and some alkylated derivatives. Observation of E and Z isomers about a free imino grouping. Bull. Des Sociétés Chim. Belg. 1983, 92, 965–993. [Google Scholar] [CrossRef]
- Chang, S.-S.; Chen, M.-H.; Wang, R.-Z.; Wu, Y.-X.; Yang, G.-H.; Dong, B.; Yu, L.-Y.; Gao, Z.-P.; Si, S.-Y. A new benzamide derivative from rice fermentation of Streptomyces sp. CPCC 202950. China J. Chin. Materia Med. 2017, 42, 2097–2101. [Google Scholar]
- Biggins, J.B.; Liu, X.F.; Feng, Z.Y.; Brady, S.F. Metabolites from the induced expression of cryptic single operons found in the genome of Burkholderia pseudomallei. J. Am. Chem. Soc. 2011, 133, 1638–1641. [Google Scholar] [CrossRef] [Green Version]
- Deng, J.J.; Lu, C.H.; Li, S.R.; Hao, H.L.; Li, Z.U.; Zhu, J.; Li, Y.Y.; Shen, Y.M. p-Terphenyl O-β-glucuronides, DNA topoisomerase inhibitors from Streptomyces sp. LZ35ΔgdmAI. Bioorganic Med. Chem. Lett. 2014, 24, 1362–1365. [Google Scholar] [CrossRef]
- Han, B.; Li, W.X.; Cui, C.B. Pteridic acid hydrate and pteridic acid C produced by Streptomyces pseudoverticillus YN17707 induce cell cycle arrest. Chin. J. Nat. Med. 2015, 13, 467–470. [Google Scholar] [PubMed]
- Ritzau, M.; Heinze, S.; Fleck, W.F.; Dahse, H.M.; Graefe, U. New macrodiolide antibiotics, 11-O-monomethyl- and 11,11’-O-dimethylelaiophylins, from Streptomyces sp. HKI-0113 and HKI-0114. J. Nat. Prod. 1998, 61, 1337–1339. [Google Scholar] [CrossRef] [PubMed]
- Xin, W.X.; Ye, X.W.; Yu, S.R.; Lian, X.Y.; Zhang, Z.Z. New capoamycin-type antibiotics and polyene acids from marine Streptomyces fradiae PTZ0025. Mar. Drugs 2012, 10, 2388–2402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, X.W.; Anjum, K.; Song, T.F.; Wang, W.L.; Yu, S.R.; Huang, H.C.; Lian, X.Y.; Zhang, Z.Z. A new curvularin glycoside and its cytotoxic and antibacterial analogues from marine actinomycete Pseudonocardia sp. HS7. Nat. Prod. Res. 2016, 30, 1156–1161. [Google Scholar] [CrossRef] [PubMed]






| No. | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 177.3, C | 177.1, C | 172.5, C | 173.1, C | 171.6, C | 171.6, C | 171.5, C | 171.7, C | 171.7, C |
| 2 | 56.9, CH | 56.9, CH | 121.9, C | 122.6, C | 126.8, C | 126.8, C | 126.8, C | 126.8, C | 126.6, C |
