Current detection methods of African swine fever virus
- 1Shandong Engineering Laboratory of Pig and Poultry Healthy Breeding and Disease Diagnosis Technology, Xiajin New Hope Liuhe Agriculture and Animal Husbandry Co., Ltd., Dezhou, China
- 2Shandong New Hope Liuhe Co., Ltd., Qingdao, China
- 3Shandong New Hope Liuhe Agriculture and Animal Husbandry Technology Co., Ltd., (NHLH Academy of Swine Research), Dezhou, China
- 4China Agriculture Research System-Yangling Comprehensive Test Station, Xianyang, China
- 5College of Veterinary Medicine, Northwest A&F University, Xianyang, China
- 6Key Laboratory of Feed and Livestock and Poultry Products Quality and Safety Control, Ministry of Agriculture and Rural Affairs, New Hope Liuhe Co., Ltd., Chengdu, China
African swine fever (ASF), caused by the African swine fever virus (ASFV), is a highly contagious and notifiable animal disease in domestic pigs and wild boars, as designated by the World Organization for Animal Health (WOAH). The effective diagnosis of ASF holds great importance in promptly controlling its spread due to its increasing prevalence and the continuous emergence of variant strains. This paper offers a comprehensive review of the most common and up-to-date methods established for various genes/proteins associated with ASFV. The discussed methods primarily focus on the detection of viral genomes or particles, as well as the detection of ASFV associated antibodies. It is anticipated that this paper will serve as a reference for choosing appropriate diagnostic methods in diverse application scenarios, while also provide direction for the development of innovative technologies in the future.
Introduction
African swine fever (ASF), caused by the African swine fever virus (ASFV), is a highly contagious and notifiable animal disease in domestic pigs and wild boars, as designated by the World Organization for Animal Health (WOAH) (1). ASFV is the only member of the Asfivirus genus within the Asfarviridae family and is the sole arthropod-borne DNA virus (2). The length of the genome exhibits variation among distinct isolates of ASFV, with a range of 170–194 kb (3) and an abundance of over 151 open reading frames (ORFs) (4–6). Notably, strains classified under the ASFV genotype II, such as the seven Polish isolates gathered from 2016 to 2017, demonstrate a high number of ORFs, specifically ranging from 187 to 190 (7). Additionally, the ASFV strain Belgium/Etalle/wb/2018, identified in wild boar in Belgium during 2018, possesses 186 ORFs (8). Based on the sequence alignment analysis of the C-terminal region of the B646L gene, ASFV can be classified into 24 distinct genotypes (9).
The first description of ASFV was done in Kenya in 1921, and then the introduction from Africa to Portugal occurred in 1957, subsequently leading to outbreaks in various European countries (10–12). In 2007, a highly virulent genotype II ASFV emerged in Georgia and rapidly disseminated throughout Eastern Europe and other geographical regions (13). Notably, in China, the detection of genotype II ASFV strains took place in 2018, followed by the identification of genotype I strains in 2021, and the emergence of recombinant strains combining genotypes I and II in 2023 (14–16).
ASFV possesses an intricate icosahedral multilayer architecture, encompassing the envelope, capsid, inner envelope, core shell, and nucleolus (17). It has the capacity to encode 68 structural proteins and more than 100 non-structural proteins (18). Investigating the structural attributes, functionalities, and molecular mechanisms of viral proteins can establish a theoretical foundation for the advancement of diagnostic kits and vaccines. The CD2v protein, which is encoded by the EP402R gene, exhibits characteristics of transmembrane proteins akin to T lymphocyte surface adhesion receptors. It is primarily situated on the external envelope of viral particles and primarily facilitates the binding of the virus to erythrocytes (19, 20). The p72 protein, encoded by the B646L gene, is positioned on the surface of the viral capsid and possesses the ability to induce the host’s production of neutralizing antibodies, as well as actively engage in the viral attachment process to host cells (21). The E183L and CP204L genes encode the p54 and p30 proteins, respectively, which are situated on the inner envelope of viral particles. The functionality of the p54 protein aligns with that of the p72 protein and primarily facilitates virus attachment (22). Conversely, the p30 protein exhibits high expression during the initial phases of infection and plays a crucial role in virus endocytosis (23). Additionally, the CP2475L gene encodes the pp220 protein, which is positioned on the core shell of viral particles and actively facilitates the packaging of the viral core (24). The pA104R and p10 proteins, which are encoded by the A104R and K78R genes, respectively, exhibit localization within the nucleolus of viral particles and play a crucial role in the replication process of the virus (25, 26). Additionally, the multi-gene family MGF (MGF110, MGF300, MGF360, MGF530/505) is associated with host specificity and immune evasion, and possesses distinct functions in determining virulence, facilitating virus replication, and establishing latency (27–29).
The effective diagnosis of ASF holds great importance in promptly controlling its spread due to its increasing prevalence and the continuous emergence of variant strains (30). According to the chapter 3.9.1 of the WOAH Terrestrial Manual (the latest edition: twelfth edition 2023) (31), the laboratory diagnostic procedures for ASF can be categorized into two groups: virus detection and serology. In terms of virus detection, there are several approaches available. Firstly, the isolation of the virus can be achieved by inoculating pig leukocyte or bone marrow cultures. Alternatively, the genomic DNA of the virus can be detected using the polymerase chain reaction (PCR) technique. Another method involves the direct fluorescent antibody test (FAT), where the antigen in smears or cryostat sections of tissues is detected. PCR techniques are particularly valuable for ASFV detection due to their exceptional sensitivity, specificity, rapidity, and applicability in diverse circumstances. Currently the PCR is the most popular technique and can detect ASFV genome from a very early stage of infection in tissues, ethylene diamine tetra-acetic acid (EDTA)-blood and serum samples. In this paper, we mainly discuss the latest research advancements of viral genome or particle detection methods, encompassing laboratory testing techniques (Table 1) such as Real-time fluorescence quantitative PCR (RT-qPCR), Propidium monoazide (PMA)-qPCR, digital polymerase chain reaction (dPCR), as well as on-site testing methods (Table 2) including recombinase polymerase amplification (RPA)/recombinase-aidamplification (RAA), loop-mediated isothermal amplification (LAMP), jumping rolling circle amplification (SRCA), cross-priming amplification (CPA), and clustered regularly interspaced short palindromic repeats (CRISPR). Additionally, as no vaccine is available, the detection of ASFV antibodies serves as a reliable indicator of past infection. Notably, these antibodies are generated within the first week of infection and endure for extended durations, making them a valuable diagnostic tool for identifying the disease, particularly in cases of subacute and chronic manifestations (31). Therefore, the research progress of antibody detection methods (Table 3) is another part of this paper, mainly including enzyme-linked immunosorbent assay (ELISA) and immunochromatographic assay. Above all, we present a comprehensive overview of the existing detection techniques employed for various genes/proteins associated with ASFV, with the objective of offering guidance for the advancement of novel technologies and serving as a point of reference for the selection of suitable diagnostic approaches in diverse application scenarios.
