Prevalence and Characterization of Cryptosporidium Species and Genotypes in Four Farmed Deer Species in the Northeast of China
- 1Chinese Center for Disease Control and Prevention, National Institute of Parasitic Diseases, Shanghai, China
- 2Chinese Center for Tropical Diseases Research, Shanghai, China
- 3WHO Collaborating Centre for Tropical Diseases, Shanghai, China
- 4National Center for International Research on Tropical Diseases, Ministry of Science and Technology, Shanghai, China
- 5Key Laboratory of Parasite and Vector Biology, Ministry of Health, Shanghai, China
- 6Department of Parasitology, Wenzhou Medical University, Wenzhou, China
Cryptosporidiosis is a major public health problem in humans and animals. Information on the prevalence and molecular diversity of Cryptosporidium in farmed deer in northeastern China is limited. In this study, the prevalence of these parasites was investigated in four farmed deer species, including 125 reindeer, 109 red deer, 86 sika deer, and 18 Siberian roe deer by nested PCR amplification of the partial small subunit of ribosomal RNA (SSU rRNA) gene. C. ubiquitum isolates were subtyped using nested PCR and sequence analysis of the 60-kDa glycoprotein (gp60) gene. The overall prevalence of Cryptosporidium was 7.1%, with 15.1% for sika deer, 4.0% for reindeer, 4.6% for red deer, and 5.6% for roe deer. C. ubiquitum (n = 4), C. xiaoi (n = 2), and Cryptosporidium deer genotype (n = 18) were identified. All four C. ubiquitum isolates belonged to the XIIa subtype (n = 4). This study confirms that Cryptosporidium deer genotype is widely occurring in deer in the investigated areas. Presence of zoonotic C. ubiquitum XIIa subtype indicates that farmed deer represent potential source of zoonotic cryptosporidia and might pose a threat to human health.
Introduction
Cryptosporidium is an important zoonotic protozoan parasite with a cosmopolitan distribution (1). The transmission routes of Cryptosporidium spp. are thought to result from fecal–oral transmission of oocysts via direct contact with infected humans or animals, or through the ingestion of contaminated water or food (2). However, the contribution of animal reservoirs to human infections remains unclear and requires clarification (3). PCR-based molecular tools for the genetic characterization of Cryptosporidium have enhanced our understanding of Cryptosporidium epidemiology, providing information on the host distribution of various species/genotypes and transmission routes/sources (4).
The genetic heterogeneity of the SSU rRNA gene has revealed the existence of ≥39 recognized species of Cryptosporidium (5, 6). Some of these species have been identified in both humans and animals (particularly farm animals, such as sheep and cattle). Contact with farmed animals is an identified risk factor for human cryptosporidiosis, and many outbreaks have been documented, often involving veterinary students and students at farm schools (7–9). The identification of Cryptosporidium species/genotypes in farmed animals has enhanced our understanding of the transmission of Cryptosporidium.
Commercially farmed deer species vary according to region, but some species such as red deer (Cervus elaphus), sika deer (Cervus nippon), and reindeer (Rangifer tarandus) are farmed across the globe (http://www.fao.org/docrep/004/X6529E/X6529E02.htm). Currently, Cryptosporidium studies in deer have focused on wild or free-ranging species rather than farmed animals (10). Genetic studies of Cryptosporidium from deer showed that eight species (C. parvum, C. hominis, C. ubiquitum, C. muris, C. andersoni, C. occultus, C. bovis, and C. ryanae) and four unnamed Cryptosporidium genotypes (deer genotype, muskrat II genotype, C. hominis-like genotype, and caribou genotype) are prevalent, suggesting that deer infection with Cryptosporidium poses a potential threat to human health (Table 1) (11–25).