| 3 | 133.1, CH | 133.6, CH | 128.9, CH | 127.5, CH | 129.7, CH | 129.7, CH | 129.7, CH | 129.7, CH | 129.7, C |
| 4 | 133.9, C | 133.5, C | 137.5, C | 137.8, C | 137.6 a, C | 137.7 a, C | 137.6 a, C | 137.7 a, C | 138.3, C |
| 4a | 13.7, CH3 | 13.6, CH3 | 21.1, CH3 | 22.1, CH3 | 16.4, CH3 | 16.4, CH3 | 16.4, CH3 | 16.4, CH3 | 21.1, CH3 |
| 5 | 167.7, C | 167.3, C | 167.6, C | 168.3, C | 168.1, C | 167.9, C | 168.1, C | 167.9, C | 168.5, C |
| 6 | 75.3, CH | 69.5, CH | 74.7, CH | 68.5, CH | 75.6, CH | 69.9, CH | 75.5, CH | 69.9, CH | 75.2, CH |
| 6a | 15.2, CH3 | 19.1, CH3 | 13.5, CH3 | 19.1, CH3 | 16.4, CH3 | 19.6, CH3 | 16.2, CH3 | 19.6, CH3 | 15.0, CH3 |
| 7 | 74.2, CH | 77.9, CH | 71.3, CH | 80.6, CH | 75.4, CH | 81.4, CH | 75.3, CH | 81.2, CH | 74.4, CH |
| 8 | 128.2, CH | 125.3, CH | 128.7, CH | 124.4, CH | 131.5, CH | 127.9, CH | 131.5, CH | 127.9, CH | 130.7, CH |
| 9 | 138.3, CH | 136.5, CH | 137.3, CH | 139.9, CH | 138.3, CH | 141.3, CH | 138.1, CH | 140.9, CH | 137.9, CH |
| 10 | 41.9, CH | 41.9, CH | 43.8, CH | 43.4, CH | 45.6, CH | 45.8, CH | 45.6, CH | 45.8, CH | 45.5, CH |
| 10a | 26.3, CH2 | 26.6, CH2 | 26.6, CH2 | 29.2, CH2 | 29.2, CH2 | 29.1, CH2 | 29.2, CH2 | 29.1, CH2 | 29.1, CH2 |
| 10b | 10.6, CH3 | 12.4, CH3 | 12.7, CH3 | 11.5, CH3 | 12.3, CH3 | 12.3, CH3 | 12.3, CH3 | 12.3, CH3 | 12.2, CH3 |
| 11 | 29.2, CH2 | 33.0, CH2 | 32.1, CH2 | 28.9, CH2 | 31.1, CH2 | 31.2, CH2 | 31.0, CH2 | 31.0, CH2 | 30.9, CH2 |
| 12 | 41.8, CH2 | 42.2, CH2 | 39.4, CH2 | 42.5, CH2 | 33.0, CH2 | 33.2, CH2 | 33.0, CH2 | 33.0, CH2 | 33.0, CH2 |
| 13 | 212.0, C | 211.2, C | 212.9, C | 212.0, C | 177.7, C | 177.8, C | 176.0, C | 176.0, C | 177.7, C |
| 14 | 130.1, C | 127.2, C | 129.9, C | 129.2, C | 113.9, CH | 113.9, CH | 113.9, CH | 113.9, CH | 113.7, CH |
| 15 | 153.0, C | 153.7, C | 158.7, C | 158.0, C | 159.8, C | 159.8, C | 159.9, C | 159.9, C | 159.5, C |
| 16 | 133.1 a, C | 133.4, C | 133.3, C | 133.7, C | 131.9 b, C | 131.9, C | 131.9, C | 131.9, C | 132.2, C |
| 16a | 17.0, CH3 | 16.9, CH3 | 17.3, CH3 | 17.2, CH3 | 16.8, CH3 | 16.8, CH3 | 16.8, CH3 | 16.8, CH3 | 16.7, CH3 |
| 17 | 131.5, CH | 132.1, CH | 131.3, CH | 130.7, CH | 131.7 b, CH | 131.6, CH | 131.6, CH | 131.6, CH | 131.7, CH |