Viral genome detection by PCR methods
qPCR
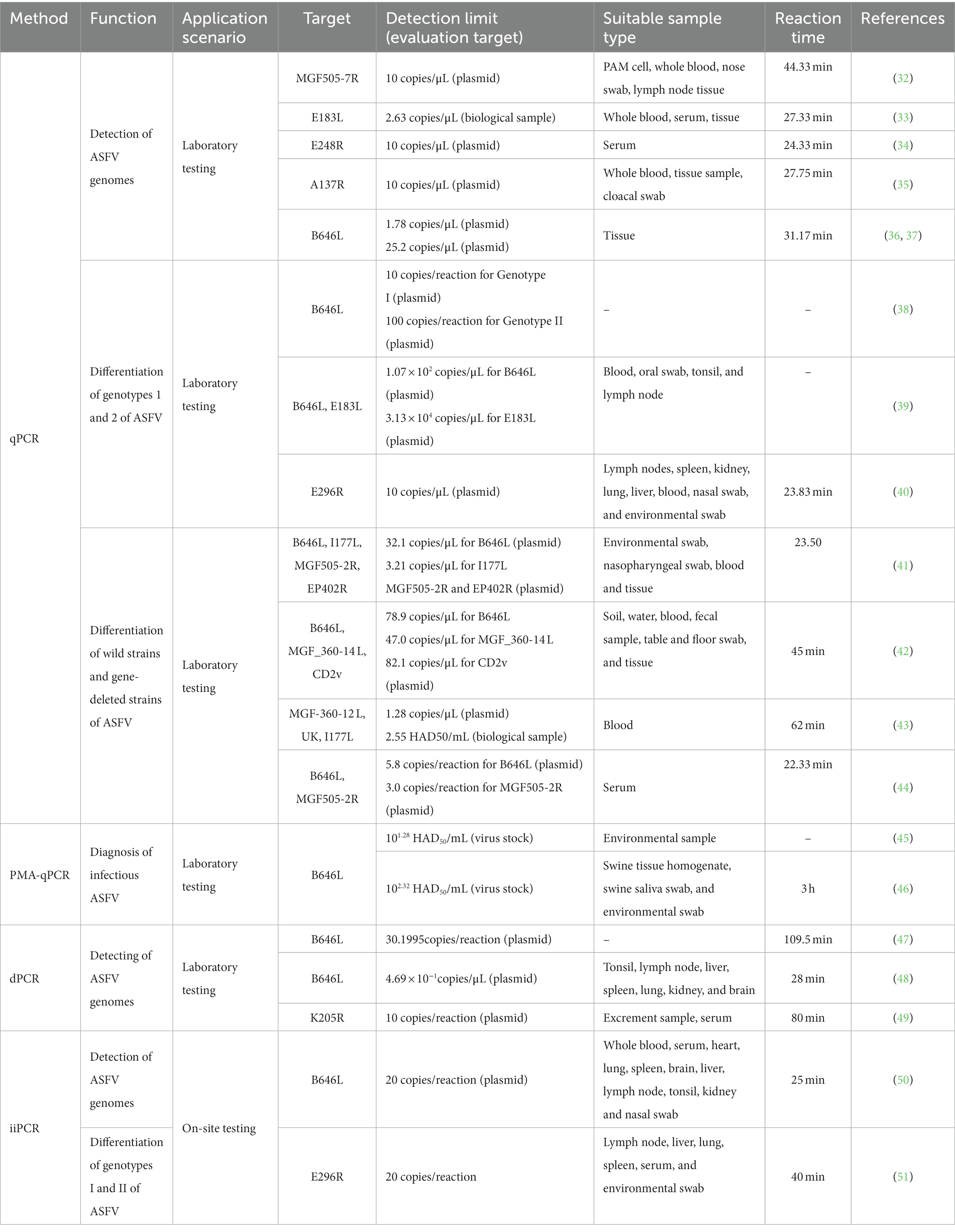
The qPCR method is a frequently employed technique and recommended by WOAH in laboratory diagnostic procedures for ASF. B646L is the target gene recommended by WOAH (109) for ASFV diagnosis and has been wildly used for many years. In the context of further gene structure analysis of ASFV, an increasing number of target genes have been identified for the purpose of ASFV detection. As presented in Table 1, various qPCR methodologies have been devised, utilizing ASFV MGF505-7R gene, E183L gene, 9GL gene, E248R gene, and A137R gene, exhibiting distinct limits of detection (LOD) ranging from 2.63 to 20 copies/μL (32–35). Furthermore, researchers have developed dual qPCR assays targeting the B646L gene, E183L gene, and E296R gene to differentiate between genotype I and genotype II strains of ASFV, in response to the clinical demand for such differentiation (38–40). Furthermore, Chen et al. (36) have successfully developed a triple RT-qPCR detection method, specifically targeting the ASFV p72 gene, which enables differentiation between ASFV, CSFV, and PRRSV. The LOD for ASFV detection using this method is determined to be 1.78 copies/μL. Similarly, Liu et al. (37) have also established a comparable approach for distinguishing the ASFV p72 gene among ASFV, CSFV, and Atypical Porcine Pestivirus, with an LOD of 25.2 copies/μL. The attention towards the CD2v, MGF, and I177L genes has been gradually increasing due to the prevalence of ASFV variant strains and the emergence of gene-deleted vaccines (110, 111). Various qPCR assays, including multiple assays targeting B646L, I177L, MGF505-2R, and EP402R genes (41), triple assays targeting B646L, MGF_360-14 L, CD2v genes (42), triple assays targeting ASFV MGF-360-12 L, United Kingdom, and I177L genes (43), and dual assays targeting B646L and MGF505-2R genes (44), possess the capability to distinguish ASFV wild-type strains from gene-deleted strains.
Propidium monoazide qPCR
The identification of ASFV infectivity has broad implications in the monitoring of epidemics, evaluating the effectiveness of disinfection measures, and eliminating false negative results (112). To achieve this, researchers have employed the use of ethidium monoazide (EMA) or PMA as a biostain to pre-treat samples. The process and mechanism of EMA/PMA pre-treatment can be described as follows. Initially, the addition of an EMA/PMA solution to samples containing both intact and membrane-compromised cells enables the selective entry of the dye solely into the compromised cells. Subsequently, the dye intercalates into nucleic acids, and the presence of an azide group facilitates a cross-linking between the dye and the DNA upon exposure to intense visible light. Thirdly, the light-induced formation of a highly reactive nitrene radical initiates a reaction with the bound DNA. Fourthly, this alteration significantly impedes the sequential DNA amplification process in polymerase chain reaction (PCR). Ultimately, upon the onset of cross-linking, the light interacts with unbound surplus dye alongside water molecules, leading to the formation of non-reactive hydroxylamine. Therefore, the DNA extracted from cells possessing intact membranes remains unmodified (113). This method has been applied in many microorganisms with membrane structures, including cells, bacteria, fungi, and enveloped viruses. ASFV is an enveloped virus so that some researchers have applied PMA pretreatments to identify infectious viruses from non-infectious ones. As shown in Table 1, Zeng et al. (45) have developed a PMA-qPCR detection method for the rapid diagnosis of infectious ASFV and the evaluation of disinfection efficacy. This method exhibits a detection sensitivity of 101.28 HAD50/mL. With heat evaluation treatments, the detection limit for ASFV is 102.28 HAD50/mL, while with chlorine disinfectants, the detection limit is 105.28 HAD50/mL. Furthermore, Liu et al. (46) has developed a Triton X-100-assisted PMA-qPCR method that allows for the assessment of ASFV infectivity in samples, with a LOD of 102.32 HAD50/mL.
Digital polymerase chain reaction
dPCR, as originally proposed by Vogelstein B, encompasses droplet dPCR and chip-based dPCR techniques, initially employed for the identification of mutations in the Ras oncogene (114). As presented in Table 1, Jia et al. (47) successfully devised a targeted chip digital PCR (cdPCR) approach for quantifying ASFV by specifically targeting the B646L gene. The LODs for both cdPCR and qPCR approved by WOAH were determined using the same set of primers and the probe with ASFV standard plasmid diluted 10 times as templates, and the results showed that the LOD of the cdPCR method was 30.1995 copies per reaction while the LOD of the qPCR assay was 1,000 copies per reaction, indicating that the cdPCR method is approximately 33-fold more sensitive than the qPCR method endorsed by the WOAH. Wu et al. (49) further advanced the field by developing a droplet digital PCR (ddPCR) method centered on the K205R gene, yielding a minimum detection limit of 10 copies/reaction. Shi et al. (48) conducted a study and engaged in the development of a multiplex dPCR assay to detect ASFV, classical swine fever virus (CSFV), and porcine reproductive and respiratory syndrome virus (PRRSV), achieving a LOD of 4.69 × 10−1 copies/μL. In addition to high sensitivity, dPCR can also provide direct absolute quantification. The process of cdPCR is to adopt a sealed chip that partitions samples into thousands of reaction wells to run independent PCR amplifications. Subsequently, the concentration of the target gene in the original sample is calculated by counting and converting positive wells, which have positive amplification of the viral target gene using the Poisson model correction coefficient (115). Therefore, the dPCR method presents itself as a promising instrument for future investigations pertaining to the detection of ASFV mutations.
Insulated isothermal PCR
The technique of iiPCR utilizing fluorescence hydrolysis probes operates on the principle of Rayleigh-Benard convection and undergoes amplification via cycles of varying temperature gradients (116). As presented in Table 1, Zou et al. (50) have devised an iiPCR approach centered around the B646L gene, exhibiting comparable sensitivity to the WOAH-recommended qPCR and only taking 25 min for on-site testing. Similarly, Song et al. (51) have successfully established an iiPCR assay targeting the E296R gene, enabling swift differentiation of ASFV genotype I and genotype II, with a detection limit of 20 copies/reaction. And the entire process takes only 40 min to complete for on-site testing. Therefore, the iiPCR method is a high-sensitivity and rapid method for ASFV onsite detection and genotype differentiation.