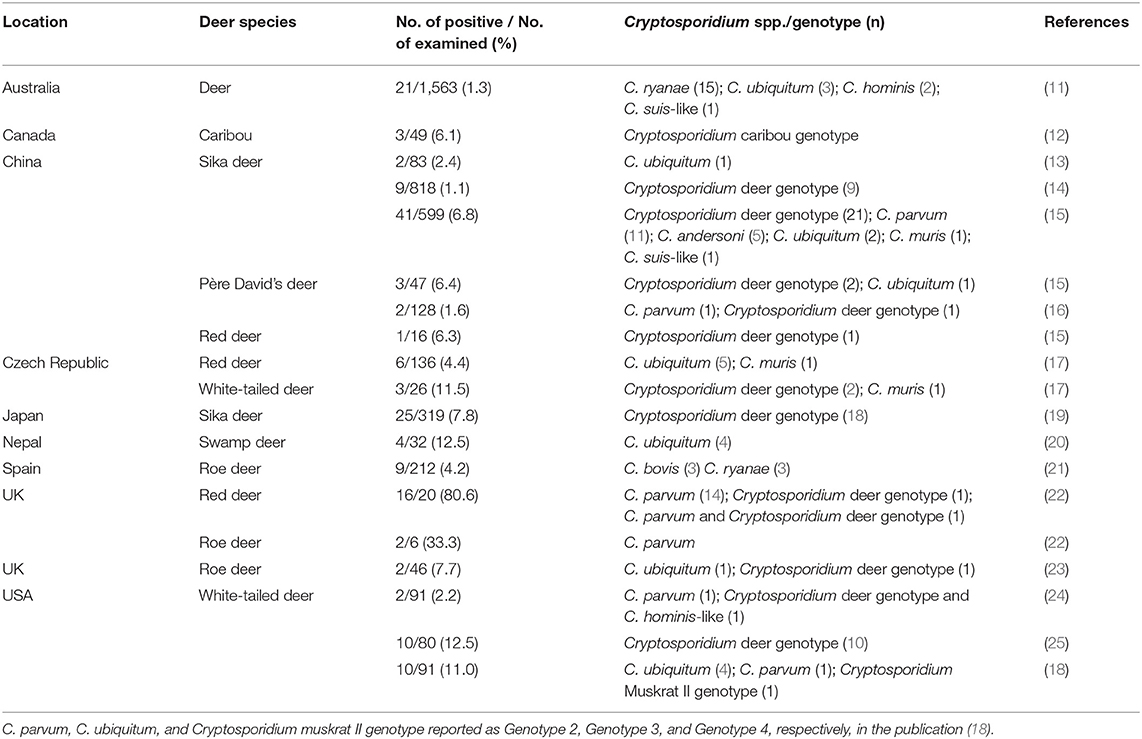
Table 1. Prevalence and distribution of Cryptosporidium species/genotypes in deer according to country.
In China, reindeer, sika deer, red deer, and Siberian roe deer (Capreolus pygargus) are commonly farmed in the northeast of China (14). However, reports on Cryptosporidium infections in these animals are limited (13–16). This study investigated the prevalence and species/genotypes distribution of Cryptosporidium in these four deer species in northeastern China.
Materials and Methods
Collection of Fecal Specimens
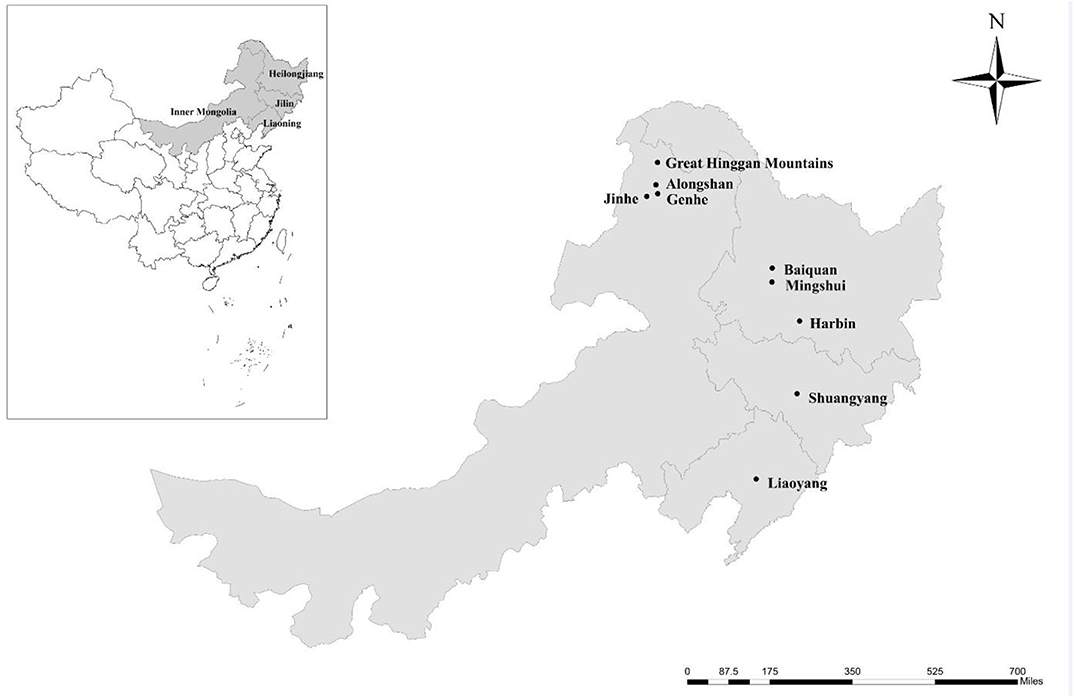
From 1 May 2012 to 31 March 2016, 338 fresh fecal specimens (approximately 10 g) were collected from four farmed deer species, including 125 reindeer, 109 red deer, 86 sika deer, and 18 Siberian roe deer from 10 farms located in nine areas of four provinces in the northeast of China (Table 1 and Figure 1). All fecal specimens were collected from the ground immediately after defecation using sterile disposable latex gloves. To avoid the contamination from the ground, only the parts that do not touch the ground were collected. The number of collected specimens accounted for ~30% of the adult or young deer on each farm. All specimens were transported to the laboratory in coolers with ice packs within 24 h and were stored at 4°C (<24 h). The source, age, and health status (with or without diarrhea) of each deer were recorded during sampling. The ages of the adults ranged from 3 to 5 years, and the ages of the young deer ranged from 1 to 6 months (no animals aged 6 months to 3 years were sampled).
