- 1United States (U.S.) Environmental Protection Agency, Office of Research and Development, Cincinnati, OH, United States
- 2U.S. Centers for Disease Control and Prevention, Atlanta, GA, United States
- 3U.S. Department of Agriculture, Agricultural Research Service (USDA, ARS), Agroecosystem Management Research, Lincoln, NE, United States
- 4USDA ARS, U.S. National Poultry Research Center, Poultry Microbiological Safety and Processing Research Unit, Athens, GA, United States
- 5Center for Food Safety and Applied Nutrition, U.S. Food and Drug Administration, College Park, MD, United States
- 6USDA, ARS, Agricultural Water Efficiency and Salinity Research Unit, Riverside, CA, United States
- 7Oak Ridge Institute for Science and Education, USDA, ARS, Beltsville, MD, United States
- 8Environmental Protection Agency, Office of Water, Washington, DC, United States
- 9Center for Veterinary Medicine, National Antimicrobial Resistance Monitoring System (NARMS), U.S. Food and Drug Administration, Laurel, MD, United States
- 10USDA, ARS Environmental Microbial and Food Safety Laboratory, Beltsville, MD, United States
- 11USDA, ARS, U.S. Meat Animal Research Center, Meat Safety and Quality, Clay Center, NE, United States
- 12USDA, ARS, US Arid-Land Agricultural Research Center, Maricopa, AZ, United States
- 13USDA, ARS Nutrition, Food Safety and Quality National Program Staff, Beltsville, MD, United States
Antimicrobial resistance (AMR) is a world-wide public health threat that is projected to lead to 10 million annual deaths globally by 2050. The AMR public health issue has led to the development of action plans to combat AMR, including improved antimicrobial stewardship, development of new antimicrobials, and advanced monitoring. The National Antimicrobial Resistance Monitoring System (NARMS) led by the United States (U.S) Food and Drug Administration along with the U.S. Centers for Disease Control and U.S. Department of Agriculture has monitored antimicrobial resistant bacteria in retail meats, humans, and food animals since the mid 1990’s. NARMS is currently exploring an integrated One Health monitoring model recognizing that human, animal, plant, and environmental systems are linked to public health. Since 2020, the U.S. Environmental Protection Agency has led an interagency NARMS environmental working group (EWG) to implement a surface water AMR monitoring program (SWAM) at watershed and national scales. The NARMS EWG divided the development of the environmental monitoring effort into five areas: (i) defining objectives and questions, (ii) designing study/sampling design, (iii) selecting AMR indicators, (iv) establishing analytical methods, and (v) developing data management/analytics/metadata plans. For each of these areas, the consensus among the scientific community and literature was reviewed and carefully considered prior to the development of this environmental monitoring program. The data produced from the SWAM effort will help develop robust surface water monitoring programs with the goal of assessing public health risks associated with AMR pathogens in surface water (e.g., recreational water exposures), provide a comprehensive picture of how resistant strains are related spatially and temporally within a watershed, and help assess how anthropogenic drivers and intervention strategies impact the transmission of AMR within human, animal, and environmental systems.
1 Introduction
Antimicrobial drugs have been widely used in human and veterinary medicine and agroecosystems for more than 80 years, with tremendous benefits to human, animal, and plant health. However, the use of antimicrobials also represents an evolutionary selective pressure on microbes (Aminov, 2010), and prolonged use and/or overuse in a particular environment can lead to alterations in the presence of antimicrobial resistant strains within a microbial community (e.g., increases or decreases of resistance naturally found in the population, evolution of new resistance, etc.). Once alterations in resistance have occurred, the genes conferring resistance can spread to other species through horizontal transfer of mobile genetic elements (MGEs) (Baharoglu et al., 2013; Marti et al., 2014), or via clonal spread of bacteria that carry the resistance element (Baker et al., 2017). In addition to antimicrobials, other stressors can mobilize MGEs, such as heavy metals, oxidative stress, and ultraviolet light. This can lead to co-selection of both antimicrobial resistance genes (ARGs) and other stress-response genes (e.g., heavy metal resistance genes) (Poole, 2012; Pal et al., 2015, 2017). Over time, these selective pressures have led to the development of highly resistant human pathogens such as Methicillin-resistant Staphylococcus aureus and extreme drug-resistant tuberculosis, that are difficult to treat (O'Neill, 2016).
As existing antimicrobials become less effective due to the emergence of antimicrobial resistant bacteria (ARB), the risks associated with bacterial infections (e.g., following surgery or chemotherapy) increase. The global burden of antimicrobial resistance (AMR) was estimated at 4.95 million deaths in 2019, with 1.27 million of those deaths directly caused by resistant infections (Murray et al., 2022). It’s predicted that the deaths attributable to AMR infections will increase to 10 million globally by 2050 (O'Neill, 2016). The World Health Organization (WHO) has also identified AMR as one of the leading global health threats (World Health Organization, 2000).
To effectively mitigate the threat of AMR, scientific researchers, health professionals, and government agencies must collaborate in new ways. The concept of One Health has been adopted to address the challenge of AMR given that the same antimicrobials are used in human and animal medicine as well as agriculture, and humans and animals can harbor the same pathogens. The One Health paradigm recognizes that human and animal health are linked to environmental health, and that there is a need to better understand the role of the environment in disease ecology and transmission. The United States (U.S.) Centers for Disease Control and Prevention (CDC) defines One Health as a collaborative, multisectoral, and transdisciplinary approach — working at the local, regional, national, and global levels — with the goal of achieving optimal health outcomes recognizing the interconnection between people, animals, plants, and their shared environment (U.S. Centers for Disease Control and Prevention (CDC), 2023). A One Health approach for AMR recognizes the need for a holistic system to combat antimicrobial resistance that encompasses human, animal, and plant health and the role of the environment in mediating the spread of AMR. This One Health approach also involves the development of collaborative systems for effectively monitoring the emergence and movement of resistance genes and resistant bacteria within and between biological compartments.
An early AMR monitoring effort was established in the U.S. in the mid 1990’s when enrofloxacin was approved for use in poultry. This use of enrofloxacin raised concerns about the transmission of fluoroquinolone resistant bacteria through the food system (Tollefson et al., 1998). As a result, in 1996, the U.S. Food and Drug Administration (FDA), the CDC, and the U.S. Department of Agriculture (USDA) collaborated to establish the National Antimicrobial Resistance Monitoring System (NARMS). NARMS was designed to detect and track AMR in foodborne and other enteric bacteria, like Salmonella, Campylobacter, E. coli, Enterococcus, etc., isolated from human and animal clinical cases, food, and food animal processing environments (U.S. Food and Drug Administration (FDA), 1994a,b, 2000). As the foundational and main system currently used to monitor AMR in the U.S. food system, NARMS provides key data on which research and policy decisions are based.
In 2000, WHO released a report drawing attention to AMR as a global health threat (World Health Organization, 2000). In 2015, WHO adopted the Global Action Plan on AMR, which urged the international community to establish national monitoring systems to assess AMR in bacteria isolated from both humans and animals and underscored the need to adopt a One Health approach (World Health Organization, 2015). Concomitant with the adoption of a One Health approach to mitigating AMR was a growing realization that understanding the ecology, evolution, and epidemiology of AMR and ARB infections requires integrating data from multiple sources and disciplines (National Academies of Sciences (NAS), Engineering, and Medicine, 2017; Topp, 2017; McEwen and Collignon, 2018).
Although the NARMS program has developed data on AMR in human and food-animal systems, information on AMR in the environment (such as surface waterways, soil, or wildlife) (Marti et al., 2014; Barrett and Bouley, 2015) is more limited. Following the 2017 FDA’s Science Board recommendation that NARMS pursue an integrated, One Health approach, a need for baseline data on AMR in the environment was identified. As a result, the establishment of a geographically representative monitoring system for AMR in the environment was added as a goal to the NARMS Strategic Plan: 2021–2025 with the intent of building off of previous work performed by the U.S. Environmental Protection Agency (EPA) analyzing select ARGs in surface waters nationwide (Keely et al., 2022). An environmental working group (EWG) coordinated by the EPA, FDA, CDC, and USDA Agricultural Research Service (ARS) was formed in 2020 to establish an initial environmentally based monitoring system within NARMS.
2 Background on environmental AMR monitoring
2.1 Current status of environmental AMR monitoring
The importance of monitoring AMR in the environment has stimulated multiple discussions and review articles on the best sampling and laboratory methods (e.g., Berendonk et al., 2015; Franklin et al., 2016; Matheu et al., 2017; Larsson et al., 2018; Ben et al., 2019; Huijbers et al., 2019; Diallo et al., 2020; Larsson and Flach, 2021; Pruden et al., 2021; Kaiser et al., 2022; Liguori et al., 2022). These reports outlined key components necessary for environmental AMR monitoring, current knowledge gaps, and limitations of the methods currently used to monitor environmental AMR. A common theme across these reviews is that the ideal environmental AMR monitoring system should be part of a larger effort that also monitors AMR in human and animal populations (i.e., a One Health approach).
A systematic literature review of publications that described AMR monitoring programs across 35 countries found that 65 of the 71 programs monitored AMR in bacteria isolated from humans, while 18 monitored AMR in bacterial isolates from animals and none monitored AMR in bacterial isolates from the environment (Diallo et al., 2020). Similarly, Kaiser et al. (2022) reviewed 25 National Action Plans (NAP) for AMR monitoring and used a One Health lens when analyzing each plan’s priorities. In general, the NAPs did not incorporate environmental monitoring, or only incorporated environmental components when they directly related to human exposures. While most environmental AMR research has been reactive to known environmental contamination, limited proactive strategies for managing ARB in the environment have been identified (Wellcome Trust, 2020; Kaiser et al., 2022). These studies highlight the fact that most existing AMR monitoring systems do not include environmental monitoring even though there is widespread consensus within the scientific community that a One Health approach is the optimal way to monitor AMR. As such, the establishment of environmental AMR monitoring systems, like the NARMS environmental monitoring effort presented herein, represent a key gap and critical need.
Even among existing human and animal AMR monitoring efforts, harmonization of methods and international collaboration is lacking (Diallo et al., 2020; Haenni et al., 2022). The sampling, laboratory methods, and data management approaches employed by different monitoring systems are not harmonized, and as a result the data produced may be difficult to compare. Sample sizes and sampling designs differ between monitoring systems, with some efforts performing selective sampling while others are sub-sampling entire populations (Chau et al., 2022). Laboratory methods similarly vary, including the type of bacterial indicator, antibiotic compounds used for susceptibility testing, and the monitored phenotypes and genotypes. In some instances, different antibiotics are used to define the same phenotype or genotype since the same genetic determinants can provide resistance to multiple antibiotics (Diallo et al., 2020). Even when the same bacterial indicators, antibiotic compounds, and/or phenotypes are monitored, different methods may be used for bacterial isolation and susceptibility testing. Lastly, data and metadata collection and management vary between monitoring efforts. Inconsistencies in the type and method of metadata collected may severely limit the international comparability of data from different monitoring systems, as well as the utility of these data for guiding public health decisions. Overall, monitoring efforts should use sampling and laboratory methods that align and provide comparable data, and, then, the data produced should be collected and managed in a manner that ensures coordination across space, time, and biological compartments, ideally within a standardized framework.