Viral genome or particle detection by multiple-polymerase amplification technologies
Recombinase polymerase amplification/recombinase-aidamplification
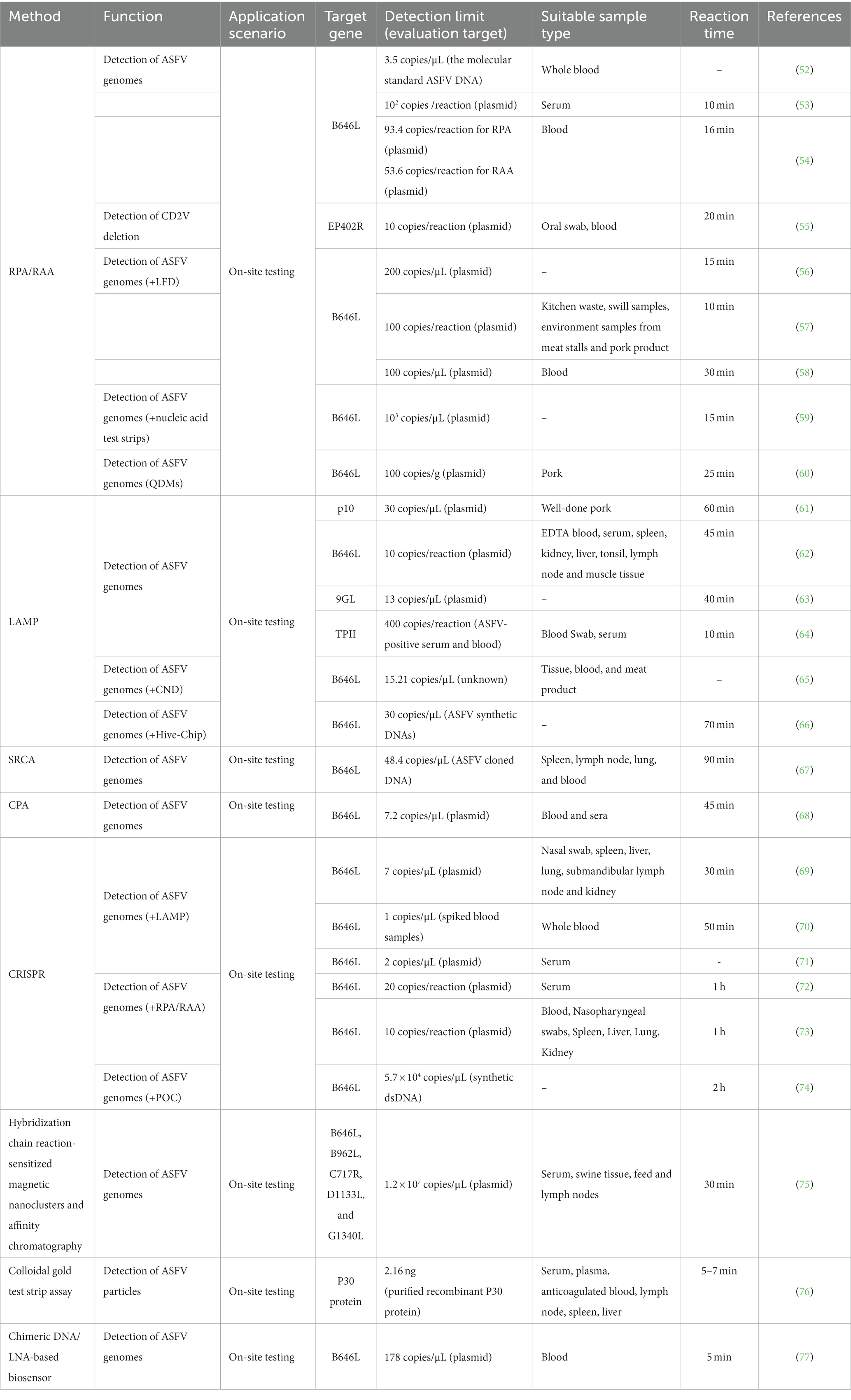
The concept of RPA technology was initially introduced in 2006 as a means to facilitate the amplification of genome in an isothermal environment, employing recombinases, polymerases, and single-stranded DNA-binding proteins (117). Depending on the specific recombinase utilized, RPA can be further classified as either RPA (T4 phage) or RAA (bacteria/fungi). As presented in Table 2, Ceruti A (52) and Wang et al. (53) have successfully developed a RPA method specifically targeting the B646L gene. This method demonstrates efficient detection capabilities, with completion within a time frame of 10–15 min. The sensitivities achieved are noteworthy, with 3.5 copies/μL and 102 copies/reaction, respectively. Additionally, Fan X (54) has contributed to the field by developing RPA/RAA methods targeting the same B646L gene, exhibiting sensitivities of 93.4 and 53.6 copies/reaction. These methods have shown promising diagnostic consistency when compared to the WOAH real-time fluorescence quantitative PCR. It is important to highlight the work of Wang ZH (55), who has developed an RAA method targeting the EP402R gene. This method allows for the detection of a minimum of 10 copies/reaction and enables the differentiation of CD2V gene deletions. Furthermore, researchers have successfully integrated RPA with various methodologies to enhance the expeditious visual identification of ASFV, thereby augmenting the efficacy of detection. Notably, the amalgamation of RPA with lateral flow chromatographic test strips (LFD) has demonstrated the capability to detect ASFV at a threshold of 1 × 102 copies/reaction (56–58). Similarly, the combination of RPA with nucleic acid test strips (Strip) has proven effective in detecting a minimum of 1 × 103 copies per microliter (μL) (59). Additionally, the integration of RPA with quantum dot microspheres (QDMs) has exhibited the ability to detect ASFV in clinical pork samples, with a sensitivity limit of 100 copies per gram (g) (60).
Loop-mediated isothermal amplification
The introduction of LAMP technology occurred in the year 2000. This technique entails the design of four primers that specifically target six regions on the desired gene, and employs DNA polymerase for amplification under constant temperature conditions (118). As presented in Table 2, various LAMP detection methods have been developed to target specific viral genes such as K78R, B646L, 9GL, and TPII (61–64), with LODs of 30 copies/μL, 10 copies/reaction, 13 copies/μL, and 400 copies/reaction, respectively. Wang et al. has devised a ladder-shaped melting temperature isothermal amplification (LMTIA) technique that specifically targets the ASFV-B646L gene. This method exhibits the same sensitivity of commercially available qPCR reagent kits with a LOD of 0.5 copies/μL (119). Furthermore, the integration of LAMP with carbon nanodots (CND) technology has facilitated the development of a fluorescent biosensor, enabling the highly sensitive detection of ASFV with a detection sensitivity of 15.21 copies/μL (65). In addition, the LOD for sample analysis using a multiplex and visual detection platform, which combines LAMP with a hive chip (Hive-Chip), is 50 copies/μL (66).
Clustered regularly interspaced short palindromic repeats
CRISPR/Cas-based nucleic acid detection technology has been developed for ASFV detection. This approach relies on the targeted cleavage of specific sequences by Cas12a/Cas13a proteins, resulting in the release of fluorescence signals from fluorescent reporter probes (120). Integration of CRISPR/Cas technology with RPA or LAMP techniques enables the conduction of the reaction in centrifuge tubes or lateral flow chromatographic test strips, thereby facilitating the visualization of the detection process and subsequent interpretation of the results. As presented in Table 2, multiple methods for ASFV detection have been developed by integrating the CRISPR-Cas12a system with the LAMP method, specifically targeting the B646L gene, with the detection limit ranging from 1 to 7 copies/μL (69–71). Furthermore, the combination of the CRISPR-Cas12a/Cas13a system with RPA/RAA methods demonstrates a detection range of 10–20 copies/μL (72, 73). Notably, He et al. (74) successfully combined CRISPR-Cas12a detection with a fluorescence-based point-of-care (POC) system for ASFV detection, which is based on a CRISPR Cas12a assay to trigger the indiscriminate ssDNA denaturation and a small and sensitive fluorescence-sensing unit with a disposable cartridge to measure the fluorescence signal. Without nucleic acid amplification, the LOD achieved to 5.7 × 104 copies/μL within 2 h. And this compact detection system is automated, integrated, small, lightweight, and inexpensive, ready to be used for on-site ASFV detection or other DNA based pathogens.
Others
Jumping rolling circle amplification (SRCA) is an emerging isothermal amplification technology that utilizes Bst DNA polymerase (121) and has been employed in the detection methodologies of Staphylococcus aureus (122) and Brucella (123). As presented in Table 2, the initial development of the SRCA technique for on-site detection was carried out by Milton AAP (67), who employed the ASFV-B646L gene as the target and achieved a LOD of 48.4 copies/μL. This characteristic renders it well-suited for on-site detection of clinical samples.
Cross-priming amplification (CPA) is an isothermal amplification technology that allows for highly specific and sensitive amplification at the constant temperature of 63°C (124). As presented in Table 2, the CPA method was initially devised by Fraczyk et al. (68) for the detection of the ASFV-B646L gene in blood and serum samples, exhibiting a LOD of 7.2 copies/μL.
Lee et al. (75) proposed an optical detection method utilizing the principle of affinity column chromatography to detect ASFV within a time frame of 30 min at ambient temperature, exhibiting a detection limit of approximately 1.2 × 107 copies/μL (Table 2). Zhang et al. (76) devised a sandwich colloidal gold test strip for swift on-site detection of ASFV, employing monoclonal antibodies targeting the P30 protein, with a LOD of 2.16 nanograms (ng) (Table 2). Gomez-Gomez et al. (77) advocated the implementation of a biosensor for the identification of ASFV in porcine blood, demonstrating a LOD of 178 copies/μL (Table 2).