DNA Extraction
Total genomic DNA from each fecal sample (~200 mg) was extracted using a QIAamp DNA Mini Stool Kit (Qiagen, Hilden, Germany) according to the manufacturer's recommendations. Eluted DNA was stored at −20°C prior to PCR analysis. All analysis of fecal DNA extractions was performed in a biosafety level 2 laboratory.
PCR Amplification of Cryptosporidium
Cryptosporidium was detected by nested PCR amplification of the SSU rRNA gene fragment of ~830 bp. Primers and cycle parameters were designed by Xiao and colleagues (26). A fragment of ~948 bp of the 60-kDa glycoprotein (gp60) gene was used to identify C. ubiquitum subtypes via nested PCR amplification using the primers described by Li et al. (27). TaKaRa Taq DNA polymerase (TaKaRa Bio Inc., Tokyo, Japan) was used for all PCRs. PCR amplifications were performed with positive (chicken-derived C. bailey DNA) and negative controls (no DNA water). PCR products were visualized on a UV transilluminator following electrophoresis on 1.5% agarose gels stained with GelStrain (Trans Gen Biotech, Beijing, China).
DNA Sequencing and Analysis
Positive PCR amplicons were transferred to Sangon Biotech Co. Ltd. (Shanghai, China) for sequencing. The accuracy of the sequencing data was confirmed by bi-directional sequencing. Species and genotypes of Cryptosporidium were identified through the comparison of the nucleotide sequences deposited at the National Center for Biotechnology Information (NCBI) using the BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Statistical Analysis
Prevalence was calculated according to deer species and age (young vs. adult deer). Categorical variables were expressed as numbers of cases (percentages), and frequencies were compared using chi-square tests. Statistical significance was established at a p ≤ 0.05. Analyses were performed using SPSS statistical software package version 19.0 (IBM Corporation, Somers, NY, USA).
Results
Prevalence of Cryptosporidium
Cryptosporidium was detected in all four deer species as assessed by nested PCR amplification of the SSU rRNA gene. In total, Cryptosporidium spp. were found in 7.1% (24/338) of deer, with 15.1% (13/86) in sika deer, 4.0% (5/125) in reindeer, 4.6% (5/109) in red deer, and 5.6% (1/18) in roe deer (Table 2). Significant differences in prevalence were observed among species as assessed by chi-square tests (χ2 = 12.8, P = 0.008). Cryptosporidium was identified in three farms for sika deer (7.7–20.1%), two farms for reindeer (1.7 and 7.3%), and two farms for red deer (6.8 and 8.3%).
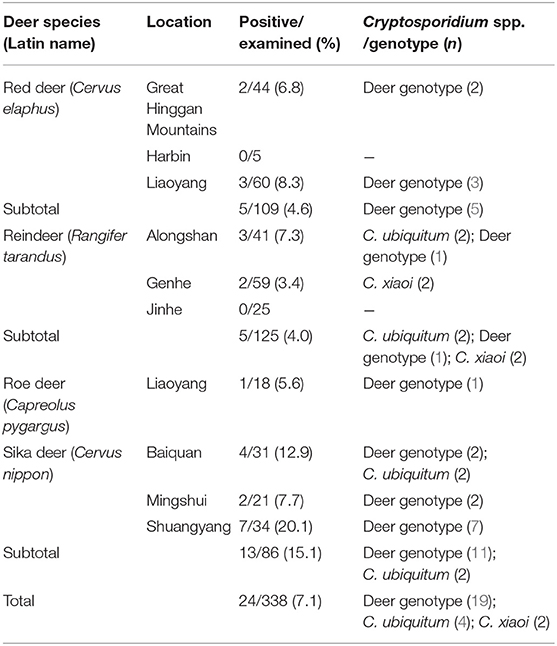
Table 2. Prevalence and species/genotypes of Cryptosporidium in the investigated deer species in China.