The NARMS EWG divided the development of the environmental monitoring effort into five areas: (i) key objectives and questions, (ii) optimal study/sampling design, (iii) selection of AMR indicators, (iv) selection of methods, and (v) development of data management/analytics/metadata plan. For each of these areas, the consensus among the scientific community and literature was reviewed and carefully considered prior to the development of this environmental monitoring program. The remainder of this paper outlines key aspects for each of these five areas, and then defines the specific implementation of the pilot environmental monitoring effort accordingly.
2.2 Current recommendations for AMR monitoring in the environment
2.2.1 Determine key objectives and questions
When designing a new monitoring system, the objectives and key questions being posed should drive the overall study design. For the development of an environmental AMR monitoring program, the role that the environment plays in AMR-related processes is key for defining these objectives and key questions. The environment can serve two primary roles in AMR-related processes; first, to disseminate already resistant bacteria and genes within and between humans and animals, and second, as a source and facilitator for the evolution of AMR (Bengtsson-Palme et al., 2023). Anthropogenic activities can actively shape and alter environmental resistomes, especially in polluted water bodies. Although evidence is sparse thus far, recent research has found that the environment can be directly attributable to human colonization by resistant bacteria (Leonard et al., 2018, 2022) as well as resistant infections in clinical settings (Stanton et al., 2022). However, the relative contributions of different environmental AMR sources (e.g., untreated human versus animal waste) to infections in humans with immediate epidemiological linkages is still unclear. Similarly, the concentrations and/or mixtures of environmental factors and pollutants (physicochemical, pharmaceutical, heavy metals) that would significantly elevate selective pressures for the maintenance of resistance in the environment is unknown. Furthermore, the levels or concentrations of AMR in surface waters that would pose an increased exposure risk to humans is still an open question (Niegowska et al., 2021).
Aligning the objectives and key questions of an environmental AMR monitoring program with current AMR monitoring programs looking at human, animal, and food systems, like NARMS, is essential for creating a One Health assessment of AMR. For example, the data gathered by NARMS from foodborne and enteric bacteria within humans, animals, and food systems can be used for source attribution of enteric illnesses, investigation of underlying genetic mechanisms of resistance, an early warning system for emerging threats, and ultimately guiding public health efforts in the prevention of resistant infections through the judicious use of antimicrobials (Karp et al., 2017). While the immediate linkages to intervention measures is more difficult within environmental systems, the purpose of environmental AMR monitoring fall into several similar categorical objectives: (i) track the rates of resistance over time in key environments and organisms, (ii) determine the sources and drivers of environmental AMR, (iii) monitor for the evolution of new resistance mechanisms, and (iv) determine the exposure risks posed to humans for colonization/infection in impacted environments. These objectives can be achieved in several different monitoring schemes, and each are dependent upon analytical methodology, budgetary constraints, and scope of the proposed monitoring system.
2.2.2 Sampling design
Once the objectives of the monitoring system are determined, the sampling design can be devised, piloted, and scaled appropriately. For example, if the objective is to characterize the baseline presence of AMR within a certain environment (e.g., river systems, soil) at a large scale (e.g., nation-wide) then a probabilistic sampling design would be adequate since it randomly selects sampling locations to represent the overall population of interest without creating sample biases. On the other hand, if the objective is to identify drivers and areas with significant AMR hazards (e.g., AMR hot spots), then environmental sampling locations need to be selected using prior knowledge of possible AMR point sources that could facilitate transmission of ARB between humans, plants, and animals. The scale of the study may also affect sampling considerations since it is much easier to implement a targeted sampling plan at a regional scale than a general sampling scheme at a national level as it requires in-depth knowledge about local processes and land uses.
A common limitation of sampling schemes designed to monitor environmental AMR is the absence of extensive, high-frequency, time-series datasets, especially in surface waters. These types of sampling designs not only establish baseline data for the examination of environmental AMR but also facilitate the identification of acute drivers of AMR through seasonality, random events, and/or other unknown factors. These timeseries datasets are best applied at critical control points where known anthropogenic inputs introduce genes and bacteria into the system. These control points include domestic and industrial wastewater treatment plants effluents (Pazda et al., 2019), hospital effluents (Paulus et al., 2019), high-density and/or older/failing septic system areas (Junaid et al., 2022), combined sewer overflow and urban runoff outfalls (Almakki et al., 2019), and high-density agricultural areas and CAFO runoff sites (Lopatto et al., 2019). In conjunction, regular monitoring of known exposure sites/routes such as impacted recreational water bodies and any groundwaters (e.g., private well users) that may be affected by these pollution sources, will allow the characterization of infection/colonization risks.
Additionally, the development of a new monitoring program (and/or research study) could be designed along a good, better, best spectrum, as outlined by (Harris et al., 2013). During the initial planning phase of a large-scale project, it may not always be clear what limitations may exist (e.g., funding availability, supply chain issues, laboratory capacity, ethical considerations, etc.). Therefore, identifying the generally accepted good, better, and best practices for each element of the study and sampling design will aid in making final decisions once funding sources, laboratory capacity, availability of supplies, etc. are known.
2.2.3 Selection of AMR indicators
For a One Health environmental monitoring system focused on ARB, selection of AMR indicators (e.g., bacteria, genes, antibiotics, etc.) should be guided by existing recommendations from the WHO and other public health organizations as well as local behaviors (e.g., which antimicrobials are commonly used to treat humans and livestock in the region where the study is being conducted). Indicators should also be selected to facilitate investigations into the transmission of AMR within and between human and animal populations, and the environment to inform possible mitigation strategies. Therefore, overall selection of indicators should be based on relevance for humans, animals, and environment in question, the feasibility of collecting and analyzing samples for that indicator, and sensitivity to change within the prescribed time frame of monitoring. To improve and broaden information about AMR in the environment, baseline lists of ARB and ARG indicators that should be used across AMR monitoring efforts have been suggested (Haenni et al., 2022). For example, a commonly suggested ARB indicator is Escherichia coli (E. coli) resistant to 3rd generation cephalosporins (3GC). The use of 3GC-resistant E. coli as an indicator for environmental AMR monitoring is supported by the WHO extended-spectrum beta-lactamases (ESBL) E. coli “Tricycle protocol” (Anjum et al., 2021; World Health Organization, 2021), and logical given the widespread use of 3GC in human and veterinary medicine (Temkin et al., 2018; U.S. Food and Drug Administration (FDA), 2018; European Medicines Agency (EMA), 2019). In addition, Enterococcus spp. (vancomycin resistant) have been proposed as a convenient gram-positive counterpart to E. coli given their extensive use as a water quality indicator for decades (U.S. Environmental Protection Agency (EPA), 2012; Holcomb and Stewart, 2020; Liguori et al., 2022). Identifying absolute and relative values (i.e., CFU/mL and percentage of resistant colonies) of these ARB provide useful information for assessing human and animal exposure rates to environmental sources of AMR and identifying hotspots in the environment.
Selection of ARGs should include clinically relevant and anthropogenically sensitive genes that commonly occur in freshwater sources and take into consideration factors such as abundance of the gene, propensity for lateral transfer, and ability of ARGs to be expressed in pathogens (Berendonk et al., 2015; Ashbolt et al., 2018; Nnadozie and Odume, 2019; U.S. Centers for Disease Control and Prevention (CDC), 2019; Keenum et al., 2022; Zhang et al., 2022). For example, blaCTX-M and vanA have been recommended as clinically relevant ARGs, since the types of resistance that these ARGs confer to pathogens are noted as “serious” concerns on the CDC threat list (U.S. Centers for Disease Control and Prevention (CDC), 2019). BlaCTX-M, which encodes for ESBL, is responsible for therapeutic problems, and vanA encodes resistance to vancomycin, a last resort antibiotic for treatment of enterococcal infections. Additionally, sulI and tetA are ARGs that tend to be associated with anthropogenic sources with sulI, typically carried by class 1 integrons, conferring resistance to sulfonamides and tetA encoding resistance to tetracyclines, a widely used antibiotic by humans and livestock (Yoshizawa et al., 2020). Besides ARGs, intI1, an integron-integrase, is commonly used as a marker of anthropogenic pressure and/or pollution with higher abundance associated with waste streams and lower in more pristine environments (Gillings et al., 2015; Lucassen et al., 2019; Keely et al., 2022). Its environmental presence, particularly in surface water, is often correlated with the presence of ARGs because integrons are genetic mechanisms that allow bacteria to adapt and evolve rapidly through the stockpiling and expression of new genes (e.g., through site-specific recombination) (Gillings et al., 2015). Coupling the analysis of intI1 with ARGs can provide insights into ARG mobility in environmental systems.
A final set of critical indicators for AMR monitoring efforts in the environment are antimicrobial compounds like antibiotics and other stressors, such as metals and pesticides (Huijbers et al., 2019). Their utility as bioactive compounds are known to create selective pressure for evolution, selection, and maintenance of AMR in bacteria, even at environmentally relevant concentrations (Sandegren, 2014). Antimicrobials in the environment also pose a potential risk to terrestrial and aquatic ecosystem health if they are present at concentrations that alter microbial community function and structure (e.g., nitrification, denitrification, anaerobic ammonium oxidation inhibition). Analysis of antimicrobials in the environment, particularly water, can provide insights into the use of antibiotics in human and animal populations and thereby allow for monitoring of its potential association with observed AMR indicators (Pärnänen et al., 2019). Simultaneous monitoring of antibiotics and AMR is recommended for ensuring continuity and comparability across efforts and maximizing data utility to end-users. Human health, animal health, and environmental health organizations each have developed lists of priority drug indicators to monitor and include fluoroquinolones, sulfonamides, tetracyclines, trimethoprim, and aminoglycosides (World Health Organization, 2018; Gomez Cortes et al., 2020; Haenni et al., 2022). However, most environmental monitoring efforts are not analyzing for antibiotics or other selective agents likely due to the number of antimicrobials that would need to be monitored, lack of technical harmonization and optimization of detection methods, difficulty detecting low levels of antimicrobials in environmental matrices, and/or costs associated with these analyses (Niegowska et al., 2021).