ASFV associated antibodies detection
ELISA
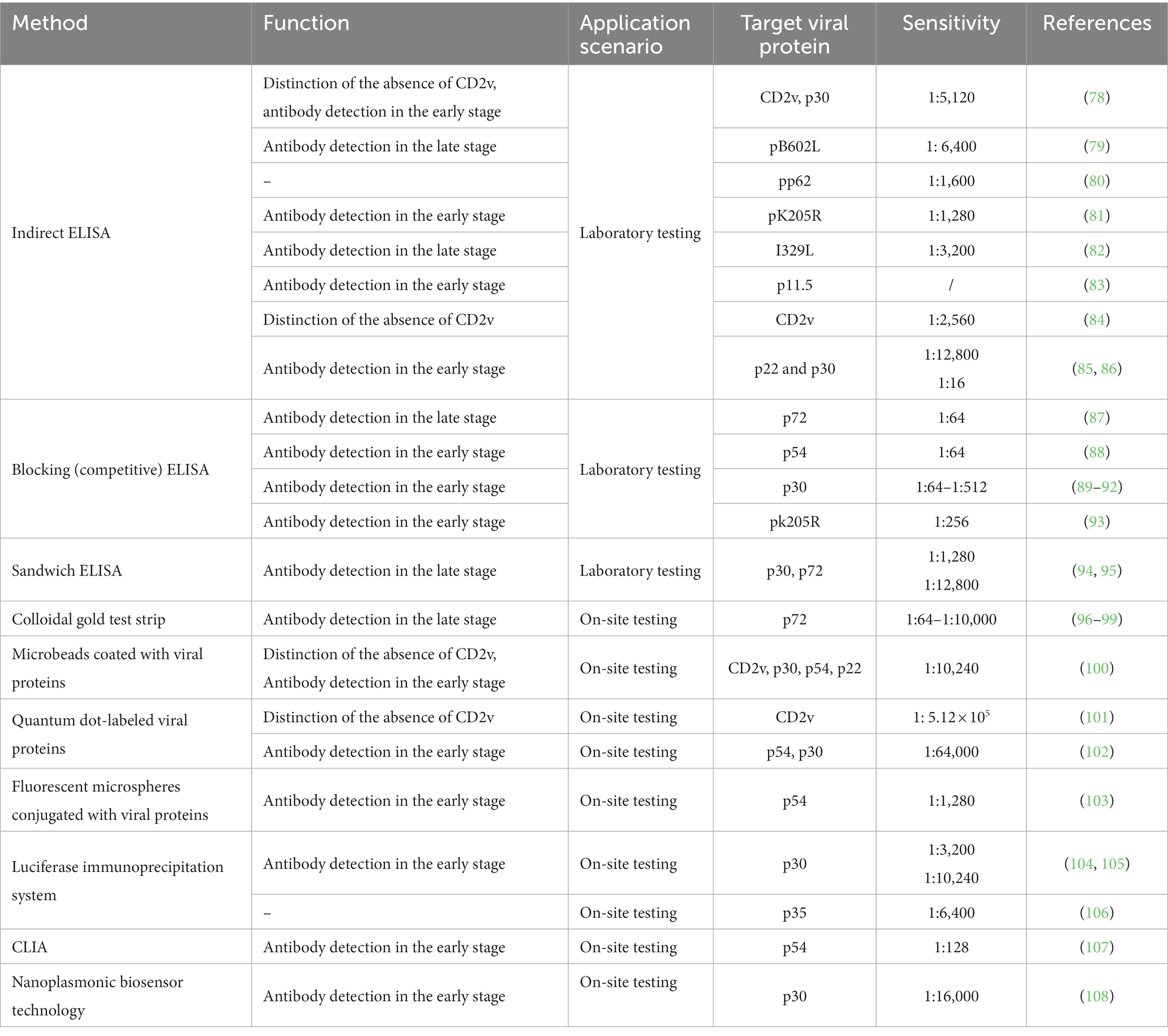
ELISA is a detection technique that relies on the specific binding of antigen and antibody. It encompasses various forms such as indirect ELISA, blocking (competitive) ELISA, sandwich ELISA, and multiplex ELISA. As presented in Table 3, in response to the emergence of low virulent ASFV strains with CD2v mutants, Lv et al. (78) developed a dual ELISA approach utilizing CD2v and p30 protein to differentiate wild-type strains from CD2v-deleted strains with low virulence. Furthermore, indirect ELISA methods based on non-structural proteins of ASFV, including pB602L (79), pp62 (80), pK205R (81), I329L (82), p11.5 (83), CD2v (84), p22 and p30 (85, 86), have demonstrated favorable detection capabilities, with an analytical sensitivity ranging from 1:1280 to 1:12800 (Table 3). In comparison to indirect ELISA, the main advantage of blocking (competitive) ELISA is its high sensitivity to compositional differences in complex antigen mixtures, even when the specific detecting antibody is present in relatively small amounts (125). Previous research has demonstrated the successful implementation of blocking (competitive) ELISA techniques based on ASFV structural proteins, including p72 (87), p54 (88), p30 (89–92) and non-structural protein pk205R (93). Wang M (94) and Wang et al. (95) have developed a double-antigen sandwich ELISA utilizing specific enzyme-conjugated antigens, namely p30 protein and p72 protein, exhibiting a sensitivity range of 1:1280 to 1:12,800 (Table 3).
The ELISA method is commonly used to detect antibodies in serum samples. However, the process of collecting serum samples from pigs can induce stress and place a significant burden on personnel. As presented in Table 3, previous research has indicated that antibodies can also be detected in oral fluids, which are easier to collect, suggesting their potential as an alternative sample to serum (126, 127). Mur et al. (128) collected saliva and serum samples from pigs that were inoculated with attenuated ASFV at various time intervals following infection. By utilizing a modified ELISA method, the presence of ASFV antibodies was successfully detected in both saliva and serum samples. Hence, utilizing the recombinant p30 protein, Gimenez-Lirola et al. (129) successfully devised an indirect ELISA method that was capable of effectively identifying antibodies against ASFV in oral fluid samples, and exhibited no significant differences when compared to serum samples.
Immunochromatographic assay
The immunochromatographic assay is a convenient and rapid technique known for its high specificity and ability to provide visualized results, making it well-suited for on-site detection. As presented in Table 3, several studies have demonstrated that the immunochromatographic assay, utilizing p72 as the labeled protein and colloidal gold as the label, exhibits an analytical sensitivity ranging from 1:64 to 1:10,000 (96–99). Sastre et al. (130) have developed a dual-flow lateral flow assay that is based on the p72 protein of ASFV and the E2 protein of CSFV. This assay specifically detects antibodies against CSFV and ASFV. The implementation of alternative carrier systems for antigen labeling has resulted in the establishment of more efficient methods of detection. For example, the utilization of microbeads coated with recombinant CD2v, p30, p54, and p22 proteins (100), quantum dot-labeled CD2v (101) or p54 and p30 proteins (102), and fluorescent microspheres conjugated with p54 protein (103), can significantly improve the detection rate and stability of positive samples. Additional fluorescent enzyme-linked antibody detection methods have been established in the field of antibody detection. One such method is the luciferase immunoprecipitation system (LIPS) assay, which employs Gaussia luciferase (GLuc) labeled p30 (104, 105) or p35 proteins (106). This assay has been specifically designed for the identification of antibodies against ASFV in pig serum, with the analytical sensitivity ranging from 1:3,200 to 1:10,240. Furthermore, a chemiluminescent immunoassay (CLIA) was developed using the ASFV protein p54 as a serum diagnostic antigen and an anti-p54 monoclonal antibody (107). This CLIA exhibits a sensitivity of 1:128 for the detection of ASFV antibodies. Zhao et al. (108) employed ASFV P30 as the focal point and nanoplasmonic biosensor technology to create an innovative device for detecting ASFV antibodies. This device exhibits a rapid reaction time of 20 min, a remarkable sensitivity of 1:16,000, and the ability to prevent cross-contamination through a simplified one-step sample addition process.
Discussion and perspectives
Currently, the identification techniques employed for ASFV primarily comprised laboratory testing and on-site rapid testing. Laboratory testing offers notable benefits such as heightened sensitivity, capacity for extensive testing, and cost-effectiveness. However, it necessitates specialized facilities and expensive equipment, rendering it more suitable for large-scale farms rather than routine monitoring in small-scale farms. In contrast, the utilization of on-site rapid testing presents notable benefits, as it allows for direct application in the field, obviating the requirement for specialized facilities or equipment. Nevertheless, it is crucial to acknowledge that the shorter the reaction time required for on-site detection technology, the lower the sensitivity of detection may be. How to improve the sensitivity of detection in a short reaction time may be a promising research direction in the future. Furthermore, it is imperative to recognize that the types of clinical samples significantly impacts the accuracy of detection outcomes. Previous studies have reported that ASFV can also be monitored daily using oral fluids (126), inguinal lymph nodes from dead pigs (131), and ear tissues (132) in addition to blood.
The detection of ASFV antibodies can be utilized as a supplementary approach to evaluate the present infection status of pigs, given the emergence of various ASFV strains (Table 3). Antibody detection targets commonly employed for ASFV infection include structural proteins p72, p54, and p30, as well as non-structural proteins pK205R and pB602L, owing to their diverse functionalities and diverse time of emergence at different stages of viral infection (133). Notably, commercial kits commonly target antibodies against p54 and p30 proteins, which manifest during the initial phases of ASFV infection. Moreover, the viremia exhibited by the attenuated strain of ASFV is characterized by intermittent occurrences (134). Consequently, relying solely on pathogen detection may result in overlooking positive pigs, necessitating the inclusion of antibody detection to overcome this constraint. In subsequent investigations, it may be imperative for researchers to undertake comprehensive examinations of ASFV proteins to devise antibody detection techniques that effectively ascertain the infection stage of pigs, particularly in identifying latent infection carriers.
In forthcoming times, a formidable undertaking persists in the realm of implementing expeditious on-site detection techniques for ASFV in circumstances encompassing import/export quarantine and inter-regional transportation of live pigs. Furthermore, the latent infection attributes of ASF remain incompletely comprehended, and extant detection methodologies lack precision in identifying pigs in a latent infection state. Consequently, the imperative development of methodologies capable of swiftly screening pigs during the latent infection period will be pivotal for the future prevention and control of ASFV.
Author contributions
ZH: Data curation, Formal analysis, Writing – original draft. XT: Data curation, Formal analysis, Writing – original draft. RL: Data curation, Formal analysis, Writing – original draft. XW: Writing – review & editing. XL: Funding acquisition, Project administration, Validation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by Taishan Industry Leadership Talent Project of Shandong Province in China, and the earmarked fund for CARS (CARS-35) and National Key R&D Program of China (2021ZD0113800).
Conflict of interest
ZH, XT, RL, XL were employed by Xiajin New Hope Liuhe Agriculture and Animal Husbandry Co., Ltd., Shandong New Hope Liuhe Co., Ltd., Shandong New Hope Liuhe Agriculture and Animal Husbandry Technology Co., Ltd., (NHLH Academy of Swine Research), and also XL was employed by New Hope Liuhe Co., Ltd.