The overall prevalence of Cryptosporidium in young deer (17.1%; 13/76) was significantly higher than that in adults (4.1%; 10/244) (χ2 = 14.70, P < 0.05). Statistical differences in prevalence were observed in sika deer between the two age groups (41.2 vs. 8.7%) (χ2 = 8.83, P < 0.05), while no significant differences were observed in reindeer (7.7 vs. 1.2%) (χ2 = 1.89, P > 0.05) and red deer (15.0 vs. 2.2%) (χ2 = 3.50, P > 0.05). No samples from young roe deer were used in this study (Table 3). All deer had no diarrhea at the time of sampling.
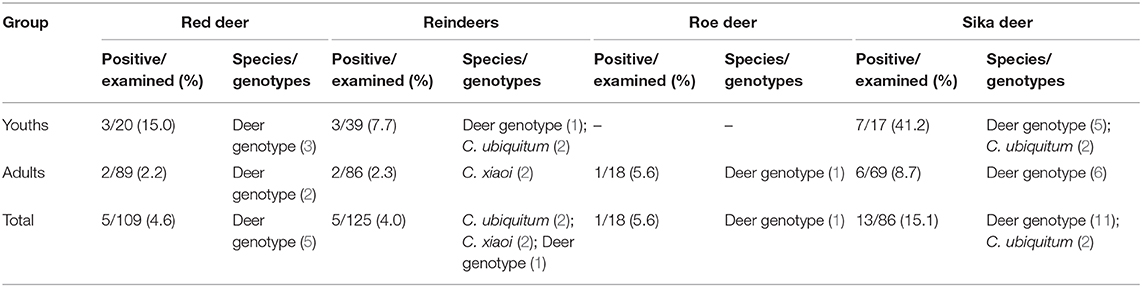
Table 3. Prevalence and Cryptosporidium species/genotypes in the four species of deer according to age.
Genotyping and Subtyping of Cryptosporidium
All 24 Cryptosporidium-positive specimens were successfully sequenced at the SSU rRNA gene. Through sequence analysis, three species/genotypes were identified: C. ubiquitum (n = 4), C. xiaoi (n = 2), and Cryptosporidium deer genotype (n = 18). All 18 sequences of Cryptosporidium deer genotype were identical to each other, showing 100% homology with those from a white-tailed deer in the Czech Republic (KR260681), and a sika deer (KX259127), a David's deer (KX259128), and a red deer (KX259129) in China. The four C. ubiquitum isolates and two C. xiaoi isolates had 100% similarity with those from feral deer in Australia (MG516762) and goats in China (KM199754), respectively. C. ubiquitum isolates were further subtyped through amplification of the gp60 gene. All four C. ubiquitum isolates were successfully amplified and sequenced, and all belonged to the XIIa subtype, sharing 100% homology with previous XIIa subtypes derived from Tibetan sheep in China (KU052815).
Cryptosporidium deer genotype was the dominant genotype (75.0%, 18/24) and had the widest host and geographical distribution, being detected in four deer species and in 6/10 of the investigated areas (Table 1). C. ubiquitum was identified in young sika deer and reindeer (two each), while C. xiaoi was found in two adult reindeer only (Table 3).
Discussion
Epidemiological investigations of Cryptosporidium in deer have been documented in over eight countries since the first detection of Cryptosporidium in deer in Scotland in 1981 (10, 28). However, the number of studies characterizing Cryptosporidium spp. in deer is low (Table 1) (11–25). In this study, the average prevalence of Cryptosporidium in the different deer species was 7.1% (15.1% for sika deer, 4.0% for reindeer, 4.6% for red deer, and 5.6% for roe deer), which was higher than previous studies in China (13–16). Meanwhile, the prevalence of Cryptosporidium was higher in young animals (17.1%) than adults (3.7%), consistent with previous studies (19, 21, 29). The assessment of Cryptosporidium infection in sika deer from Japan showed that fawns had a higher prevalence (16.1%) than yearlings (6.4%) and adults (4.7%) (19). Likewise, studies from Norway demonstrated that 6.2% of roe deer were infected with Cryptosporidium, with prevalence in calves significantly higher than in yearlings and adults (29). Cryptosporidium infection was more prevalent in juvenile deer compared to roe deer in Spain, although the differences were not significant (21). The prevalence of Cryptosporidium in young deer was higher than those of adults, most likely due to the underdeveloped immune systems of the young animals.