2.2.4 Selection of methods
Once appropriate AMR indicators are selected, analytical methods need to be identified. A combination of culture-based and culture-independent methods provide a comprehensive analysis of AMR in the environment (Franklin et al., 2016, 2021; Niegowska et al., 2021; Pruden et al., 2021). Culturing bacteria and performing standardized in vitro antimicrobial susceptibility testing has been a cornerstone of AMR monitoring since the beginning of the antibiotic era in medicine. This methodology feeds directly into the goals of a One Health approach for AMR by detecting and characterizing ARB that can potentially cause human and animal disease. However, when looking at environmental microbiomes for a comprehensive picture of resistance, this approach is inadequate. Only a small subset of environmental bacteria can be cultured in a laboratory setting, and determination of phenotypic resistance for environmental bacteria is limited by what susceptibility testing can be performed (e.g., availability of validated methods, laboratory capacity, etc.). Furthermore, the diversity of the gene pool for environmental bacteria is much larger compared to bacteria associated with humans or domestic animals, creating a wider array of genetic traits, including novel ARGs (Panthee et al., 2022). The inclusion of molecular analysis of AMR (targeted gene analysis, metagenomics, and whole genome sequencing) can provide information about the entire bacterial population and the environmental resistome of each sample that would otherwise be missed with culture-based analysis alone. A comprehensive molecular method approach can identify and/or quantify known ARGs and MGEs through targeted gene analysis as well as discover emerging or novel forms of resistance with non-targeted techniques like metagenomics and/or whole genome sequencing (Franklin et al., 2021). If monitoring E. coli and Enterococcus bacteria and fecal indicator genes both culture and molecular analysis can also be used to measure fecal contamination, which provides information about the potential for transmission and evolution of AMR (Liguori et al., 2022).
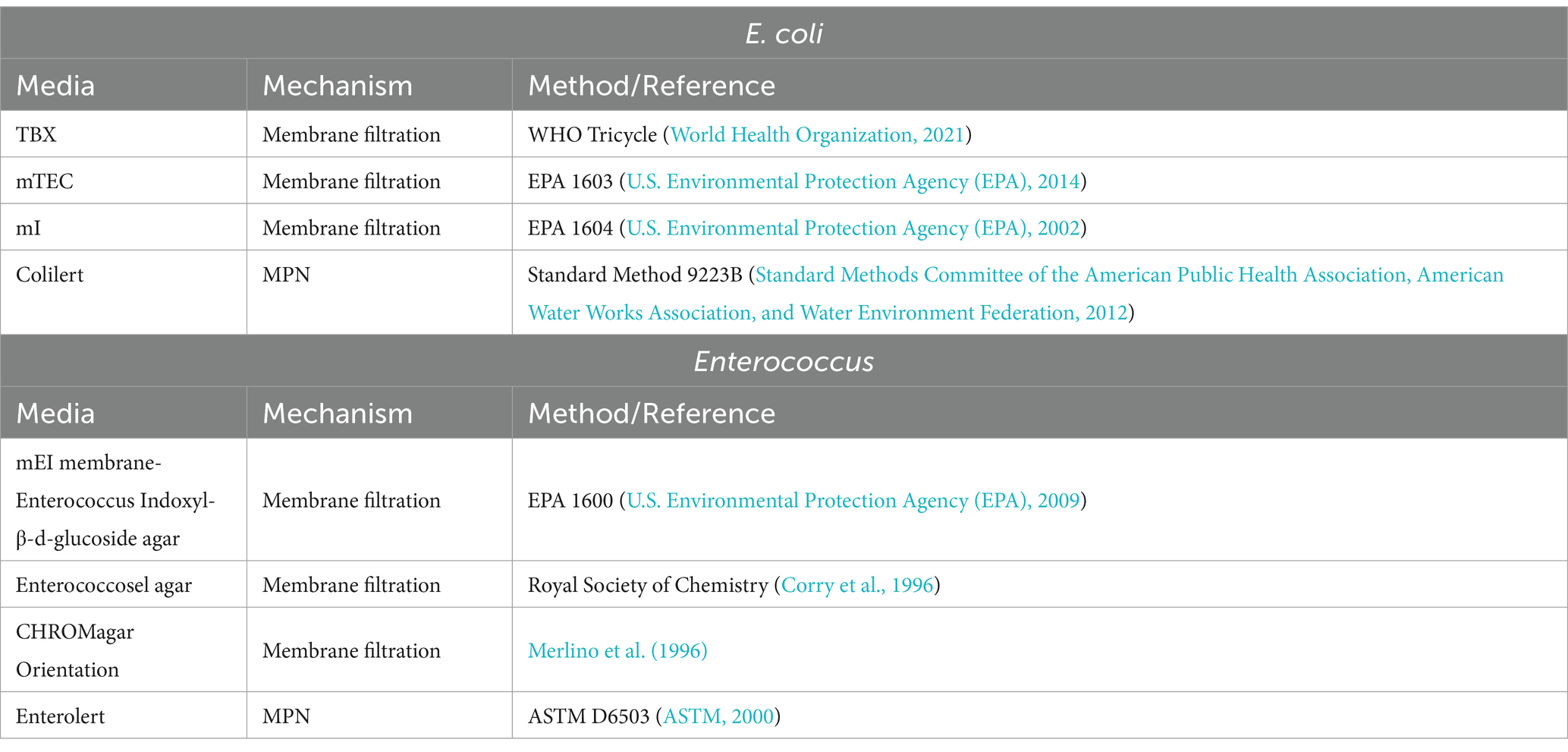
The use of standard methods within and across multiple monitoring efforts is needed to ensure consistency across laboratories (Berendonk et al., 2015; Franklin et al., 2016; Liguori et al., 2022) so that results will be comparable across studies and monitoring efforts. While standard methods are readily available for analysis of AMR in human and animal clinical samples, these methods are not always compatible with the complex matrices of environmental samples. Several recommendations from governmental and non-governmental groups on the best methods to use in detecting certain indicators have been proposed. For example, the WHO is currently recommending the Tricycle protocol for analyzing ESBL E. coli in surface waters, wastewaters, human, and animal samples (World Health Organization, 2021). While a recent U.S. effort funded by the Water Research Foundation has recommended a modified mTEC method (modification of EPA standard method 1603, U.S. Environmental Protection Agency (EPA), 2014; Liguori et al., 2022) and a modified mEI method (modification of EPA standard method 1600, U.S. Environmental Protection Agency (EPA), 2009; Davis et al., 2022) for the analysis of resistant E. coli and Enterococci, respectively, in surface waters, wastewaters, and reused waters.
2.2.5 Development of data management/analytics/metadata plan
Obtaining pertinent key metadata is crucial for interpreting AMR data as well as for use in subsequent models to determine key drivers and risks of AMR in environmental, human, and animal sectors. Metadata is broadly defined as the contextual information about data, but for most biological studies, this refers to basic descriptive information like geographic location, sample type, and sampling date. The type of metadata collected and the method of collection need to be carefully considered when establishing any monitoring effort. The specific metadata that should be collected is dependent on the system being analyzed or monitored. Key metadata categories have been deemed important for environmental efforts, such as climate information, water quality, geographical information, watershed information, and sampling methodologies (Sano et al., 2020). Currently the curation of metadata and knowledge from monitoring systems and published literature is a challenge in the assessment of AMR and the ability to compare across systems (McArthur and Wright, 2015). Therefore, having clear, standardized metadata management, including metadata collection, cleaning, storage, and nomenclature is important for sharing data across studies and time frames.
First and foremost, metadata collection ensures the preservation of contextual information. Careful management and stewardship also ensure accuracy, consistency, privacy/confidentiality concerns, and access to metadata. Indeed, a sampling site’s GPS coordinates are considered critical metadata; if the coordinate reference system for the coordinates is not recorded and linked to the GPS data then those coordinates cannot be reliably used for linking the water quality data with other spatial metadata or when using the GPS coordinates for follow-on meta-analyses. Similarly, slight variations in the way a given parameter is measured by different studies can affect comparability; for instance, data generated by studies that use total suspended solids to track sediment levels are not comparable to data generated by studies that measure turbidity. These considerations are particularly important as there is an increasing interest in reusing data (and associated metadata) for meta-analysis and other research outside the scope for which the data were originally collected. In the context of this evolving interest, it is paramount that metadata collection and management is standardized and harmonized in a way that facilitates re-use and is amenable to the use of machine learning, artificial intelligence, and other big data analytical approaches. This impetus was a driving factor behind the establishment of the FAIR (Findable, Accessible, Interoperable, Reusable) guiding principles (Wilkinson et al., 2016).
Just as research studies can be designed along a good, better, best spectrum (Harris et al., 2013), the same principles can be applied to metadata. A good metadata system would be comprehensive, while a better system would be standardized and contain controlled vocabularies and taxonomies. Controlled vocabularies and taxonomies can be thought of as pick lists of terms that are accepted for a certain variable (Hedden, 2010). The best system would be one that has maximum re-use potential, conveying rich contextual data in a structured, machine-readable format. Ontologies are formal and standardized terms that describe objects or data in a particular setting, similar to controlled vocabularies, and additionally their relationship to each other, in a hierarchical system. Ontologies can also and often do share vocabularies, thereby further connecting and layering contextual information across studies and disciplines. This additional layer, or layers, of information enable even more complex queries of research data.
One approach to managing data is the inclusion of data management or stewardship plans, which are becoming more common and increasingly required by funding agencies. Metadata standards serve an analogous purpose for metadata. These standards, or schema, establish a structured and organized way to manage metadata. A growing list of metadata standard packages and models are available, with some disciplines offering several choices (Yilmaz et al., 2011; Delgado et al., 2018; Harrison et al., 2018).
3 Development of the surface water antimicrobial resistance monitoring system (SWAM)
3.1 SWAM study design: objectives and sampling plan
Surface waters were selected as the preferred matrices to monitor and profile AMR since water creates a conduit for environmental transmission of AMR microbes between humans, animals, and the other environments. The overall objective of the newly designated Surface Water Antimicrobial Resistance Monitoring System (SWAM) was to profile AMR in bacteria from freshwater surface waters (i.e., a watershed) as an initial environmental component within a One Health focused NARMS program. The EWG defined four main primary uses for these data: (i) generate baseline data on AMR in U.S. surface waters, (ii) perform quantitative risk assessment for AMR associated with various water uses (e.g., recreational, drinking, agricultural), (iii) characterize drivers of AMR occurrence and selective pressures that facilitate the emergence, spread, and persistence of AMR, and (iv) identify critical control points for managing AMR hazards in surface water systems.
To coordinate the establishment of a national surface water monitoring system, task-oriented subgroups were formed from the EWG membership to develop study designs, standardized sampling, laboratory and data management decisions and protocols, and data use plans (Figure 1; Table 1). For example, the End Use of the Data Group provided an interface with the NARMS program, which helped resolve issues related to integration with existing NARMS reporting structures and ensured that the data collected met user needs. Specifically, the End Use of the Data Group aimed to answer (i) what are the key insights and outputs desired from SWAM, (ii) how will and could the SWAM data be used to support modeling and quantitative risk assessment, (iii) how do the SWAM data link with data collected by other monitoring programs, such as NARMS and the National Rivers and Streams Assessment (NRSA), an EPA program that monitors water quality.