| 18 | 133.0 a, C | 134.0, C | 131.2, C | 130.1, C | 122.8, C | 122.8, C | 122.7, C | 122.8, C | 123.2, C |
| 19 | 75.4, C | 75.8, C | 134.8, C | 136.4, C | 137.4 a, C | 137.4 a, C | 137.4 a, C | 137.4 a, C | 139.6, C |
| 20 | 164.6, C | 164.5, C | 154.8, C | 155.1, C | 154.0, C | 154.0, C | 154.0, C | 154.0, C | 154.5, C |
| 21 | 105.1, CH | 105.3 CH | 106.0, CH | 105.8, CH | 106.3, CH | 106.4, CH | 106.3, CH | 106.4, CH | 106.0, CH |
| 22 | 185.6, C | 185.9, C | 187.4, C | 187.0, C | 186.5, C | 186.4, C | 186.4, C | 186.4, C | 186.7, C |
| 23 | 129.7, C | 131.1, C | 129.3, C | 128.4, C | 131.9 b, C | 131.9, C | 131.9, C | 131.9, C | 131.7, C |
| 24 | - | - | - | - | - | - | 52.1, CH3 | 52.2, CH3 | - |
| No. | 5 | 6 | 7 | 8 |
|---|---|---|---|---|
| 2 | 4.06, d (11.1) | 4.04, d (11.1) | - | - |
| 3 | 6.09, dd (11.1, 1.8) | 6.10, dd (11.1, 1.4) | 6.90, s | 6,81, s |
| 4a | 2.06, d (1.8) | 2.03, s | 2.21, s | 2.19, s |
| 6 | 4.65, m | 3.67, m | 4.87 a, m | 3.69, m |
| 6a | 1.15, d (6.4) | 1.02, d (6.7) | 0.91, d (6.0) | 0.95, d (6.4) |
| 7 | 3.90, dd (6.8, 2.8) | 5.11, t (5.0) | 3.86, m | 4.97, t (5.8) |
| 8 | 4.92, dd (16.0, 6.8) | 5.26, dd (16.1, 5.0) | 4.59, d (15.1) | 4.41, dd (15.6, 6.6) |
| 9 | 5.50, dd (16.0, 5.5) | 5.06, dd (16.1, 6.4) | 5.30, dd (15.1, 9.2) | 5.36, dd (15.6, 5.8) |
| 10 | 1.84, m | 1.69, m | 1.55, m | 1.56, m |
| 10a | 1.49, m; 1.23, m | 1.32, m | 1.48, m; 0.92, m | 1.30, m; 1.14, m |
| 10b | 0.71, t (7.3) | 0.82, t (7.4) | 0.69, t (7.2) | 0.76, t (7.2) |
| 11 | 1.45, m; 1.19, m | 1.55, m; 1.38, m | 1.28, m; 1.18, m | 1.53, m; 1.35, m |
| 12 | 2.71, m; 2.59, m | 2.67, m; 2.49, m | 2.75, d (11.6) | 2.81, dd (16.8, 8.5); 2.43, dd (16.8, 10.0) |
| 16a | 2.29, s | 2.27, s | 2.21, s | 2.15, s |
| 17 | 7.22, s | 7.28, s | 7.57, s | 7.65, s |
| 21 | 5.87, s | 5.93, s | 5.85, s | 5.81, s |
| No. | 9 | 10 | 11 | 12 | 13 |
|---|---|---|---|---|---|
| 3 | 7.53, s | 7.54, s | 7.51, s | 7.54, d (1.4) | 7.54, s |
| 4a | 1.92, s | 1.92, s | 1.92, s | 1.93, d (1.4) | 2.24, s |
| 6 | 5.05, m | 3.93, m | 5.06, m | 3.92, m | 4.84, m |
| 6a | 1.34, d (6.4) | 1.24, d (6.6) | 1.33, d (6.4) | 1.24, d (6.4) | 1.14, d (6.4) |
| 7 | 4.19, t (6.3) | 5.23, t (6.6) | 4.19, t (6.1) | 5.24, t (6.3) | 4.03, t (6.0) |