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Liu, Y , Zhang, X , Qi, W , Yang, Y , Liu, Z , An, T, et al. Prevention and control strategies of African swine fever and Progress on pig farm repopulation in China. Viruses. (2021) 13:2552. doi: 10.3390/v13122552
2. Galindo, I , and Alonso, C . African swine fever virus: a review. Viruses. (2017) 9:1255. doi: 10.3390/v9050103
3. Alonso, C , Borca, M , Dixon, L , Revilla, Y , Rodriguez, F , Escribano, JM, et al. ICTV virus taxonomy profile: asfarviridae. J Gen Virol. (2018) 99:613–4. doi: 10.1099/jgv.0.001049
4. Li, Z , Chen, W , Qiu, Z , Li, Y , Fan, J , Wu, K, et al. African swine fever virus: a review. Life (Basel). (2022) 12:1255. doi: 10.3390/life12081255
5. Alejo, A , Matamoros, T , Guerra, M , and Andrés, G . A proteomic atlas of the African swine fever virus particle. J Virol. (2018) 92:e01293–18. doi: 10.1128/jvi.01293-18
6. Torresi, C , Fiori, M , Bertolotti, L , Floris, M , Colitti, B , Giammarioli, M, et al. The evolution of African swine fever virus in Sardinia (1978-2014) as revealed by whole-genome sequencing and comparative analysis. Transbound Emerg Dis. (2020) 00:1–10. doi: 10.1111/tbed.13540
7. Mazur-Panasiuk, N , Woźniakowski, G , and Niemczuk, K . The first complete genomic sequences of African swine fever virus isolated in Poland. Sci Rep. (2019) 9:4556. doi: 10.1038/s41598-018-36823-0
8. Gilliaux, G , Garigliany, M , Licoppe, A , Paternostre, J , Lesenfants, C , Linden, A, et al. Newly emerged African swine fever virus strain Belgium/Etalle/Wb/2018: complete genomic sequence and comparative analysis with reference P72 genotype ii strains. Transbound Emerg Dis. (2019) 66:2566–91. doi: 10.1111/tbed.13302
9. Bastos, AD , Penrith, ML , Cruciere, C , Edrich, JL , Hutchings, G , Roger, F, et al. Genotyping field strains of African swine fever virus by partial P72 gene characterisation. Arch Virol. (2003) 148:693–706. doi: 10.1007/s00705-002-0946-8
10. Costard, S , Mur, L , Lubroth, J , Sanchez-Vizcaino, JM , and Pfeiffer, DU . Epidemiology of African swine fever virus. Virus Res. (2013) 173:191–7. doi: 10.1016/j.virusres.2012.10.030
11. Dixon, LK , Sun, H , and Roberts, H . African swine fever. Antivir Res. (2019) 165:34–41. doi: 10.1016/j.antiviral.2019.02.018
12. Eustace, MR . On a form of swine fever occurring in British East Africa (Kenya Colony). J. Comp Pathol Ther. (1921) 34:159–91. doi: 10.1016/S0368-1742(21)80031-4
13. Oganesyan, AS , Petrova, ON , Korennoy, FI , Bardina, NS , Gogin, AE , and Dudnikov, SA . African swine fever in the Russian Federation: Spatio-temporal analysis and epidemiological overview. Virus Res. (2013) 173:204–11. doi: 10.1016/j.virusres.2012.12.009
14. Zhou, X , Li, N , Luo, Y , Liu, Y , Miao, F , Chen, T, et al. Emergence of African swine fever in China, 2018. Transbound Emerg Dis. (2018) 65:1482–4. doi: 10.1111/tbed.12989
15. Sun, E , Huang, L , Zhang, X , Zhang, J , Shen, D , Zhang, Z, et al. Genotype I African swine fever viruses emerged in domestic pigs in China and caused chronic infection. Emerg Microbes Infect. (2021) 10:2183–93. doi: 10.1080/22221751.2021.1999779
16. Zhao, D , Sun, E , Huang, L , Ding, L , Zhu, Y , Zhang, J, et al. Highly lethal genotype I and ii recombinant African swine fever viruses detected in pigs. Nat Commun. (2023) 14:3096. doi: 10.1038/s41467-023-38868-w
17. Liu, S , Luo, Y , Wang, Y , Li, S , Zhao, Z , Bi, Y, et al. Cryo-EM structure of the African swine fever virus. Cell Host Microbe. (2019) 26:836–843.e3. doi: 10.1016/j.chom.2019.11.004
18. Wang, G , Xie, M , Wu, W , and Chen, Z . Structures and functional diversities of ASFV proteins. Viruses. (2021) 13:2124. doi: 10.3390/v13112124
19. Borca, MV , Kutish, GF , Afonso, CL , Irusta, P , Carrillo, C , Brun, A, et al. An African swine fever virus gene with similarity to the T-lymphocyte surface antigen Cd2 mediates Hemadsorption. Virology. (1994) 199:463–8. doi: 10.1006/viro.1994.1146
20. Goatley, LC , and Dixon, LK . Processing and localization of the African swine fever virus CD2v transmembrane protein. J Virol. (2011) 85:3294–305. doi: 10.1128/JVI.01994-10
21. Gomez-Puertas, P , Rodriguez, F , Oviedo, JM , Ramiro-Ibanez, F , Ruiz-Gonzalvo, F , Alonso, C, et al. Neutralizing antibodies to different proteins of African swine fever virus inhibit both virus attachment and internalization. J Virol. (1996) 70:5689–94. doi: 10.1128/JVI.70.8.5689-5694.1996
22. Rodriguez, F , Alcaraz, C , Eiras, A , Yanez, RJ , Rodriguez, JM , Alonso, C, et al. Characterization and molecular basis of heterogeneity of the African swine fever virus envelope protein P54. J Virol. (1994) 68:7244–52. doi: 10.1128/JVI.68.11.7244-7252.1994
23. Gomez-Puertas, P , Rodriguez, F , Oviedo, JM , Brun, A , Alonso, C , and Escribano, JM . The African swine fever virus proteins P54 and P30 are involved in two distinct steps of virus attachment and both contribute to the antibody-mediated protective immune response. Virology. (1998) 243:461–71. doi: 10.1006/viro.1998.9068
24. Andres, G , Alejo, A , Salas, J , and Salas, ML . African swine fever virus polyproteins Pp220 and Pp62 assemble into the Core Shell. J Virol. (2002) 76:12473–82. doi: 10.1128/jvi.76.24.12473-12482.2002
25. Frouco, G , Freitas, FB , Coelho, J , Leitao, A , Martins, C , and Ferreira, F . DNA-binding properties of African swine fever virus Pa104r, a histone-like protein involved in viral replication and transcription. J Virol. (2017) 91:e02498-16. doi: 10.1128/JVI.02498-16
26. Nunes-Correia, I , Rodriguez, JM , Eulalio, A , Carvalho, AL , Citovsky, V , Simoes, S, et al. African swine fever virus P10 protein exhibits nuclear import capacity and accumulates in the nucleus during viral infection. Vet Microbiol. (2008) 130:47–59. doi: 10.1016/j.vetmic.2007.12.010
27. O'Donnell, V , Holinka, LG , Sanford, B , Krug, PW , Carlson, J , Pacheco, JM, et al. African swine fever virus Georgia isolate harboring deletions of 9gl and Mgf360/505 genes is highly attenuated in swine but does not confer protection against parental virus challenge. Virus Res. (2016) 221:8–14. doi: 10.1016/j.virusres.2016.05.014
28. Burrage, TG , Lu, Z , Neilan, JG , Rock, DL , and Zsak, L . African swine fever virus multigene family 360 genes affect virus replication and generalization of infection in Ornithodoros porcinus ticks. J Virol. (2004) 78:2445–53. doi: 10.1128/jvi.78.5.2445-2453.2004
29. Afonso, CL , Piccone, ME , Zaffuto, KM , Neilan, J , Kutish, GF , Lu, Z, et al. African swine fever virus multigene family 360 and 530 genes affect host interferon response. J Virol. (2004) 78:1858–64. doi: 10.1128/jvi.78.4.1858-1864.2004
30. Qiu, Z , Li, Z , Yan, Q , Li, Y , Xiong, W , Wu, K, et al. Development of diagnostic tests provides technical support for the control of African swine fever. Vaccine. (2021) 9:343. doi: 10.3390/vaccines9040343
31. World Organisation for Animal Health . (2023). Available at: https://www.Woah.Org/En/What-We-Do/Standards/Codes-and-Manuals/Terrestrial-Manual-Online-Access/ (Accessed October 25, 2023).