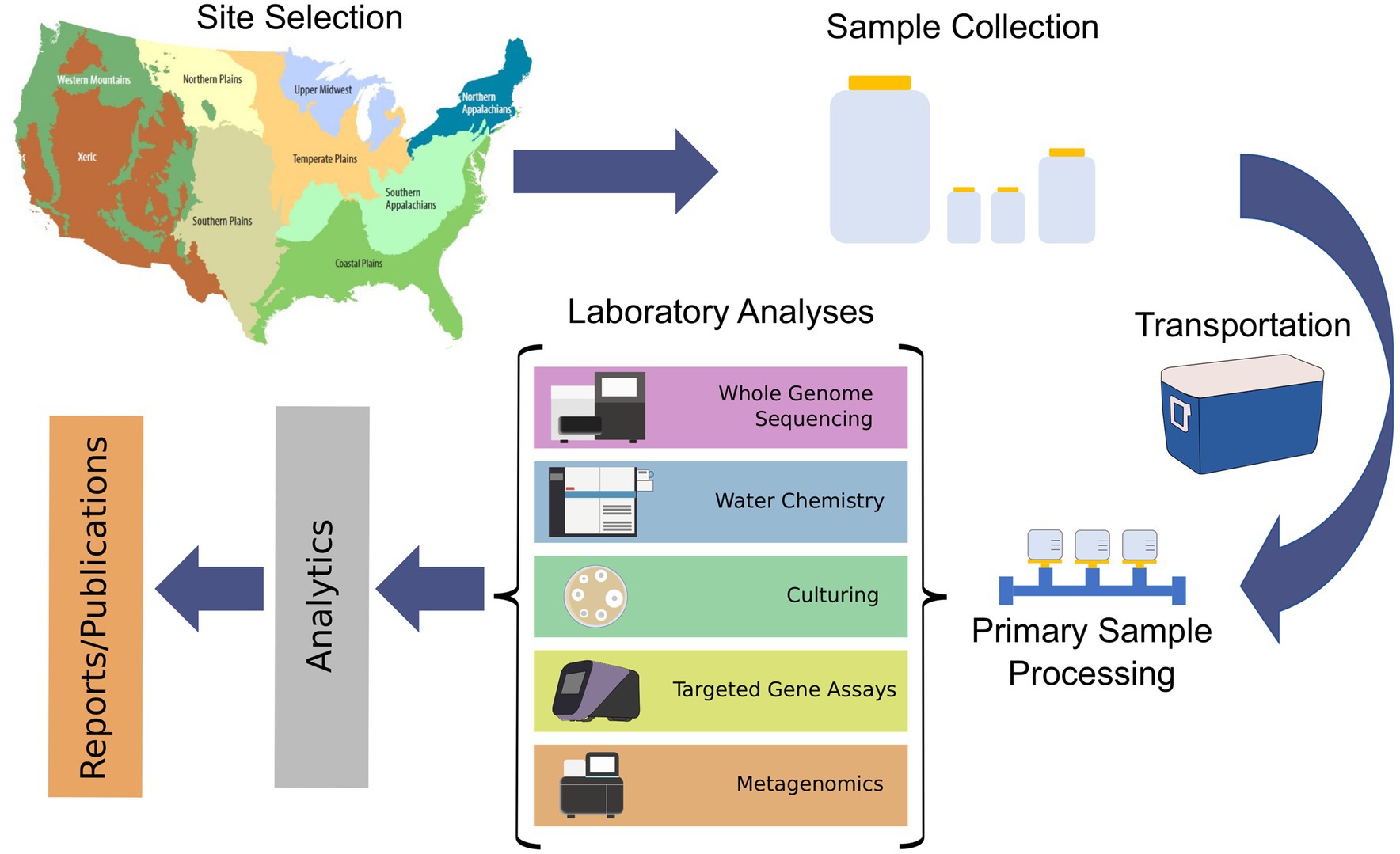
Figure 1. Schematic of an environmental monitoring effort for antimicrobial resistance in the environment.

Table 1. List of subgroups and their respective tasks for developing an environmental monitoring effort for antimicrobial resistance.
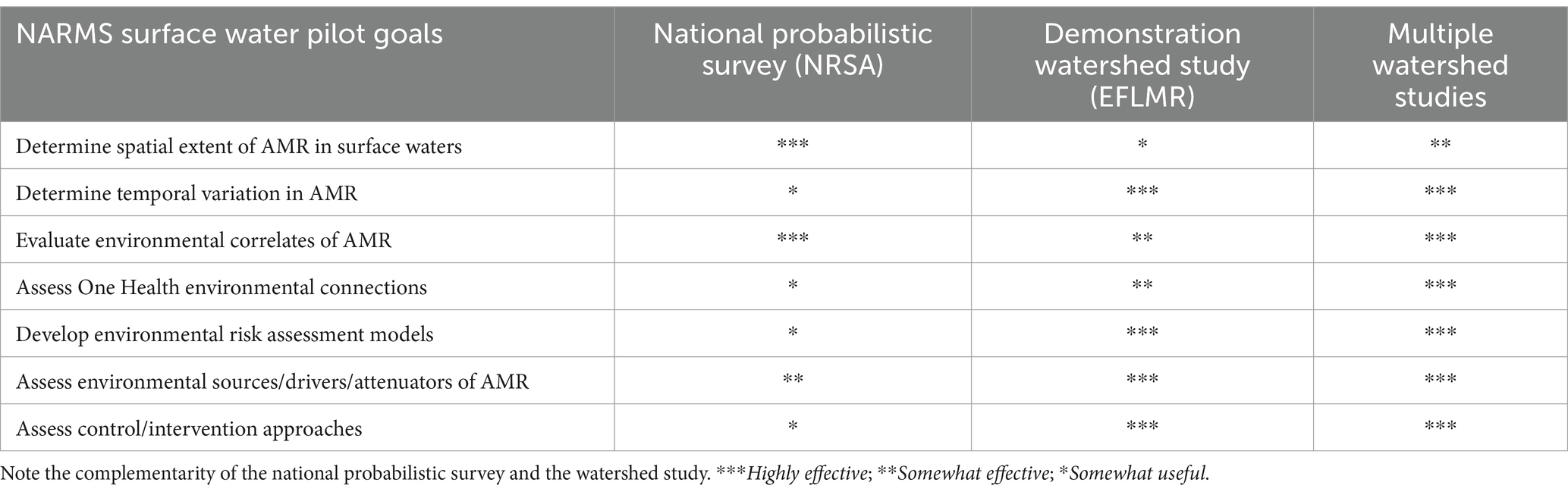
After an initial planning period, the EWG convened a summit so that each subgroup could share their proposals for the respective elements of the new national monitoring system. A specific focus of these proposals was the ability to provide robust data on environmental AMR that aligned with NARMS priorities and data reporting. Overall, this meeting provided an integrated assessment of the system’s scope and needs, including what data and metadata needed to be collected and how this data would be managed and used. Given the large scale of the SWAM effort, a phased approach was adopted for implementing the national monitoring system. The five phases were (i) Method Development Evaluation and In-Lab Validation; (ii) Field Validation of Methods in a Single Watershed Pilot Study; (iii) a Probabilistic National Study; (iv) Finalized National Monitoring Program; and (v) Additional Focused Studies to Address Specific Research Needs (see Table 2 for objectives of each phase).
Multiple sampling designs were evaluated to determine which could best fit the proposed goals of the surface water pilot (see Table 3 for surface water pilot goals). However, no single study could adequately capture the requirements for providing a quantitative assessment of AMR at a national scale while also providing insight into local scale dynamics, including AMR drivers and selection pressures needed to inform risk models and mitigation strategies. To circumvent these problems, a “hybrid” sampling design was selected, entailing both extensive national sampling and intensive watershed scale sampling, which would provide insight on both national trends and watershed scale dynamics. As suggested by World Health Organization (2021) and others, design of both the national-scale and watershed scale components aimed to leverage existing environmental monitoring programs for cost efficiency and to ensure that they provide contextual environmental data. Various national monitoring programs that were explored, which included U.S. Geological Survey’s National Water Quality Assessment (NAWQA; Gilliom et al., 1995), National Science Foundation’s National Ecological Observatory Network (NEON, 2011), USDA’s Conservation Effects Assessment Project (CEAP; Duriancik et al., 2008), EPA’s NRSA (U.S. Environmental Protection Agency (EPA), 2020), and US Army Corps of Engineers’ Water Quality Program for reservoirs (Medina et al., 2019). These programs were evaluated for a variety of factors related to AMR monitoring, including the sample population, the sampling density and frequency, their ability to integrate AMR sampling methods, and associated costs.
The EPA’s NRSA was chosen for the national scale study because it utilizes a spatially stratified probabilistic design with the objective of providing an unbiased population assessment of rivers and streams across the 48 contiguous states and 9 distinct ecoregions. With over 1,800 sites included in the survey, the target sampling locations include a wide range of perennial flowing waters from headwater streams to the largest rivers and catchments in the U.S., representing over 1.2 million river and stream miles. Given the natural variation in biological and chemical water quality indicators across the country, an integral part of the study design is the demarcation of strata (state, ecoregion, and river and stream size) which allows for the identification of least-disturbed reference sites that are regionally relevant (U.S. Environmental Protection Agency (EPA), 2020). These reference sites can then be used to identify drivers of environmental AMR at the national scale and across macroecological boundaries. Of note, while Alaska and Hawaii are not included in the overall study design due to differing climates, shipping limitations, and monetary restrictions, smaller scale projects may be performed in those states.
The East Fork Little Miami River (EFLMR) in southeastern Ohio was selected for the pilot watershed study because an established surface water monitoring study was already in place since 2006 to assess nutrient inputs and management (Peed et al., 2011; Schenck et al., 2015; Scown et al., 2017) and it is within proximity of EPA’s research facility in Cincinnati, OH. The watershed encompasses 1,295 km2 and is primarily agricultural (64%) but grades into suburban and urban areas closer to Cincinnati. Septic systems, many failing, are abundant in rural areas while wastewater treatment plants of varying capacities are situated near smaller population centers (Ohio EPA, 2021). Harsha Lake, an 8 km2 reservoir that includes two recreational beaches and the intake for a drinking water plant, is downstream of many of these effluents. Since any one watershed can only possess a subset of characteristics that are important for characterizing AMR, it is important to build out a series of watershed studies over time to complement the national probabilistic survey. For example, it will be important to capture watersheds with inputs from more concentrated livestock operations and highly urbanized landscapes to build a more complete picture of watershed-scale AMR dynamics. Therefore, a primary objective for the East Fork Little Miami pilot watershed study, apart from understanding of watershed scale AMR dynamics in this system, is to establish measurement protocols, sampling design parameters, and reporting guidelines that will facilitate data aggregation across studies as more watersheds are assessed.