| 8 | 5.54, dd (15.5, 6.3) | 5.57, m | 5.52, dd (15.5, 6.1) | 5.56, m | 5.30, dd (15.5, 6.0) |
| 9 | 5.48, dd (15.5, 8.6) | 5.57, m | 5.45, dd (15.5, 8.6) | 5.57, m | 5.24, dd (15.5, 8.8) |
| 10 | 1.95, m | 1.95, m | 1.90, m | 1.96, m | 1.71, m |
| 10a | 1.43, m; 1.28, m | 1.49, m; 1.31, m | 1.45, m; 1.26, m | 1.48, m; 1.29, m | 1.26, m; 1.18, m |
| 10b | 0.87, t (7.1) | 0.86, t (7.3) | 0.86, t (7.2) | 0.86, t (7.4) | 0.76, t (7.4) |
| 11 | 1.74, m; 1.46, m | 1.76, m; 1.47, m | 1.72, m; 1.41, m | 1.77, m; 1.52, m | 1.57, m; 1.09, m |
| 12 | 2.30, m; 2.21, m | 2.30, m; 2.23, m | 2.28, m; 2.24, m | 2.30, m; 2.26, m | 2.10, m; 2.02, m |
| 14 | 7.43, s | 7.42, s | 7.43, s | 7.43, s | 7.46, s |
| 16a | 2.24, s | 2.21, s | 2.23, s | 2.21, s | 2.28, s |
| 17 | 7.42, s | 7.39, s | 7.40, s | 7.40, s | 6.83, s |
| 21 | 5.88, s | 5.88, s | 5.87, s | 5.88, s | 5.88, s |
| 24 | - | - | 3.57, s | 3.65, s | - |
| No. | δC, Type | δH, Multi. (J in Hz) | No. | δC, Type | δH, Multi. (J in Hz) |
|---|---|---|---|---|---|
| 1 | 171.8, C | - | 10 | 180.1, C | - |
| 2 | 37.7, CH2 | 2.90, d (7.2) | 10a | 138.6, C | - |
| 3 | 125.5, CH | 5.83, td (7.2, 1.2) | 11 | 208.5, C | - |
| 4 | 136.4, C | - | 12 | 42.5, CH2 | 2.76, m; 2.67, m |
| 4a | 130.1, C | - | 13 | 29.7, CH2 | 1.95, m; 1.67, m |
| 4b | 21.4, CH3 | 2.13, s | 14 | 45.2, CH | 2.00, m |
| 5 | 185.4, C | - | 14a | 29.2, CH2 | 1.49, m; 1.28, m |
| 5a | 131.8, C | - | 14b | 12.3, CH3 | 0.90, t (7.5) |
| 6 | 124.3, C | - | 15 | 138.6, CH | 5.47, dd (15.8, 6.8) |
| 7 | 158.8, C | - | 16 | 131.3 a, CH | 5.43, dd (15.8, 6.3) |
| 8 | 133.0, C | - | 17 | 78.6, CH | 3.76, t (6.3) |
| 8a | 17.0, CH3 | 2.35, s | 18 | 72.0, CH | 3.53, m |
| 9 | 131.3 a, CH | 7.92, s | 18a | 19.4, CH3 | 1.05, d (6.3) |
| 9a | 131.2 a, C | - |
| No. | 18 | No. | 23 | ||
|---|---|---|---|---|---|
| δC, Type | δH, Multi. (J in Hz) | δC, Type | δH, Multi. (J in Hz) | ||
| 1 | 169.3, C | - | 1 | 135.6, C | - |
| 2 | 36.4, CH2 | 2.83, d (7.1) | 2 | 136.6, C | - |
| 3 | 123.7, CH | 5.79, t (7.1) | 3 | 125.3, CH | 7.41, d (7.5) |
| 4 | 134.2, C | - | 4 | 126.4 b, CH | 7.13, m |
| 4a | 127.8, C | - | 5 | 126.3 b, CH | 7.12, m |
| 4b | 20.8, CH3 | 2.09, s | 6 | 130.7, CH | 7.15, m |
| 5 | 184.2, C | - | 7 | 37.5, CH2 | 3.08, dd (14.1, 3.6); 2.64, dd (14.1, 9.3) |