In this study, Cryptosporidium deer genotype, C. ubiquitum, and C. xiaoi were identified. Cryptosporidium deer genotype had the highest frequency and widest distribution. A number of lines of evidence supported the observation that deer are the only animal host for Cryptosporidium deer genotype (15). This genotype has been identified in sika deer from China and Japan, white-tailed deer from the Czech Republic and USA, roe deer from the UK, red deer from China and the UK, and Père David's deer from China (13–16, 19, 22, 23).
Cryptosporidium ubiquitum, previously named Cryptosporidium cervine genotype, has a broad host range that includes carnivores, rodents, primates, and domestic and wild ruminants (13, 27). To date, C. ubiquitum has been detected in sika deer in China, white-tailed deer in the USA, swamp deer in Nepal, roe deer in the UK, red deer in the Czech Republic, and deer (sambar deer, red deer, and fallow deer) in Australia (11, 13, 17, 18, 20). Human infections have been documented in the UK, Slovenia, the USA, Canada, Spain, and New Zealand (13, 27). Cryptosporidium ubiquitum is also one of the most common Cryptosporidium species in drinking water in China (30). In this study, four C. ubiquitum isolates were identified as zoonotic XIIa subtype, which were identified in domestic and wild ruminants, rodents, humans, and water samples (5, 30–32). The facts above indicate that the deer infected with this subtype represent infection reservoir and might potentially pose a threat to human health.
Cryptosporidium xiaoi (previously named as C. bovis-like or C. bovis) from sheep was initially identified by Chalmers and colleagues in 2002 and was formally described as a species in 2009, which is genetically distinct but closely related to C. bovis (33, 34). Cryptosporidium xiaoi primarily infected sheep and appeared asymptomatic, but has also been reported in yaks, goats, fish, and kangaroos (35–38). Up to now, only two cases of cryptosporiosis caused by C. xiaoi have been reported in HIV/AIDS patients from Ethiopia (39). Cryptosporidium xiaoi was detected as a dominant species in small ruminants in some other African countries including Egypt and Tanzania, in addition to Asian countries including Bangladesh and China (3, 5, 40, 41). Similarly, C. xiaoi was the major Cryptosporidium species in small ruminants in Europe countries including France, Greece, Poland and Norway, and Australia (40, 42–44). Our findings of C. xiaoi in reindeer indicate that this species might have a more extensive host spectrum than previously expected. The source of C. xiaoi infection and its transmission dynamics require further investigation to elucidate the cross-species transmission potential of C. xiaoi in deer and other animals, including humans in China.
Conclusions
This study demonstrates the prevalence and species/genotypes distribution of Cryptosporidium in four deer species in the northeast of China. Two species (C. ubiquitum and C. xiaoi) and one genotype (Cryptosporidium deer genotype) of Cryptosporidium were identified. Additionally, the zoonotic subtype of C. ubiquitum XIIa subtype was found in the reindeer and sika deer. Presence of the zoonotic subtype C. ubiquitum XIIa in reindeer and sika deer suggests the importance of deer as a potential source of zoonotic cryptosporidia in the environment.
Data Availability Statement
All 24 sequences in the article are 100% homology with some sequences in the GenBank database. These data were included in the article.
Ethics Statement
The study protocol was approved by the Laboratory Animal Welfare and Ethics Committee (LAWEC), National Institute of Parasitic Diseases, Chinese Center for Disease Control and Prevention, China (reference no. 2012-12). Prior to initiating the study, permission was obtained from farm owners or managers. No deer were harmed during specimen collection.
Author Contributions
BZ and YS conceived, designed the experiments, and revised the manuscript. WZ performed the experiments and wrote the paper. WZ, JX, MX, and YJ analyzed the data. JC and HH contributed reagents and material analysis tools. All authors read and approved the final version of the manuscript.