3.2 AMR indicators for SWAM effort
The types of analyses that will be employed for the SWAM effort include a combination of culture-based and molecular-based techniques with indicator selection based on importance and relevance for human, animal, and environmental health. For culture analysis, E. coli, Enterococcus spp., and Salmonella spp. were selected as priority organisms for AMR monitoring in water based on what NARMS already assesses for food, animals, and humans as well as their environmental relevance (Nyirabahizi et al., 2020; Zhao et al., 2020; Yin et al., 2021). E. coli and Enterococcus are recommended fecal indicators for surface waters (U.S. Environmental Protection Agency (EPA), 2009, 2014), as well as sentinel organisms used by NARMS to monitor carriage and emergence of ARGs that could be transferred to both gram-negative and gram-positive pathogens (Ge et al., 2020). Salmonella is an important zoonotic pathogen (Alakomi and Saarela, 2009) that is systematically monitored by NARMS in human clinical isolates, outbreaks, retail meats, and food-producing animals.
Quantitative concentrations of ARB (counts or most probably number (MPN)) were deemed necessary since they add significant value to the analysis of AMR in surface waters for those indicators that are anticipated to be at sufficient density for quantification. Knowing the number of cultivable ARB can be used to: (1) compare magnitudes across sites/studies, (2) determine elevated risk with respect to background levels, (3) quantify risk using QMRA models; and (4) characterize gradients across land use. Therefore, E. coli and Enterococcus analysis will include colony counts and quantification of both total isolates and isolates resistant to select antibiotics (cefotaxime for E. coli and vancomycin for Enterococcus). A subset of resistant isolates will undergo species confirmation and subsequent whole genome sequencing (WGS). Given the variable and typically low numbers of Salmonella found in surface waters, a selective enrichment method will be utilized to determine presence or absence of Salmonella and to obtain isolates in pure culture in the presence of other bacteria. All Salmonella isolates will undergo WGS and, if possible, NARMS standard susceptibility testing. Quantification of antimicrobial susceptible E. coli and Enterococcus and obtaining isolates of Salmonella, E. coli and Enterococcus from surface waters will allow the SWAM effort to fit within the existing NARMS reporting framework as an environmental component moving toward a One Health assessment of AMR.
While culturing select priority organisms fits within the typical NARMS framework, given the complexity and diversity of the environmental microbiome, the inclusion of targeted molecular techniques can be used to provide a more expansive characterization of AMR in surface waters. The molecular methods to analyze environmental AMR will consist of quantification of ARGs, intI1, fecal source indicators, and other related genes and bacterial isolates using quantitative polymerase chain reaction (qPCR)/droplet digital PCR (ddPCR), metagenomics, and WGS. qPCR/ddPCR data will provide a quantitative assessment of ARGs, intI1, fecal indicators, and other genes of interest that are present across a microbial population which can inform models that impart information about AMR trends, hot spots, and/or reservoirs within surface waters. Furthermore, for those research efforts that cannot conduct extensive culture-based approaches, qPCR/ddPCR methods allow for the exploration of relationships between molecular fecal indicators and ARGs within a particular environment/microbial population.
Similarly, metagenomics will help identify types and sources of AMR contamination (animal production, agriculture, health care/human, etc.) by characterizing the resistome of the entire microbial community in surface waters (Mendes et al., 2017; de Abreu et al., 2020; Franklin et al., 2021). Metagenomics is also valuable for possibly selecting additional culture and/or molecular indicators, providing a more robust characterization of baseline contamination levels and differentiating risky ARGs from the background endogenous resistome. WGS together with in silico characterization of ARGs, plasmids, sequence types, and virulence factors can be employed to describe bacterial characteristics with much greater breadth and precision than phenotypic analysis alone (McDermott and Davis, 2021). WGS is also critical for detecting relatedness among isolates from different sampling locations including potential source or exposure areas, and it can be used to associate resistance with virulence and mobility traits to support risk assessment. Together, this array of methodologies will support a robust assessment of AMR dynamics at both the watershed and national scales for risk assessment as well as integration into existing NARMS monitoring programs.
Analysis of antibiotics was also considered as an important element to the evaluation of AMR and possible drivers of AMR in surface waters. The selection of antibiotics to analyze within surface waters should be based on antibiotic usage in humans and animals with a focus on high priority antibiotics like fluoroquinolones, sulfonamides, tetracyclines, trimethoprim, and aminoglycosides. While beta-lactams and the bacteria resistant to them (e.g., ESBL E. coli) are of highest priority and deemed critically important in human medicine, these antibiotic compounds are highly unstable in the environment and rarely found intact, especially in surface waters, due to the beta-lactam ring that can be opened by beta-lactamases (enzymes carried by certain bacteria) and/or by chemical hydrolysis (Christian et al., 2003; Huijbers et al., 2019). Even though the importance of analyzing for antibiotics was highlighted and discussed during the development of the SWAM effort, it was not included in the final designs of the watershed and national scale studies due to various reasons (e.g., cost and manpower constraints, concerns of what antibiotics to select, etc.), but may be revisited later.
3.3 Analytical method selection for SWAM AMR indicators
For method development and evaluation, utilization of standard methods when possible was deemed a high priority to ensure comparability of this effort with similar environmental monitoring efforts (World Health Organization, 2021; Liguori et al., 2022). Since various sampling and laboratory methods are used by different researchers, the SWAM environmental working group aimed to determine optimal method(s) for AMR analysis in surface waters that will provide comparative data across studies. As a result, standard methods were compared with those methods commonly used for analysis of AMR in surface waters, with final selection of methods based on their adaptability within the requirements and limitations of the SWAM effort as well as how well they aligned with similar water monitoring projects to create consistency across efforts. Any modifications to these methods occurred because they were deemed beneficial and/or necessary to support study objectives.
Development of culture methods included evaluation of methods for the quantification of total and resistant E. coli and Enterococcus spp. and isolation of Salmonella spp. The culture methods that were considered for E. coli and Enterococcus spp. consisted of those commonly used and recommended for the quantification and isolation of these bacteria in surface waters, including standard methods recommended by WHO, EPA, and ASTM International (Table 4) with EPA 1603 and EPA 1600 selected for E. coli and Enterococcus spp., respectively. These methods were modified to perform susceptibility testing with cefotaxime for E. coli and vancomycin for Enterococcus spp. Method evaluation for Salmonella included considerations of different water volumes and comparisons of filtration and/or concentration techniques to optimize the recovery of low and sporadic levels of these bacteria in surface waters (Sharma et al., 2020; Kraft et al., 2023). Additionally, different selective enrichments, agars, and identification methods for Salmonella isolates (culture recovery versus rapid screening) were compared. The Salmonella method selected for this effort was based off the modified Standard Method 9260.B2, which has been used extensively to analyze surface waters in the southeastern U.S. (Meinersmann et al., 2008; Cho et al., 2022; Kraft et al., 2022). This method involves filtration utilizing perlite (in place of diatomaceous earth) to capture the bacterial cells, a general enrichment to revive injured cells, selective enrichments, and plating on selective media (Figure 2).
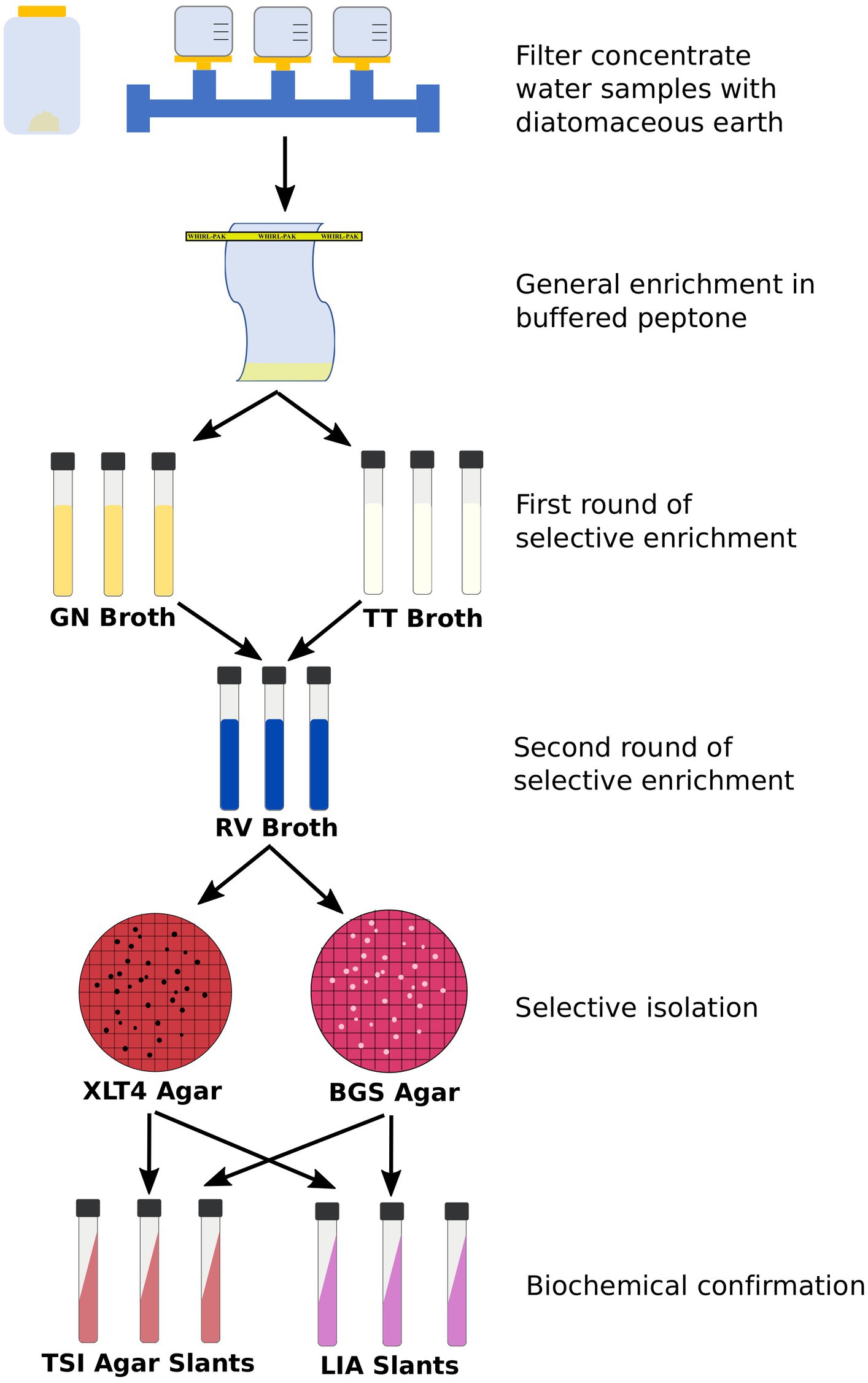
Figure 2. General flow chart of Salmonella enrichment and isolation procedure. Protocols for the filtration via the Modified Standard Method 9260.B2 and selective enrichment can be found at: dx.doi.org/10.17504/protocols.io.rm7vzy72xlx1/v2 and dx.doi.org/10.17504/protocols.io.kxygxz5q4v8j/v1, respectively. GN, Gram negative; TT, Tetrathionate; RV, Rappaport Vassiliadis; XLT4, Xylose lysine tergitol 4; BGS, Brilliant Green Sulfa; TSI, Triple Sugar Iron; LIA, Lysine Iron Agar.