32. Qi, C , Zhang, Y , Wang, Z , Li, J , Hu, Y , Li, L, et al. Development and application of a TaqMan-based real-time PCR method for the detection of the ASFV Mgf505-7r gene. Front Vet Sci. (2023) 10:1093733. doi: 10.3389/fvets.2023.1093733
33. Trinh, TBN , Truong, T , Nguyen, VT , Vu, XD , Dao, LA , Nguyen, TL, et al. Development of a novel real-time PCR assay targeting P54 gene for rapid detection of African swine fever virus (ASFV) strains circulating in Vietnam. Vet Med Sci. (2021) 7:2268–72. doi: 10.1002/vms3.605
34. Li, L , Du, N , Chen, J , Zhang, K , Tong, W , Zheng, H, et al. Establishment and application of a quantitative PCR method for E248r gene of African swine fever virus. Vet Sci. (2022) 9:417. doi: 10.3390/vetsci9080417
35. Yin, D , Geng, R , Lv, H , Bao, C , Shao, H , Ye, J, et al. Development of real-time PCR based on A137r gene for the detection of African swine fever virus. Front Vet Sci. (2021) 8:753967. doi: 10.3389/fvets.2021.753967
36. Chen, Y , Shi, K , Liu, H , Yin, Y , Zhao, J , Long, F, et al. Development of a multiplex QRT-PCR assay for detection of African swine fever virus, classical swine fever virus and porcine reproductive and respiratory syndrome virus. J Vet Sci. (2021) 22:e87. doi: 10.4142/jvs.2021.22.e87
37. Liu, H , Shi, K , Zhao, J , Yin, Y , Chen, Y , Si, H, et al. Development of a one-step multiplex QRT-PCR assay for the detection of African swine fever virus, classical swine fever virus and atypical porcine Pestivirus. BMC Vet Res. (2022) 18:43. doi: 10.1186/s12917-022-03144-4
38. Cao, S , Lu, H , Wu, Z , and Zhu, S . A duplex fluorescent quantitative PCR assay to distinguish the genotype I and II strains of African swine fever virus in Chinese epidemic strains. Front Vet Sci. (2022) 9:998874. doi: 10.3389/fvets.2022.998874
39. Gao, Q , Feng, Y , Yang, Y , Luo, Y , Gong, T , Wang, H, et al. Establishment of a dual real-time PCR assay for the identification of African swine fever virus genotypes I and II in China. Front Vet Sci. (2022) 9:882824. doi: 10.3389/fvets.2022.882824
40. Li, X , Hu, Y , Liu, P , Zhu, Z , Liu, P , Chen, C, et al. Development and application of a duplex real-time PCR assay for differentiation of genotypes I and II African swine fever viruses. Transbound Emerg Dis. (2022) 69:2971–9. doi: 10.1111/tbed.14459
41. Zhao, K , Shi, K , Zhou, Q , Xiong, C , Mo, S , Zhou, H, et al. The development of a multiplex real-time quantitative PCR assay for the differential detection of the wild-type strain and the Mgf505-2r, Ep402r and I177l gene-deleted strain of the African swine fever virus. Animals. (2022) 12:1754. doi: 10.3390/ani12141754
42. Yang, H , Peng, Z , Song, W , Zhang, C , Fan, J , Chen, H, et al. A triplex real-time PCR method to detect African swine fever virus gene-deleted and wild type strains. Front Vet Sci. (2022) 9:943099. doi: 10.3389/fvets.2022.943099
43. Velazquez-Salinas, L , Ramirez-Medina, E , Rai, A , Pruitt, S , Vuono, EA , Espinoza, N, et al. Development real-time PCR assays to genetically differentiate vaccinated pigs from infected pigs with the Eurasian strain of African swine fever virus. Front Vet Sci. (2021) 8:768869. doi: 10.3389/fvets.2021.768869
44. Guo, Z , Li, K , Qiao, S , Chen, XX , Deng, R , and Zhang, G . Development and evaluation of duplex TaqMan real-time PCR assay for detection and differentiation of wide-type and Mgf505-2r gene-deleted African swine fever viruses. BMC Vet Res. (2020) 16:428. doi: 10.1186/s12917-020-02639-2
45. Zeng, D , Qian, B , Li, Y , Zong, K , Ding, L , Wang, M, et al. Quickly assessing disinfection effectiveness to control the spread of African swine fever virus. Appl Microbiol Biotechnol. (2023) 107:4947–59. doi: 10.1007/s00253-023-12611-3
46. Liu, H , Meng, F , Nyaruaba, R , He, P , Hong, W , Jiang, M, et al. A triton X-100 assisted PMAxx-qPCR assay for rapid assessment of infectious African swine fever virus. Front Microbiol. (2022) 13:1062544. doi: 10.3389/fmicb.2022.1062544
47. Jia, R , Zhang, G , Liu, H , Chen, Y , Zhou, J , Liu, Y, et al. Novel application of nanofluidic chip digital PCR for detection of African swine fever virus. Front Vet Sci. (2020) 7:621840. doi: 10.3389/fvets.2020.621840
48. Shi, K , Chen, Y , Yin, Y , Long, F , Feng, S , Liu, H, et al. A multiplex crystal digital PCR for detection of African swine fever virus, classical swine fever virus, and porcine reproductive and respiratory syndrome virus. Front Vet Sci. (2022) 9:926881. doi: 10.3389/fvets.2022.926881
49. Wu, X , Xiao, L , Lin, H , Chen, S , Yang, M , An, W, et al. Development and application of a droplet digital polymerase chain reaction (DDPCR) for detection and investigation of African swine fever virus. Can J Vet Res. (2018) 82:70–4.
50. Zou, T , Deng, J , Li, X , Zhang, S , Chen, L , Hao, L, et al. Development of a fluorescent probe hydrolysis-insulated isothermal PCR for rapid and sensitive on-site detection of African swine fever virus. Virol Sin. (2022) 37:462–4. doi: 10.1016/j.virs.2022.03.002
51. Song, R , Liu, P , Yang, Y , Lee, HS , Chen, C , Wu, X, et al. Development of a duplex insulated isothermal PCR assay for rapid on-site detection and differentiation of genotypes 1 and 2 of African swine fever virus. Front Cell Infect Microbiol. (2022) 12:948771. doi: 10.3389/fcimb.2022.948771
52. Ceruti, A , Kobialka, RM , Ssekitoleko, J , Okuni, JB , Blome, S , Abd El Wahed, A, et al. Rapid extraction and detection of African swine fever virus DNA based on isothermal recombinase polymerase amplification assay. Viruses. (2021) 13:1731. doi: 10.3390/v13091731
53. Wang, J , Wang, J , Geng, Y , and Yuan, W . A recombinase polymerase amplification-based assay for rapid detection of African swine fever virus. Can J Vet Res. (2017) 81:308–12.
54. Fan, X , Li, L , Zhao, Y , Liu, Y , Liu, C , Wang, Q, et al. Clinical validation of two recombinase-based isothermal amplification assays (RPA/RAA) for the rapid detection of African swine fever virus. Front Microbiol. (2020) 11:1696. doi: 10.3389/fmicb.2020.01696
55. Wang, ZH , Li, P , Lin, X , Jia, H , Jiang, YT , Wang, XJ, et al. Application of portable real-time recombinase-aided amplification (Rt-RAA) assay in the clinical diagnosis of ASFV and prospective diva diagnosis. Appl Microbiol Biotechnol. (2021) 105:3249–64. doi: 10.1007/s00253-021-11196-z
56. Li, JS , Hao, YZ , Hou, ML , Zhang, X , Zhang, XG , Cao, YX, et al. Development of a recombinase-aided amplification combined with lateral flow dipstick assay for the rapid detection of the African swine fever virus. Biomed Environ Sci. (2022) 35:133–40. doi: 10.3967/bes2022.018
57. Zhai, Y , Ma, P , Fu, X , Zhang, L , Cui, P , Li, H, et al. A recombinase polymerase amplification combined with lateral flow dipstick for rapid and specific detection of African swine fever virus. J Virol Methods. (2020) 285:113885. doi: 10.1016/j.jviromet.2020.113885
58. Wang, Z , Yu, W , Xie, R , Yang, S , and Chen, A . A strip of lateral flow gene assay using gold nanoparticles for point-of-care diagnosis of African swine fever virus in limited environment. Anal Bioanal Chem. (2021) 413:4665–72. doi: 10.1007/s00216-021-03408-2
59. Wu, K , Zhang, Y , Zeng, S , Liu, X , Li, Y , Li, X, et al. Development and application of RAA nucleic acid test strip assay and double RAA gel electrophoresis detection methods for ASFV and CSFV. Front Mol Biosci. (2021) 8:811824. doi: 10.3389/fmolb.2021.811824
60. Wen, X , Xie, Q , Li, J , Pei, Y , Bai, Y , Liu, F, et al. Rapid and sensitive detection of African swine fever virus in pork using recombinase aided amplification combined with QDMs-based test strip. Anal Bioanal Chem. (2022) 414:3885–94. doi: 10.1007/s00216-022-04030-6
61. Wang, D , Yu, J , Wang, Y , Zhang, M , Li, P , Liu, M, et al. Development of a real-time loop-mediated isothermal amplification (Lamp) assay and visual Lamp assay for detection of African swine fever virus (ASFV). J Virol Methods. (2020) 276:113775. doi: 10.1016/j.jviromet.2019.113775
62. Wang, Y , Dai, J , Liu, Y , Yang, J , Hou, Q , Ou, Y, et al. Development of a potential Penside colorimetric Lamp assay using neutral red for detection of African swine fever virus. Front Microbiol. (2021) 12:609821. doi: 10.3389/fmicb.2021.609821
63. Wang, S , Shen, H , Lin, Q , Huang, J , Zhang, C , Liu, Z, et al. Development of a cleaved probe-based loop-mediated isothermal amplification assay for rapid detection of African swine fever virus. Front Cell Infect Microbiol. (2022) 12:884430. doi: 10.3389/fcimb.2022.884430
64. Mee, PT , Wong, S , O'Riley, KJ , da Conceicao, F , da Costa, B , Jong, J, et al. Field verification of an African swine fever virus loop-mediated isothermal amplification (Lamp) assay during an outbreak in Timor-Leste. Viruses. (2020) 12:1444. doi: 10.3390/v12121444