| 5a | 132.9, C | - | 8 | 71.9, CH | 3.92, ddd (9.3, 6.2, 3.6) |
| 6 | 112.1, CH | 7.19, s | 9 | 176.0, C | - |
| 7 | 165.6 a, C | - | 10 | 128.6, CH | 6.74, dd (15.8, 1.8) |
| 8 | 131.1, C | - | 11 | 126.7, CH | 6.09, dq (15.8, 6.5) |
| 8a | 16.3, CH3 | 2.19, s | 12 | 18.7, CH3 | 1.86, dd (6.5, 1.8) |
| 9 | 129.1, CH | 7.75, s | OH-8 | - | 5.42, d (6.2) |
| 9a | 119.6 a, C | - | NH2-9 | - | 7.25, s; 7.16, s |
| 10 | 177.4, C | - | |||
| 10a | 137.6, C | - | |||
| OH-7 | - | 9.34, 1H, s | |||
| Compounds | Glioma Cells (IC50: μM) | Microorganisms (MIC: μg/mL) | ||
|---|---|---|---|---|
| U87MG | U251 | MRSA | Escherichia coli | |
| 1 | 0.16 ± 0.01 | 0.35 ± 0.01 | NA | NA |
| 2 | 0.39 ± 0.04 | 2.63 ± 0.47 | NA | NA |
| 4 | 0.57 ± 0.09 | 7.33 ± 0.20 | NA | NA |
| 8 | 8.17 ± 0.17 | 7.04 ± 0.28 | 15 | 8 |
| 9 | NA | NA | 24 | 20 |
| 11 | 8.81 ± 0.80 | 10.46 ± 0.27 | NA | NA |
| 12 | 8.32 ± 0.38 | 7.86 ± 0.26 | 9 | 16 |
| 17 | NA | NA | 44 | 25 |
| 18 | 34.68 ± 0.58 | >50 | 3 | 6 |
| 21 | NA | NA | 10 | 16 |
| 22 | 6.18 ± 0.18 | 8.13 ± 0.56 | 3 | 8 |
| 26 | NA | NA | 5 | 12 |
| 27 | 11.18 ± 0.92 | 14.64 ± 1.73 | 6 | 28 |
| 28 | 19.39 ± 0.67 | 13.42 ± 1.71 | 8 | 48 |
| 30 | 1.64 ± 0.06 | 1.35 ± 0.05 | NA | NA |
| Doxorubicin | 0.43 ± 0.01 | 4.18 ± 0.39 | NT | NT |
| Vancomycin | NT | NT | 0.25 | NT |
| Gentamicin | NT | NT | 0.50 | 0.25 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yi, W.; Newaz, A.W.; Yong, K.; Ma, M.; Lian, X.-Y.; Zhang, Z. New Hygrocins K–U and Streptophenylpropanamide A and Bioactive Compounds from the Marine-Associated Streptomyces sp. ZZ1956. Antibiotics 2022, 11, 1455. https://doi.org/10.3390/antibiotics11111455
Yi W, Newaz AW, Yong K, Ma M, Lian X-Y, Zhang Z. New Hygrocins K–U and Streptophenylpropanamide A and Bioactive Compounds from the Marine-Associated Streptomyces sp. ZZ1956. Antibiotics. 2022; 11(11):1455. https://doi.org/10.3390/antibiotics11111455
Chicago/Turabian StyleYi, Wenwen, Asif Wares Newaz, Kuo Yong, Mingzhu Ma, Xiao-Yuan Lian, and Zhizhen Zhang. 2022. "New Hygrocins K–U and Streptophenylpropanamide A and Bioactive Compounds from the Marine-Associated Streptomyces sp. ZZ1956" Antibiotics 11, no. 11: 1455. https://doi.org/10.3390/antibiotics11111455