Method development for molecular techniques included comparisons of different water volumes, filtration techniques, DNA extraction kits, whole cell standards, and DNA standards. Having sufficient volumes of water and a DNA extraction kit that provided adequate amounts of high-quality DNA was deemed a high priority for the success of subsequent molecular work. Due to the lack of standardization for molecular techniques, including qPCR, ddPCR, WGS, and metagenomics, this work focused on having quality control measures at each step of sample processing to account for any processing variability. QA/QC guidelines will follow the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines (Bustin et al., 2009).
Although recommendations and guidelines for WGS and shotgun metagenomic data are currently limited for environmental studies, factors that are important across all next generation sequencing (NGS) approaches include data quality metrics such as average Q scores, sequence complexity distributions, contamination, number of ambiguous bases, sequence length, coverage and N50s for assembly. A minimum coverage, ranging from 30X for Salmonella to 40X for E. coli, will be targeted for all WGS experiments (Timme et al., 2020). For metagenomic studies, ‘coverage’ is a far more complicated subject because hundreds or thousands of distinct genomes may be present in any particular sample. Recommendations for depth of sequencing will vary by matrix and the overall aim of the study (Rodriguez-R and Konstantinidis, 2014). For taxonomic composition and AMR gene profiling, work has shown that the required depth of sequencing varies significantly by matrix (Gweon et al., 2019). The complexity and diversity of microbiomes in a sample, the interest to characterize the less abundant organisms, and sequencing cost were considered in deciding the depth of sequencing for the shotgun metagenomic sequencing. A shotgun metagenomics approach will be used to characterize the microbiome and to index the full complement of environmental AMR genes in the surface water samples. In addition to shotgun metagenomic sequencing, sequencing of culture enrichments from surface waters, known as quasimetagenomics, will be performed to characterize ARGs present in less abundant organisms. Preliminary studies by this NARMS surface water sampling initiative (Ottesen et al., 2022; Kocurek et al., 2024) have demonstrated that quasimetagenomic data could identify as many as 30% of critically important AMR genes (Table 5) from surface water samples while metagenomic data without enrichment only detected 1% of these AMR genes in the same samples at the same sequencing depth.
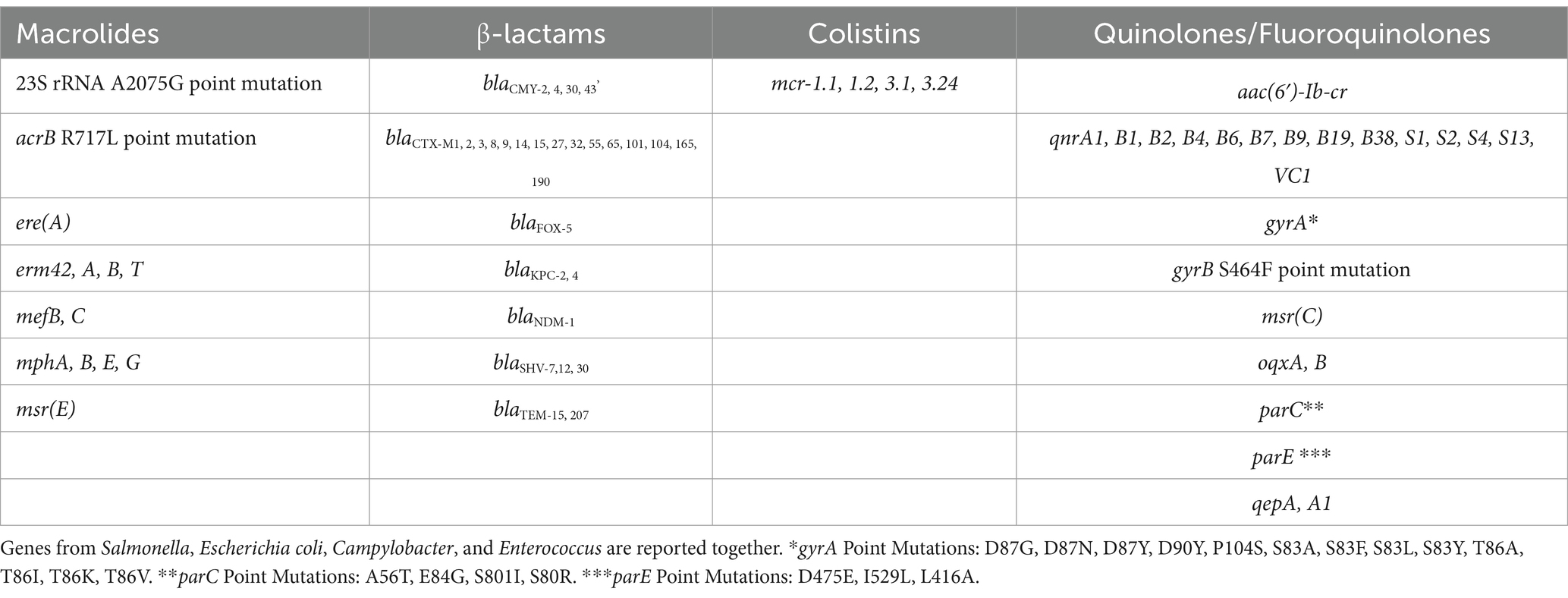
Table 5. List of genes encoding resistance to critically important antimicrobial agents identified by the National Antimicrobial Resistance Monitoring System (NARMS).
All sequencing reads will go through quality control steps to remove adaptors, low quality and complexity sequences using Trimmomatic (Bolger et al., 2014) prior to analysis. A combination of read-based/assembly-free and assembly-based approaches will be used for taxonomic and resistome profiling. For screening environmental metagenomes for ARGs, standalone databases containing functionally verified genes, such as CARD, NCBI’s AMRFinderPlus, and ResFinder (Feldgarden et al., 2019; Bortolaia et al., 2020; Feldgarden et al., 2021; Alcock et al., 2023) and predictive models, like DeepARG (Arango-Argoty et al., 2018) were examined for maximum coordination of gene nomenclature. Overall, the success of the molecular analysis is dependent on important considerations, like consistent quality control measures, metadata, data storage and sharing, as well as coordination of results from PCR, metagenomic, and WGS data.
3.4 Data management for SWAM effort
Given the large scope of the SWAM effort, the planning and management of metadata needed to be carefully considered. To identify and guide metadata needs, a conceptual schematic of the project scope was developed (Figure 1). The project was divided into the following categories based on setting and activity: Sample site, in situ measurements, weather and climate, sample collection, sample transportation, primary sample processing, culturing, metagenomics, targeted gene assays, water chemistry, and isolate WGS. This categorized approach was helpful as it segregated the development of the metadata standard into manageable sections.
Since this environmental study will include metagenomic and microbiome sequence data, the MIxS metadata standard, which is implemented by NCBI, will be used to facilitate ease of data submission. The MIxS, or Minimum Information about any (x) Sequence, standard is a metadata framework established and maintained by the Genomic Standards Consortium (Yilmaz et al., 2011). MIxS provides a standardized format for annotation of sample attributes through a series of environmental packages, including core terms as well as setting-specific checklists. One of the main points of emphasis within the MIxS framework is the re-use of existing terms from other environmental packages, when appropriate, to promote interoperability as well as to minimize metadata term maintenance efforts. Therefore, current MIxS environmental packages were examined to identify terms that could be reused for the metadata standard associated with this study, and currently includes 24 reused MIxS terms. A draft metadata sheet is presented in the Appendix 1.
To maximize the impact of the contextual information contained in this research study, ontological terms and definitions were utilized whenever possible. The current metadata standard draft includes 12 ontological terms. These terms include geographic location descriptors from Gazetteer ontology (GAZ), general biological and microbiological terms from the National Cancer Institute Thesaurus (NCIT), and phenotypic and microbiological terms from the Ontology of Prokaryotic Phenotypic and Metabolic Characters (MICRO) to name a few. The Ontology Lookup Service, maintained by EMBL-EBI and the Open Biological and Biomedical Ontology (OBO) Foundry were invaluable in finding existing ontological terms to define and describe certain attributes in the metadata standard.
4 Discussion
4.1 Next steps
With the preliminary planning, decision-making, method development, and method evaluation for the SWAM effort complete, the next steps for this effort are completion of the yearlong watershed scale study and the national scale study that will span a two-year time frame. During the East Fork Watershed study, thirty-five sites throughout the watershed will be sampled every three weeks with a few locations upstream and downstream of point sources being sampled weekly. This study will not only provide an opportunity to test the culture and molecular methods with a variety of sampling locations during base flow and various weather conditions (rain events, snow, snow melt, etc.) but will also provide information about the temporal variation of AMR, assist in assessing possible drivers of AMR, inform exposure risk assessment, and/or identify critical control points at a watershed scale.
The national-scale study for the SWAM effort will utilize the U.S. EPA NRSA survey that will be executed in 2023–2024. This national scale assessment of rivers and streams occurs every five years over a two-year time frame (sampling during May – September) and includes approximately 2,000 sites (about 1,000 sites per year). Sites are sampled only during base flow conditions, and most sites are only visited once, except for 10% that are revisited as a quality control measure. Since the NRSA survey collects a wide variety of water quality indicators to assess the ecological condition of surface waters nation-wide, this national-scale study will build off the trends previously identified by Keely et al. (2022) and provide additional information about the spatial variation of AMR across the nation as well as how water quality parameters may correlate with AMR indicators.
4.2 Future directions for SWAM
Once the watershed scale and national scale studies are completed, the SWAM effort will have generated a library of isolates (Salmonella, Enterococcus, and E. coli) that will be compared and cross-referenced with the NARMS isolate libraries to explore interconnections between human, animal, and environmental compartments at local, regional, and national scales. Assessments of what was successful and/or feasible during the watershed- and national-scale studies will guide the development of the national environmental monitoring program as well as recommendations for how to perform additional watershed-scale studies. Other needs or questions that remain to be addressed can be added during subsequent watershed-based and national-scale studies. Having validated standard frameworks for environmental monitoring of AMR will facilitate data aggregation across these studies as additional watershed- and national-scale studies are performed.