65. Cao, G , Qiu, Y , Long, K , Xiong, Y , Shi, M , Yang, J, et al. Carbon Nanodots combined with loop-mediated isothermal amplification (Lamp) for detection of African swine fever virus (ASFV). Mikrochim Acta. (2022) 189:1–9. doi: 10.1007/s00604-022-05390-7
66. Zhu, YS , Shao, N , Chen, JW , Qi, WB , Li, Y , Liu, P, et al. Multiplex and visual detection of African swine fever virus (Asfv) based on hive-Chip and Direct loop-mediated isothermal amplification. Anal Chim Acta. (2020) 1140:30–40. doi: 10.1016/j.aca.2020.10.011
67. Milton, AAP , Das, S , Khan, S , Momin, KM , Prasad, CB , Kylla, H, et al. Novel sensitive isothermal-based diagnostic technique for the detection of African swine fever virus. Arch Virol. (2023) 168:79. doi: 10.1007/s00705-023-05702-z
68. Fraczyk, M , Wozniakowski, G , Kowalczyk, A , Niemczuk, K , and Pejsak, Z . Development of cross-priming amplification for direct detection of the African swine fever virus, in pig and wild boar blood and sera samples. Lett Appl Microbiol. (2016) 62:386–91. doi: 10.1111/lam.12569
69. Yang, B , Shi, Z , Ma, Y , Wang, L , Cao, L , Luo, J, et al. Lamp assay coupled with Crispr/Cas12a system for portable detection of African swine fever virus. Transbound Emerg Dis. (2022) 69:e216–23. doi: 10.1111/tbed.14285
70. Qian, S , Chen, Y , Peng, C , Wang, X , Wu, H , Che, Y, et al. Dipstick-based rapid nucleic acids purification and Crispr/Cas12a-mediated isothermal amplification for visual detection of African swine fever virus. Talanta. (2022) 242:123294. doi: 10.1016/j.talanta.2022.123294
71. Tao, D , Liu, J , Nie, X , Xu, B , Tran-Thi, TN , Niu, L, et al. Application of Crispr-Cas12a enhanced fluorescence assay coupled with nucleic acid amplification for the sensitive detection of African swine fever virus. ACS Synth Biol. (2020) 9:2339–50. doi: 10.1021/acssynbio.0c00057
72. Wang, X , Ji, P , Fan, H , Dang, L , Wan, W , Liu, S, et al. Crispr/Cas12a technology combined with Immunochromatographic strips for portable detection of African swine fever virus. Commun Biol. (2020) 3:62. doi: 10.1038/s42003-020-0796-5
73. Wei, N , Zheng, B , Niu, J , Chen, T , Ye, J , Si, Y, et al. Rapid detection of genotype II African swine fever virus using CRISPR Cas13a-based lateral flow strip. Viruses. (2022) 14:179. doi: 10.3390/v14020179
74. He, Q , Yu, D , Bao, M , Korensky, G , Chen, J , Shin, M, et al. High-throughput and all-solution phase African swine fever virus (ASFV) detection using Crispr-Cas12a and fluorescence based point-of-care system. Biosens Bioelectron. (2020) 154:112068. doi: 10.1016/j.bios.2020.112068
75. Lee, H , Lee, S , Park, C , Yeom, M , Lim, JW , Vu, TTH, et al. Rapid visible detection of African swine fever virus using hybridization chain reaction-sensitized magnetic nanoclusters and affinity chromatography. Small. (2023) 19:e2207117. doi: 10.1002/smll.202207117
76. Zhang, X , Liu, X , Wu, X , Ren, W , Zou, Y , Xia, X, et al. A colloidal gold test strip assay for the detection of African swine fever virus based on two monoclonal antibodies against P30. Arch Virol. (2021) 166:871–9. doi: 10.1007/s00705-020-04915-w
77. Biagetti, M , Cuccioloni, M , Bonfili, L , Cecarini, V , Sebastiani, C , Curcio, L, et al. Chimeric DNA/Lna-based biosensor for the rapid detection of African swine fever virus. Talanta. (2018) 184:35–41. doi: 10.1016/j.talanta.2018.02.095
78. Lv, C , Zhao, Y , Jiang, L , Zhao, L , Wu, C , Hui, X, et al. Development of a dual Elisa for the detection of Cd2v-unexpressed lower-virulence mutational ASFV. Life. (2021) 11:1214. doi: 10.3390/life11111214
79. Yang, Y , Xia, Q , Sun, Q , Zhang, Y , Li, Y , Ma, X, et al. Detection of African swine fever virus antibodies in serum using a Pb602l protein-based indirect Elisa. Front Vet Sci. (2022) 9:971841. doi: 10.3389/fvets.2022.971841
80. Zhong, K , Zhu, M , Yuan, Q , Deng, Z , Feng, S , Liu, D, et al. Development of an indirect Elisa to detect African swine fever virus Pp62 protein-specific antibodies. Front Vet Sci. (2021) 8:798559. doi: 10.3389/fvets.2021.798559
81. Li, L , Qiao, S , Liu, J , Zhou, Y , Tong, W , Dong, S, et al. A highly efficient indirect Elisa and Monoclonal antibody established against African swine fever virus Pk205r. Front Immunol. (2022) 13:1103166. doi: 10.3389/fimmu.2022.1103166
82. Shen, Z , Qiu, W , Luan, H , Sun, C , Cao, X , Wang, G, et al. I329l protein-based indirect Elisa for detecting antibodies specific to African swine fever virus. Front Cell Infect Microbiol. (2023) 13:1150042. doi: 10.3389/fcimb.2023.1150042
83. Watanabe, M , Kitamura, T , Nagata, K , Ikezawa, M , Kameyama, KI , Masujin, K, et al. Development of a novel indirect Elisa for the serological diagnosis of African swine fever using P11.5 protein as a target antigen. Pathogens. (2023) 12:774. doi: 10.3390/pathogens12060774
84. Jiang, W , Jiang, D , Li, L , Wan, B , Wang, J , Wang, P, et al. Development of an indirect Elisa for the identification of African swine fever virus wild-type strains and Cd2v-deleted strains. Front Vet Sci. (2022) 9:1006895. doi: 10.3389/fvets.2022.1006895
85. Li, J , Jiao, J , Liu, N , Ren, S , Zeng, H , Peng, J, et al. Novel P22 and P30 dual-proteins combination based indirect Elisa for detecting antibodies against African swine fever virus. Front Vet Sci. (2023) 10:1093440. doi: 10.3389/fvets.2023.1093440
86. Nah, JJ , Kwon, OK , Choi, JD , Jang, SH , Lee, HJ , Ahn, DG, et al. Development of an indirect Elisa against African swine fever virus using two recombinant antigens, partial P22 and P30. J Virol Methods. (2022) 309:114611. doi: 10.1016/j.jviromet.2022.114611
87. Caixia, W , Songyin, Q , Ying, X , Haoyang, Y , Haoxuan, L , Shaoqiang, W, et al. Development of a blocking Elisa kit for detection of ASFV antibody based on a monoclonal antibody against full-length P72. J AOAC Int. (2022) 105:1428–36. doi: 10.1093/jaoacint/qsac050
88. Gao, Y , Xia, T , Bai, J , Zhang, L , Zheng, H , and Jiang, P . Preparation of monoclonal antibodies against the viral P54 protein and a blocking Elisa for detection of the antibody against African swine fever virus. Viruses. (2022) 14:2335. doi: 10.3390/v14112335
89. Zhou, J , Ni, Y , Wang, D , Fan, B , Zhu, X , Zhou, J, et al. Development of a competitive enzyme-linked immunosorbent assay targeting the-P30 protein for detection of antibodies against African swine fever virus. Viruses. (2023) 15:154. doi: 10.3390/v15010154
90. Yu, X , Zhu, X , Chen, X , Li, D , Xu, Q , Yao, L, et al. Establishment of a blocking Elisa detection method for against African swine fever virus P30 antibody. Front Vet Sci. (2021) 8:781373. doi: 10.3389/fvets.2021.781373
91. Zhao, J , Zhu, J , Wang, Y , Yang, M , Zhang, Q , Zhang, C, et al. A simple Nanobody-based competitive Elisa to detect antibodies against African swine fever virus. Virol Sin. (2022) 37:922–33. doi: 10.1016/j.virs.2022.09.004
92. Yuan, F , Petrovan, V , Gimenez-Lirola, LG , Zimmerman, JJ , Rowland, RRR , and Fang, Y . Development of a blocking enzyme-linked immunosorbent assay for detection of antibodies against African swine fever virus. Pathogens. (2021) 10:760. doi: 10.3390/pathogens10060760
93. Zhang, A , Wu, S , Duan, X , Zhao, H , Dong, H , Ren, J, et al. K205r specific Nanobody-horseradish peroxidase fusions as reagents of competitive Elisa to detect African swine fever virus serum antibodies. BMC Vet Res. (2022) 18:321. doi: 10.1186/s12917-022-03423-0
94. Wang, M , Song, J , Sun, J , Du, Y , Qin, X , Xia, L, et al. Development of an effective double antigen Sandwich Elisa based on P30 protein to detect antibodies against African swine fever virus. Viruses. (2022) 14:2170. doi: 10.3390/v14102170
95. Wang, L , Li, D , Liu, Y , Zhang, L , Peng, G , Xu, Z, et al. Development of an effective one-step double-antigen Sandwich Elisa based on P72 to detect antibodies against African swine fever virus. Front Vet Sci. (2023) 10:1160583. doi: 10.3389/fvets.2023.1160583
96. Wan, Y , Shi, Z , Peng, G , Wang, L , Luo, J , Ru, Y, et al. Development and application of a colloidal-gold dual Immunochromatography strip for detecting African swine fever virus antibodies. Appl Microbiol Biotechnol. (2022) 106:799–810. doi: 10.1007/s00253-021-11706-z
97. Zhang, X , Guo, J , Wang, L , Li, Z , Liu, Y , Tian, L, et al. Development and evaluation of multi-epitope protein P72 (Mep72) for the Serodiagnosis of African swine fever. Acta Virol. (2021) 65:273–8. doi: 10.4149/av_2021_304
98. Zhu, W , Meng, K , Zhang, Y , Bu, Z , Zhao, D , and Meng, G . Lateral flow assay for the detection of African swine fever virus antibodies using gold nanoparticle-labeled acid-treated P72. Front Chem. (2021) 9:804981. doi: 10.3389/fchem.2021.804981
99. Aira, C , Monedero, A , Hernandez-Anton, S , Martinez-Cano, J , Camunas, A , Casado, N, et al. Improving African swine fever surveillance using fluorescent rapid tests. Pathogens. (2023) 12:811. doi: 10.3390/pathogens12060811