Funding
This work was supported by the National Science and Technology Major Program of China (Nos. 2018ZX107134-404 to BZ and 2018ZX10713001-004 to YS) and the Open Subject of Key Laboratory of Parasite and Vector Biology, MOPH (WSBKFKT-201802 to WZ). The funders had no role in the study design, data collection, data interpretation, or the decision to submit the work for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Checkley W, White AC Jr, Jaganath D, Arrowood MJ, Chalmers RM, Chen XM, et al. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for Cryptosporidium. Lancet Infect Dis. (2015) 15:85–94. doi: 10.1016/S1473-3099(14)70772-8
2. Ryan U, Fayer R, Xiao L. Cryptosporidium species in humans and animals: current understanding and research needs. Parasitology. (2014) 141:1667–85. doi: 10.1017/S0031182014001085
3. Ryan U, Zahedi A, Paparini A. Cryptosporidium in humans and animals-a one health approach to prophylaxis. Parasite Immunol. (2016) 38:535–47. doi: 10.1111/pim.12350
4. Ryan U, Paparini A, Oskam C. New technologies for detection of enteric parasites. Trends Parasitol. (2017) 33:532–46. doi: 10.1016/j.pt.2017.03.005
5. Feng Y, Ryan UM, Xiao L. Genetic diversity and population structure of Cryptosporidium. Trends Parasitol. (2018) 34:997–1011. doi: 10.1016/j.pt.2018.07.009
6. Holubová N, Zikmundová V, Limpouchová Z, Sak B, Konečný R, Hlásková L, et al. Cryptosporidium proventriculi sp. n. (apicomplexa: cryptosporidiidae) in psittaciformes birds. Eur J Protistol. (2019) 69:70–87. doi: 10.1016/j.ejop.2019.03.001
7. Cacci,ò SM, Sannella AR, Mariano V, Valentini S, Berti F, Tosini F, et al. A rare Cryptosporidium parvum genotype associated with infection of lambs and zoonotic transmission in Italy. Vet Parasitol. (2013) 191:128–31. doi: 10.1016/j.vetpar.2012.08.010
8. Chalmers RM, Smith RP, Hadfield SJ, Elwin K, Giles M. Zoonotic linkage and variation in Cryptosporidium parvum from patients in the United Kingdom. Parasitol Res. (2011) 108:1321–5. doi: 10.1007/s00436-010-2199-x
9. Conrad CC, Stanford K, Narvaez-Bravo C, Callaway T, McAllister T. Farm fairs and petting zoos: a review of animal contact as a source of zoonotic enteric disease. Foodborne Pathog Dis. (2017) 14:59–73. doi: 10.1089/fpd.2016.2185
10. Kv,áč M, McEvoy J, Stenger B, Clark M. Cryptosporidiosis in other vertebrates. In: Cacciò SM, Widmer G, editors. Cryptosporidium: Parasite and Disease. Vienna: Springer (2014). p. 237–323. doi: 10.1007/978-3-7091-1562-6_5
11. Koehler AV, Haydon SR, Jex AR, Gasser RB. Cryptosporidium and Giardia taxa in faecal samples from animals in catchments supplying the city of Melbourne with drinking water (2011 to 2015). Parasites Vect. (2016) 9:315. doi: 10.1186/s13071-016-1607-1
12. Siefker C, Rickard LG, Pharr GT, Simmons JS, O'Hara TM. Molecular characterization of Cryptosporidium sp. isolated from northern Alaskan caribou (Rangifer tarandus). J Parasitol. (2002) 88:213–6. doi: 10.1645/0022-3395(2002)088[0213:MCOCSI]2.0.CO;2
13. Wang R, Wang J, Sun M, Dang H, Feng Y, Ning C, et al. Molecular characterization of the Cryptosporidium cervine genotype from a sika deer (Cervus nippon Temminck) in Zhengzhou, China and literature review. Parasitol Res. (2008) 103:865–9. doi: 10.1007/s00436-008-1069-2
14. Tao WF, Ni HB, Du HF, Jiang J, Li J, Qiu HY, et al. Molecular detection of Cryptosporidium and Enterocytozoon bieneusi in dairy calves and sika deer in four provinces in Northern China. Parasitol Res. (2020) 119:105–14. doi: 10.1007/s00436-019-06498-1
15. Huang J, Zhang Z, Zhang Y, Yang Y, Zhao J, Wang R, et al. Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in deer in Henan and Jilin, China. Parasites Vect. (2018) 11:239. doi: 10.1186/s13071-018-2813-9