The SWAM effort will be a significant step forward for environmental monitoring and the assessment of AMR from a One Health perspective, allowing direct comparison of surface water isolates and metagenomes with existing NARMS isolate libraries. This effort will produce standard measurement protocols, sampling design parameters, and reporting guidelines for monitoring AMR in surface waters at both the watershed and national scale. The protocols from this effort could also be utilized by other researchers in their own surface water studies (e.g., additional watershed scale studies), which can then be integrated into larger assessments/meta-analyses to address deeper questions about AMR dynamics. Overall, the unique data set on surface waters produced by this effort will provide a One Health assessment of AMR to support the NARMS monitoring program and create a framework for environmental monitoring programs at national and international scales.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
AF: Validation, Investigation, Writing – review & editing, Writing – original draft, Supervision, Methodology, Conceptualization. DW: Writing – review & editing, Writing – original draft, Conceptualization. LD: Methodology, Writing – review & editing, Writing – original draft, Conceptualization. MB: Writing – review & editing, Writing – original draft, Conceptualization. BD: Writing – review & editing, Visualization. JF: Writing – original draft, Conceptualization. CG: Writing – review & editing, Writing – original draft, Data curation, Conceptualization. AI: Writing – review & editing, Methodology, Conceptualization. MJ: Writing – original draft, Conceptualization. SK: Writing – original draft, Methodology, Conceptualization. AK: Writing – original draft, Methodology, Conceptualization. BM: Writing – original draft, Methodology, Conceptualization. RM: Writing – original draft, Conceptualization. AO: Writing – review & editing, Writing – original draft, Methodology, Conceptualization. MS: Writing – original draft, Methodology, Conceptualization. ES: Writing – original draft, Conceptualization. DT: Writing – review & editing, Writing – original draft, Methodology, Conceptualization. HT: Writing – original draft, Conceptualization. JW: Writing – original draft, Methodology, Conceptualization. CW: Writing – original draft, Methodology, Conceptualization. KC: Writing – review & editing, Project administration, Conceptualization. CK: Writing – review & editing, Writing – original draft, Project administration, Conceptualization. PM: Writing – review & editing, Writing – original draft, Project administration, Conceptualization. JG: Writing – review & editing, Project administration, Conceptualization.
Environmental Working Group
Mark Bagley (EPA), Kathe Bjork (USDA), Laura Boczek (EPA), Nichole Brinkman (EPA), Kim Cook (USDA), Benjamin Davis (EPA), Lisa Durso (USDA), Jonathan Frye (USDA), Jay Garland (EPA), Susan Gerber (CDC), Christopher Grim (FDA), Jason Folster (CDC), Alison Franklin (EPA), Heather Harbottle (FDA), Vincent Hill (CDC), Andrew Huang (CDC), Wesley Hunter (FDA), Mark Ibekwe (USDA), Michael A. Jahne (EPA), Claudia Kabera (FDA), Scott Keely (EPA), Amy Kirby (CDC), David Knaebel (USDA), Autumn Kraft (USDA), Roy Martin (EPA), Cita Medalla (CDC), Betty McConn (USDA), Patrick F. McDermott (FDA), Richard Mitchell (EPA), Shatavia Morrison (CDC), Roxann Motroni (USDA), Chris Nietch (EPA), Epiphanie Nyirabahizi (FDA), Andrea Ottesen (FDA), Padmini Ramachandran (FDA), Manan Sharma (USDA), Errol Strain (FDA), Daniel Tadesse (FDA), Heather Tate (FDA), Ruth Timme (FDA), Hatti Webb (CDC), Daniel Weller (CDC), Jim Wells (USDA), Clinton Williams (USDA), Yu Ye (FDA), Jie Zheng (FDA).
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Author disclaimer
U.S. Environmental Protection Agency: The United States Environmental Protection Agency through its Office of Research and Development funded and managed the research described here. It has been subjected to Agency’s administrative review and approved for publication. The views expressed in this article are those of the author(s) and do not necessarily represent the views or policies of the U.S. Environmental Protection Agency. Mention of trade names or commercial products does not constitute endorsement or recommendation for use. U.S. Food & Drug Administration: The views expressed in this presentation are those of the author and do not necessarily reflect the official policy of the Department of Health and Human Services, the U.S. Food and Drug Administration, or the U.S. Government. U.S. Centers for Disease Control and Prevention: The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention. U.S. Department of Agriculture – Agricultural Research Service: Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frwa.2024.1359109/full#supplementary-material
References
Alakomi, H.-L., and Saarela, M. (2009). Salmonella importance and current status of detection and surveillance methods. Qual. Assur. Safety Crops Food. 1, 142–152. doi: 10.1111/j.1757-837X.2009.00032.x
Alcock, B. P., Huynh, W., Chalil, R., Smith, K. W., Raphenya, A. R., Wlodarski, M. A., et al. (2023). CARD 2023: expanded curation, support for machine learning, and resistome prediction at the comprehensive antibiotic resistance database. Nucleic Acids Res. 51, D690–D699. doi: 10.1093/nar/gkac920
Almakki, A., Jumas-Bilak, E., Marchandin, H., and Licznar-Fajardo, P. (2019). Antibiotic resistance in urban runoff. Sci. Total Environ. 667, 64–76. doi: 10.1016/j.scitotenv.2019.02.183
Aminov, R. I. (2010). A brief history of the antibiotic era: lessons learned and challenges for the future. Front. Microbiol. 1:134. doi: 10.3389/fmicb.2010.00134
Anjum, M. F., Schmitt, H., Borjesson, S., and Berendonk, T. U. (2021). The potential of using E. coli as an indicator for the surveillance of antimicrobial resistance (AMR) in the environment. Curr. Opin. Micro. 64, 152–158. doi: 10.1016/j.mib.2021.09.011
Arango-Argoty, G., Garner, E., Pruden, A., Heath, L. S., Vikesland, P., and Zhang, L. (2018). Deep ARG: a deep learning approach for predicting antibiotic resistance genes from metagenomic data. Microbiome 6, 1–15. doi: 10.1186/s40168-018-0401-z
Ashbolt, N. J., Pruden, A., Miller, J. H., Riquelme, M. V., and Maile-Moskowitz, A. (2018). “Antimicrobial resistance: fecal sanitation strategies for combatting a global public health threat” in Global water pathogens project (part 3 Bacteria) (Lansing, MI: UNESCO, Michigan State University)
Baharoglu, Z., Garriss, G., and Mazel, D. (2013). Multiple pathways of genome plasticity leading to development of antibiotic resistance. Antibiotics 2, 288–315. doi: 10.3390/antibiotics2020288
Baker, S., Thomson, N., Weill, F.-X., and Holt, K. E. (2017). Genomic insights into the emergence and spread of antimicrobial-resistant pathogens. Science 360, 733–738. doi: 10.1126/science.aar3777
Barrett, M. A., and Bouley, T. A. (2015). Need for enhanced environmental representation in the implementation of one health. EcoHealth 12, 212–219. doi: 10.1007/s10393-014-0964-5
Ben, Y., Fu, C., Hu, M., Liu, L., Wong, M. H., and Zheng, C. (2019). Human health risk assessment of antibiotic resistance associated with antibiotic residues in the environment: a review. Environ. Res. 169, 483–493. doi: 10.1016/j.envres.2018.11.040
Bengtsson-Palme, J., Abramova, A., Berendonk, T. U., Coelho, L. P., Forslund, S. K., Gschwind, R., et al. (2023). Towards monitoring of antimicrobial resistance in the environment: for what reasons, how to implement it, and what are the data needs? Environ. Int. 178:108089. doi: 10.1016/j.envint.2023.108089
Berendonk, T. U., Manaia, C. M., Merlin, C., Fatta-Kassinos, D., Cytryn, E., Walsh, F., et al. (2015). 2015 tackling antibiotic resistance: the environmental framework. Nat. Rev. Microbiol. 13, 310–317. doi: 10.1038/nrmicro3439
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bortolaia, V., Kaas, R. S., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., et al. (2020). Res finder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75, 3491–3500. doi: 10.1093/jac/dkaa345
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Chau, K. K., Barker, L., Budgell, E. P., Vihta, K. D., Sims, N., Kasprzyk-Hordern, B., et al. (2022). Systematic review of wastewater surveillance of antimicrobial resistance in human populations. Environ. Int. 162:107171. doi: 10.1016/j.envint.2022.107171
Cho, S., Hiott, L. M., House, S. L., Woodley, T. A., Mcmillan, E. A., Sharma, P., et al. (2022). Analysis of Salmonella enterica isolated from a mixed-use watershed in Georgia, USA: antimicrobial resistance, serotype diversity, and genetic relatedness to human isolates. Appl. Environ. Microbiol. 88:e0039322. doi: 10.1128/aem.00393-22
Christian, T., Schneider, R. J., Färber, H. A., Skutlarek, D., Meyer, M. T., and Goldbach, H. E. (2003). Determination of antibiotic residues in manure, soil, and surface waters. Acta Hydrochim. Hydrobiol. 31, 36–44. doi: 10.1002/aheh.200390014
Corry, J. E. L., Curtis, G. D. W., and Baird, R. M. (1996). Handbook of culture Media for Food and Water Microbiology, 3rd Edn. London: Royal Society of Chemistry.
Davis, B. C., Keenum, I., Calarco, J., Liguori, K., Milligan, E., Pruden, A., et al. (2022). Towards the standardization of Enterococcus culture methods for waterborne antibiotic resistance monitoring: a critical review of trends across studies. Water Research X 17:100161. doi: 10.1016/j.wroa.2022.100161
de Abreu, V. A. C., Perdigao, J., and Almeida, S. (2020). Metagenomic approaches to analyze antimicrobial resistance: an overview. Front. Genet. 11:575592. doi: 10.3389/fgene.2020.575592
Delgado, J., Naro, D., Llorente, S., Gelpi, J. L., and Royo, R. (2018). Metadata to describe genomic information. A: "building continents of knowledge in oceans of data: The future of co-created eHealth". Amsterdam: IOS Press, p. 621–625.
Diallo, O. O., Baron, S. A., Abat, C., Colson, P., Chaudet, H., and Rolain, J. M. (2020). Antibiotic resistance surveillance systems: a review. J. Glob. Antimicrob. Resist. 23, 430–438. doi: 10.1016/j.jgar.2020.10.009
Duriancik, L. F., Bucks, D., Dobrowolski, J. P., Tom Drewes, S., Eckles, D., Jolley, L., et al. (2008). The first five years of the conservation effects assessment project. J. Soil Water Conserv. 63, 185A–197A. doi: 10.2489/jswc.63.6.185A
European Medicines Agency (EMA). (2019). European surveillance of veterinary antimicrobial consumption. Sales of veterinary antimicrobial agents in 31 European countries in 2017. European Medicines Agency: Amsterdam.