100. Li, C , Zou, Z , Lv, C , Zhao, Y , Han, P , Sun, X, et al. Flow cytometry-based multiplexing antibody detection for diagnosis of African swine fever virus. Anal Chim Acta. (2022) 1225:340244. doi: 10.1016/j.aca.2022.340244
101. Niu, Y , Zhang, G , Zhou, J , Liu, H , Chen, Y , Ding, P, et al. Differential diagnosis of the infection caused by wild-type or Cd2v-deleted ASFV strains by quantum dots-based Immunochromatographic assay. Lett Appl Microbiol. (2022) 74:1001–7. doi: 10.1111/lam.13691
102. Li, J , Bai, Y , Li, F , Zhang, Y , Xie, Q , Zhang, L, et al. Rapid and ultra-sensitive detection of African swine fever virus antibody on site using QDM based-ASFV immunosensor (Qais). Anal Chim Acta. (2022) 1189:339187. doi: 10.1016/j.aca.2021.339187
103. Li, C , He, X , Yang, Y , Gong, W , Huang, K , Zhang, Y, et al. Rapid and visual detection of African swine fever virus antibody by using fluorescent Immunochromatography test strip. Talanta. (2020) 219:121284. doi: 10.1016/j.talanta.2020.121284
104. Ding, J , Yang, J , Jiang, D , Zhou, Y , Li, C , and Li, Y . Development of a highly sensitive Gaussia luciferase immunoprecipitation assay for the detection of antibodies against African swine fever virus. Front Cell Infect Microbiol. (2022) 12:988355. doi: 10.3389/fcimb.2022.988355
105. Liu, H , He, P , Meng, F , Jiang, M , Xiong, J , Li, J, et al. A Semiautomated luciferase immunoprecipitation assay for rapid and easy detection of African swine fever virus antibody. J Clin Microbiol. (2021) 59:e0099021. doi: 10.1128/JCM.00990-21
106. Wang, Q , Tian, Z , Yang, J , Gao, S , Du, J , Zhang, H, et al. An improved luciferase immunosorbent assay for ultrasensitive detection of antibodies against African swine fever virus. Front Microbiol. (2022) 13:1013678. doi: 10.3389/fmicb.2022.1013678
107. Yang, Y , Lv, C , Fan, J , Zhao, Y , Jiang, L , Sun, X, et al. Development of a Chemiluminescence immunoassay to accurately detect African swine fever virus antibodies in serum. J Virol Methods. (2021) 298:114269. doi: 10.1016/j.jviromet.2021.114269
108. Zhao, Y , Li, R , Lv, C , Zhang, Y , Zhou, H , Xia, X, et al. One-step rapid and sensitive ASFV P30 antibody detection via nanoplasmonic biosensors. Microbiol Spectr. (2022) 10:e0234322. doi: 10.1128/spectrum.02343-22
109. Fernández-Pinero, J , Gallardo, C , Elizalde, M , Robles, A , Gómez, C , Bishop, R, et al. Molecular diagnosis of African swine fever by a new real-time Pcr using universal probe library. Transbound Emerg Dis. (2013) 60:48–58. doi: 10.1111/j.1865-1682.2012.01317.x
110. Borca, MV , Ramirez-Medina, E , Silva, E , Vuono, E , Rai, A , Pruitt, S, et al. Development of a highly effective African swine fever virus vaccine by deletion of the I177l gene results in sterile immunity against the current epidemic Eurasia strain. J Virol. (2020) 94:e02017-19. doi: 10.1128/JVI.02017-19
111. Portugal, R , Coelho, J , Hoper, D , Little, NS , Smithson, C , Upton, C, et al. Related strains of African swine fever virus with different virulence: genome comparison and analysis. J Gen Virol. (2015) 96:408–19. doi: 10.1099/vir.0.070508-0
112. Zeng, D , Qian, B , Li, Y , Zong, K , Peng, W , Liao, K, et al. Prospects for the application of infectious virus detection technology based on Propidium Monoazide in African swine fever management. Front Microbiol. (2022) 13:1025758. doi: 10.3389/fmicb.2022.1025758
113. Zeng, D , Chen, Z , Jiang, Y , Xue, F , and Li, B . Advances and challenges in viability detection of foodborne pathogens. Front Microbiol. (2016) 7:1833. doi: 10.3389/fmicb.2016.01833
114. Vogelstein, B , and Kinzler, KW . Digital PCR. Proc Natl Acad Sci U S A. (1999) 96:9236–41. doi: 10.1073/pnas.96.16.9236
115. Quan, PL , Sauzade, M , and Brouzes, E . dPCR: a technology review. Sensors (Basel). (2018) 18:1271. doi: 10.3390/s18041271
116. Krishnan, M , Ugaz, VM , and Burns, MA . PCR in a Rayleigh-Benard convection cell. Science. (2002) 298:793. doi: 10.1126/science.298.5594.793
117. Piepenburg, O , Williams, CH , Stemple, DL , and Armes, NA . DNA detection using recombination proteins. PLoS Biol. (2006) 4:e204. doi: 10.1371/journal.pbio.0040204
118. Notomi, T , Okayama, H , Masubuchi, H , Yonekawa, T , Watanabe, K , Amino, N, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. (2000) 28:63e–663e. doi: 10.1093/nar/28.12.e63
119. Wang, Y , Wang, B , Xu, D , Zhang, M , Zhang, X , and Wang, D . Development of a ladder-shape melting temperature isothermal amplification (LMTIA) assay for detection of African swine fever virus (ASFV). J Vet Sci. (2022) 23:e51. doi: 10.4142/jvs.22001
120. Wang, M , Zhang, R , and Li, J . Crispr/Cas systems redefine nucleic acid detection: principles and methods. Biosens Bioelectron. (2020) 165:112430. doi: 10.1016/j.bios.2020.112430
121. Zyrina, NV , Zheleznaya, LA , Dvoretsky, EV , Vasiliev, VD , Chernov, A , and Matvienko, NI . N.Bspd6i DNA nickase strongly stimulates template-independent synthesis of non-palindromic repetitive DNA by Bst DNA polymerase. Biol Chem. (2007) 388:367–72. doi: 10.1515/BC.2007.043
122. Yang, Q , Zhang, Y , Li, S , Lu, X , Yuan, Y , and Zhang, W . Saltatory rolling circle amplification for sensitive visual detection of Staphylococcus aureus in Milk. J Dairy Sci. (2019) 102:9702–10. doi: 10.3168/jds.2019-16724
123. Milton, AAP , Momin, KM , Srinivas, K , Priya, GB , Ghatak, S , Das, S, et al. Development of a novel visual isothermal amplification assay for rapid detection of Brucella Spp. J Microbiol Methods. (2023) 207:106695. doi: 10.1016/j.mimet.2023.106695
124. Xu, G , Hu, L , Zhong, H , Wang, H , Yusa, S , Weiss, TC, et al. Cross priming amplification: mechanism and optimization for isothermal DNA amplification. Sci Rep. (2012) 2:246. doi: 10.1038/srep00246
125. Gan, SD , and Patel, KR . Enzyme immunoassay and enzyme-linked immunosorbent assay. J Invest Dermatol. (2013) 133:e12:1–3. doi: 10.1038/jid.2013.287
126. Prickett, JR , and Zimmerman, JJ . The development of Oral fluid-based diagnostics and applications in veterinary medicine. Anim Health Res Rev. (2010) 11:207–16. doi: 10.1017/S1466252310000010
127. Kittawornrat, A , Panyasing, Y , Goodell, C , Wang, C , Gauger, P , Harmon, K, et al. Porcine reproductive and respiratory syndrome virus (PRRSV) surveillance using pre-weaning Oral fluid samples detects circulation of wild-type PRRSV. Vet Microbiol. (2014) 168:331–9. doi: 10.1016/j.vetmic.2013.11.035
128. Mur, L , Gallardo, C , Soler, A , Zimmermman, J , Pelayo, V , Nieto, R, et al. Potential use of Oral fluid samples for serological diagnosis of African swine fever. Vet Microbiol. (2013) 165:135–9. doi: 10.1016/j.vetmic.2012.12.034
129. Gimenez-Lirola, LG , Mur, L , Rivera, B , Mogler, M , Sun, Y , Lizano, S, et al. Detection of African swine fever virus antibodies in serum and Oral fluid specimens using a recombinant protein 30 (P30) dual matrix indirect Elisa. PLoS One. (2016) 11:e0161230. doi: 10.1371/journal.pone.0161230
130. Sastre, P , Perez, T , Costa, S , Yang, X , Raber, A , Blome, S, et al. Development of a duplex lateral flow assay for simultaneous detection of antibodies against African and classical swine fever viruses. J Vet Diagn Invest. (2016) 28:543–9. doi: 10.1177/1040638716654942
131. Goonewardene, KB , Onyilagha, C , Goolia, M , Le, VP , Blome, S , and Ambagala, A . Superficial inguinal lymph nodes for screening dead pigs for African swine fever. Viruses. (2022) 14:83. doi: 10.3390/v14010083
132. Lee, HS , Bui, VN , Dao, DT , Bui, NA , Le, TD , Kieu, MA, et al. Pathogenicity of an African swine fever virus strain isolated in Vietnam and alternative diagnostic specimens for early detection of viral infection. Porcine Health Manage. (2021) 7:36. doi: 10.1186/s40813-021-00215-0
133. Kollnberger, SD , Gutierrez-Castaneda, B , Foster-Cuevas, M , Corteyn, A , and Parkhouse, RME . Identification of the principal serological immunodeterminants of African swine fever virus by screening a virus CDNA library with antibody. J Gen Virol. (2002) 83:1331–42. doi: 10.1099/0022-1317-83-6-1331
Keywords: African swine fever virus, detection method, viral genomes, antibodies, laboratory testing, on-site testing
Citation: Hu Z, Tian X, Lai R, Wang X and Li X (2023) Current detection methods of African swine fever virus. Front. Vet. Sci. 10:1289676. doi: 10.3389/fvets.2023.1289676
Edited by:
Francisco Javier Salguero, UK Health Security Agency (UKHSA), United KingdomReviewed by:
Martin Ashby, The Pirbright Institute, United KingdomJean Nepomuscene Hakizimana, Sokoine University of Agriculture, Tanzania
Copyright © 2023 Hu, Tian, Lai, Wang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaowen Li, lxw8272@163.com
†These authors have contributed equally to this work
 Zhiqiang Hu1,2,3,4†
Zhiqiang Hu1,2,3,4†  Xinglong Wang
Xinglong Wang Xiaowen Li
Xiaowen Li