16. Xie F, Zhang Z, Zhao A, Jing B, Qi M, Wang R. Molecular characterization of Cryptosporidium and Enterocytozoon bieneusi in Père David's deer (Elaphurus davidianus) from Shishou, China. Int J Parasitol Parasites Wildl. (2019) 10:184–7. doi: 10.1016/j.ijppaw.2019.09.001
17. Kotkova M, Nemejc K, Sak B, Hanzal V, Kvetonova D, Hlaskova L, et al. Cryptosporidium ubiquitum, C. muris and Cryptosporidium deer genotype in wild cervids and caprines in the Czech Republic. Folia Parasitologica. (2016) 63:2016. doi: 10.14411/fp.2016.003
18. Perz JF, Le Blancq SM. Cryptosporidium parvum infection involving novel genotypes in wildlife from lower New York State. Appl Environ Microbiol. (2001) 67:1154–62. doi: 10.1128/AEM.67.3.1154-1162.2001
19. Kato S, Yanagawa Y, Matsuyama R, Suzuki M, Sugimoto C. Molecular identification of the Cryptosporidium deer genotype in the Hokkaido sika deer (Cervus nippon yesoensis) in Hokkaido, Japan. Parasitol Res. (2016) 115:1463–71. doi: 10.1007/s00436-015-4880-6
20. Feng Y, Karna SR, Dearen TK, Singh DK, Adhikari LN, Shrestha A, et al. Common occurrence of a unique Cryptosporidium ryanae variant in zebu cattle and water buffaloes in the buffer zone of the Chitwan National Park, Nepal. Vet Parasitol. (2012) 185:309–14. doi: 10.1016/j.vetpar.2011.09.025
21. García-Presedo I, Pedraza-Díaz S, González-Warleta M, Mezo M, Gómez-Bautista M, Ortega-Mora LM, et al. The first report of Cryptosporidium bovis, C. ryanae and Giardia duodenalis sub-assemblage A-II in roe deer (Capreolus capreolus) in Spain. Vet Parasitol. (2013) 197:658–64. doi: 10.1016/j.vetpar.2013.07.002
22. Wells B, Shaw H, Hotchkiss E, Gilray J, Ayton R, Green J, et al. Prevalence, species identification and genotyping Cryptosporidium from livestock and deer in a catchment in the Cairngorms with a history of a contaminated public water supply. Parasites Vect. (2015) 8:66. doi: 10.1186/s13071-015-0684-x
23. Robinson G, Chalmers RM, Stapleton C, Palmer SR, Watkins J, Francis C, et al. A whole water catchment approach to investigating the origin and distribution of Cryptosporidium species. J Appl Microbiol. (2011) 111:717–30. doi: 10.1111/j.1365-2672.2011.05068.x
24. Jellison KL, Lynch AE, Ziemann JM. Source tracking identifies deer and geese as vectors of human-infectious Cryptosporidium genotypes in an urban/suburban watershed. Environ Sci Technol. (2009) 43:4267–72. doi: 10.1021/es900081m
25. Santin M, Fayer R. Enterocytozoon bieneusi, giardia, and Cryptosporidium infecting white-tailed deer. J Eukaryot Microbiol. (2015) 62:34–43. doi: 10.1111/jeu.12155
26. Xiao L, Escalante L, Yang C, Sulaiman I, Escalante AA, Montali RJ, et al. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl Environ Microbiol. (1999) 65:1578–83. doi: 10.1128/AEM.65.4.1578-1583.1999
27. Li N, Xiao L, Alderisio K, Elwin K, Cebelinski E, Chalmers R, et al. Subtyping Cryptosporidium ubiquitum a zoonotic pathogen emerging in humans. Emerg Infect Dis. (2014) 20:217–24. doi: 10.3201/eid2002.121797
28. Tzipori S, Angus KW, Campbell I, Sherwood D. Diarrhea in young red deer associated with infection with Cryptosporidium. J Infect Dis. (1981) 144:170–5. doi: 10.1093/infdis/144.2.170
29. Hamnes IS, Gjerde B, Robertson L, Vikøren T, Handeland K. Prevalence of Cryptosporidium and Giardia in free-ranging wild cervids in Norway. Vet Parasitol. (2006) 141:30–41. doi: 10.1016/j.vetpar.2006.05.004
30. Huang C, Hu Y, Wang L, Wang Y, Li N, Guo Y, et al. Environmental transport of emerging human-pathogenic Cryptosporidium species and subtypes through combined sewer overflow and wastewater. Appl Environ Microbiol. (2017) 83:e00682–17. doi: 10.1128/AEM.00682-17
31. Baroudi D, Hakem A, Adamu H, Amer S, Khelef D, Adjou K, et al. Zoonotic Cryptosporidium species and subtypes in lambs and goat kids in Algeria. Parasites Vect. (2018) 11:582. doi: 10.1186/s13071-018-3172-2
32. Zhao W, Wang J, Ren G, Yang Z, Yang F, Zhang W, et al. Molecular characterizations of Cryptosporidium spp. and Enterocytozoon bieneusi in brown rats (Rattus norvegicus) from Heilongjiang Province, China. Parasites Vect. (2018) 11:313. doi: 10.1186/s13071-018-2892-7
33. Chalmers RM, Elwin K, Reilly WJ, Irvine H, Thomas AL, Hunter PR, et al. Cryptosporidium in farmed animals: the detection of a novel isolate in sheep. Int J Parasitol. (2002) 32:21–6. doi: 10.1016/S0020-7519(01)00309-5
34. Fayer R, Santín M. Cryptosporidium xiaoi n. sp (Apicomplexa: Cryptosporidiidae) in sheep (Ovis aries). Vet Parasitol. (2009) 164:192–200. doi: 10.1016/j.vetpar.2009.05.011
35. Reid A, Lymbery A, Ng J, Tweedle S, Ryan U. Identification of novel and zoonotic Cryptosporidium species in marine fish. Vet Parasitol. (2010) 168:190–5. doi: 10.1016/j.vetpar.2009.11.015
36. Díaz P, Quílez J, Robinson G, Chalmers RM, Díez-Baños P, Morrondo P. Identification of Cryptosporidium xiaoi in diarrhoeic goat kids (Capra hircus) in Spain. Vet Parasitol. (2010) 172:132–4. doi: 10.1016/j.vetpar.2010.04.029
37. Li P, Cai J, Cai M, Wu W, Li C, Lei M, et al. Distribution of Cryptosporidium species in Tibetan sheep and yaks in Qinghai, China. Vet Parasitol. (2016) 215:58–62. doi: 10.1016/j.vetpar.2015.11.009
38. Yang R, Fenwick S, Potter A, Ng J, Ryan U. Identification of novel Cryptosporidium genotypes in kangaroos from Western Australia. Vet Parasitol. (2011) 179:22–7. doi: 10.1016/j.vetpar.2011.02.011
39. Adamu H, Petros B, Zhang G, Kassa H, Amer S, Ye J, et al. Distribution and clinical manifestations of Cryptosporidium species and subtypes in HIV/AIDS patients in Ethiopia. PLoS Negl Trop Dis. (2014) 8:e2831. doi: 10.1371/journal.pntd.0002831
40. Feng Y. Cryptosporidium in wild placental mammals. Exp Parasitol. (2010) 124:128–37. doi: 10.1016/j.exppara.2008.11.005
41. Feng Y, Xiao L. Molecular Epidemiology of cryptosporidiosis in China. Front Microbiol. (2017) 8:1701. doi: 10.3389/fmicb.2017.01701
42. Rieux A, Paraud C, Pors I, Chartier C. Molecular characterization of Cryptosporidium spp. in pre-weaned kids in a dairy goat farm in western France. Vet Parasitol. (2013) 192:268–72. doi: 10.1016/j.vetpar.2012.11.008
43. Robertson LJ, Gjerde BK, Furuseth HE. The zoonotic potential of Giardia and Cryptosporidium in Norwegian sheep: a longitudinal investigation of 6 flocks of lambs. Vet Parasitol. (2010) 171:140–5. doi: 10.1016/j.vetpar.2010.03.014
Keywords: Cryptosporidium, deer, zoonotic, genetic characterization, human
Citation: Zhao W, Xu J, Xiao M, Cao J, Jiang Y, Huang H, Zheng B and Shen Y (2020) Prevalence and Characterization of Cryptosporidium Species and Genotypes in Four Farmed Deer Species in the Northeast of China. Front. Vet. Sci. 7:430. doi: 10.3389/fvets.2020.00430
Received: 14 February 2020; Accepted: 15 June 2020;
Published: 10 August 2020.
Edited by:
David Modrý, University of Veterinary and Pharmaceutical Sciences Brno, CzechiaReviewed by:
Xiangye Liu, Xuzhou Medical University, ChinaXianyong Liu, China Agricultural University, China
Copyright © 2020 Zhao, Xu, Xiao, Cao, Jiang, Huang, Zheng and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yujuan Shen, amyshyj12@163.com; Bin Zheng, zhengbin@nipd.chinacdc.cn
 Wei Zhao
Wei Zhao Jie Xu1,2,3,4,5
Jie Xu1,2,3,4,5  Jianping Cao
Jianping Cao Yanyan Jiang
Yanyan Jiang Huicong Huang
Huicong Huang Yujuan Shen
Yujuan Shen