Feldgarden, M., Brover, V., Gonzalez-Escalona, N., Frye, J. G., Haendiges, J., Haft, D. H., et al. (2021). AMRFinderPlus and the reference gene catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep. 11:12728. doi: 10.1038/s41598-021-91456-0
Feldgarden, M., Brover, V., Haft, D. H., Prasad, A. B., Slotta, D. J., Tolstoy, I., et al. (2019). Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob. Agents Chemother. 64, e00483–e00419. doi: 10.1128/AAC.00483-19
Franklin, A. M., Aga, D. S., Cytryn, E., Durso, L. M., McLain, J. E., Pruden, A., et al. (2016). Antibiotics in agroecosystems: introduction to the special section. J. Environ. Qual. 45, 377–393. doi: 10.2134/jeq2016.01.0023
Franklin, A. M., Brinkman, N. E., Jahne, M. A., and Keely, S. P. (2021). Twenty-first century molecular methods for analyzing antimicrobial resistance in surface waters to support one health assessments. J. Micro Methods 184:106174. doi: 10.1016/j.mimet.2021.106174
Ge, B., Domesle, K. J., Gaines, S. A., Lam, C., Bodeis Jones, S. M., Yang, Q., et al. (2020). Prevalence and antimicrobial susceptibility of indicator organisms Escherichia coli and Enterococcus spp. isolated from U.S. animal food, 2005-2011. Microorganisms 8:1048. doi: 10.3390/microorganisms8071048
Gillings, M., Gaze, W., Pruden, A., Smalla, K., Tiedje, J. M., Zhu, Y.-G., et al. (2015). Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. ISME J. 9, 1269–1279. doi: 10.1038/ismej.2014.226
Gilliom, R. J., Alley, W. M., and Gurtz, M. E. (1995). Design of the National Water-Quality Assessment Program; occurrence and distribution of water-quality conditions. USGS Circular 112:1112. doi: 10.3133/cir1112
Gomez Cortes, L., Marinov, D., Sanseverino, I., Navarro Cuenca, A., Niegowska, M., Porcel Roderiguez, E., et al. (2020). Selection of substances for the 3rd watch list under the water framework directive publications. Office of the European Union: Luxembourg.
Gweon, H. S., Shaw, L. P., Swann, J., De Maio, N., AbuOun, M., Niehus, R., et al. (2019). The impact of sequencing depth on the inferred taxonomic composition and AMR gene content of metagenomic samples. Environ. Microb. 14:7. doi: 10.1186/s40793-019-0347-1
Haenni, M., Dagot, C., Chesneau, O., Bibbal, D., Labanowski, J., Vialette, M., et al. (2022). Environmental contamination in a high-income country (France) by antibiotics, antibiotic-resistant bacteria, and antibiotic resistance genes: status and possible causes. Environ. Int. 159, 107047–104120. doi: 10.1016/j.envint.2021.107047
Harris, L. J., Berry, E. D., Blessington, T., Erickson, M., Jay-Russell, M., Jiang, X., et al. (2013). A framework for developing research protocols for evaluation of microbial hazards and controls during production that pertain to the application of untreated soil amendments of animal origin on land used to grow produce that may be consumed raw. J. Food Prot. 76, 1062–1084. doi: 10.4315/0362-028X.JFP-13-007
Harrison, X. A., Donaldson, L., Correa-Cano, M. E., Evans, J., Fisher, D. N., Goodwin, C. E. D., et al. (2018). A brief introduction to mixed effects modelling and multi-model inference in ecology. Peer J. 6:e4794. doi: 10.7717/peerj.4794
Hedden, H. (2010). Taxonomies and controlled vocabularies best practices for metadata. J. Digit. Asset Manag. 6, 279–284. doi: 10.1057/dam.2010.29
Holcomb, D. A., and Stewart, J. R. (2020). Microbial indicators of fecal pollution: recent progress and challenges in assessing water quality. Curr. Environ. Health Rep. 7, 311–324. doi: 10.1007/s40572-020-00278-1
Huijbers, P. M. C., Flach, C.-F., and Larsson, D. G. J. (2019). A conceptual framework for the environmental surveillance of antibiotics and antibiotic resistance. Environ. Int. 130:104880. doi: 10.1016/j.envint.2019.05.074
Junaid, M., Zainab, S. M., Xu, N., Sadaf, M., Malik, R. N., and Wang, J. (2022). Antibiotics and antibiotic resistant genes in urban aquifers. Curr. Opin. Environ. Sci. Health 26:100324. doi: 10.1016/j.coesh.2021.100324
Kaiser, R. A., Taking, L., and Bhatia, H. (2022). Antimicrobial resistance and environmental health: a water stewardship framework for global and national action. Antibiotics 11:63. doi: 10.3390/antibiotics11010063
Karp, B. E., Tate, H., Plumblee, J. R., Dessai, U., Whichard, J. M., Thacker, E. L., et al. (2017). National antimicrobial resistance monitoring system: two decades of advancing public health through integrated surveillance of antimicrobial resistance. Foodborne Pathog. Dis. 14, 545–557. doi: 10.1089/fpd.2017.2283
Keely, S. P., Brinkman, N. E., Wheaton, E. A., Jahne, M. A., Siefring, S. D., Varma, M., et al. (2022). Geospatial patterns of antimicrobial resistance genes in the US EPA National Rivers and streams assessment survey. Environ. Sci. Technol. 56, 14960–14971. doi: 10.1021/acs.est.2c00813
Keenum, I., Liguori, K., Calarco, J., Davis, B. C., Milligan, E., Harwood, V. J., et al. (2022). A framework for standardized qPCR-targets and protocols for quantifying antibiotic resistance in surface water, recycled water and wastewater. Crit. Rev. Environ. Sci. Technol. 52, 4395–4419. doi: 10.1080/10643389.2021.2024739
Kocurek, B., Behling, S., Martin, G., Ramachandran, P., Reed, E., Grim, C., et al. (2024). Metagenomic survey of antimicrobial resistance (AMR) in Maryland surface waters differentiated by high and low human impact. Microbiol. Resour. Announc. 13:e0047723. doi: 10.1128/MRA.00477-23
Kraft, A. L., Sharma, M., Frye, J. G., and Wells, J. E. (2022). Protocol for modified standard method 9260.B2 for the isolation of Salmonella from surface water V.2. Washington, DC: USDA Agricultural Research Service.
Kraft, A. L., Wells, J. E., Frye, J. G., Ibekwe, A. M., Durso, L. M., Hiott, L., et al. (2023). A comparison of methods to detect low levels of Salmonella enterica in surface waters to support antimicrobial resistance surveillance efforts performed in multiple laboratories. Sci. Total Environ. 905:167189. doi: 10.1016/j.scitotenv.2023.167189
Larsson, D. G. J., Andremont, A., Bengtsson-Palme, J., Brandt, K. K., de Roda Husman, A. M., Fagerstedt, P., et al. (2018). Critical knowledge gaps and research needs related to the environmental dimensions of antibiotic resistance. Environ. Int. 117, 132–138. doi: 10.1016/j.envint.2018.04.041
Larsson, D. G. J., and Flach, C.-F. (2021). Antibiotic resistance in the environment. Nat. Rev. Microbiol. 20, 257–269. doi: 10.1038/s41579-021-00649-x
Leonard, A. F., Morris, D., Schmitt, H., and Gaze, W. H. (2022). Natural recreational waters and the risk that exposure to antibiotic resistant bacteria poses to human health. Curr. Opin. Microbiol. 65, 40–46. doi: 10.1016/j.mib.2021.10.004
Leonard, A. F., Zhang, L., Balfour, A. J., Garside, R., Hawkey, P. M., Murray, A. K., et al. (2018). Exposure to and colonisation by antibiotic-resistant E. coli in UK coastal water users: environmental surveillance, exposure assessment, and epidemiological study (beach bum survey). Environ. Int. 114, 326–333. doi: 10.1016/j.envint.2017.11.003
Liguori, K., Keenum, I., Davis, B. C., Calarco, J., Milligan, E., Harwood, V. J., et al. (2022). Antimicrobial resistance monitoring of water environments: a framework for standardized methods and quality control. ES&T 56, 9149–9160. doi: 10.1021/acs.est.1c08918
Lopatto, E., Choi, J., Colina, A., Ma, L., Howe, A., and Hinsa-Leasure, S. (2019). Characterizing the soil microbiome and quantifying antibiotic resistance gene dynamics in agricultural soil following swine CAFO manure application. PLoS One 14:e0220770. doi: 10.1371/journal.pone.0220770
Lucassen, R., Rehberg, L., Heyden, M., and Bockmühl, D. (2019). Strong correlation of total phenotypic resistance of samples from household environments and the prevalence of class 1 integrons suggests for the use of the relative prevalence of int I1 as a screening tool for multi-resistance. PLoS One 14:e0218277. doi: 10.1371/journal.pone.0218277
Marti, E., Variatza, E., and Balcazar, J. L. (2014). The role of aquatic ecosystems as reservoirs of antibiotic resistance. Trends Microbiol. 22, 36–41. doi: 10.1016/j.tim.2013.11.001
Matheu, J., Aidara-Kane, A., and Andremont, A. (2017). The ESBL tricycle AMR surveillance project: A simple, one health approach to global surveillance. Global Health Dynamics, Suffolk, United Kingdom. Available at: http://resistancecontrol.info/2017/the-esbl-tricycle-amr-surveillance-project-a-simple-one-health-approach-to-global-surveillance/ (Accessed August 31, 2023).
McArthur, A. G., and Wright, G. D. (2015). Bioinformatics of antimicrobial resistance in the age of molecular epidemiology. Curr. Opin. Micro. 27, 45–50. doi: 10.1016/j.mib.2015.07.004
McDermott, P. F., and Davis, J. J. (2021). Predicting antimicrobial susceptibility from the bacterial genome: a new paradigm for one health resistance monitoring. J. Vet. Pharmacol. Ther. 44, 223–237. doi: 10.1111/jvp.12913
McEwen, S. A., and Collignon, P. J. (2018). Antimicrobial resistance: a one health perspective. Microbiol. Spectr. 6:25. doi: 10.1128/9781555819804.ch25
Medina, V., Butler, A., Emery, E., and Clyde, G. (2019). Evaluation of water quality/environmental management surveys on USACE managed reservoirs and waterways evaluation of water quality/environmental management surveys on USACE managed reservoirs and waterways. Defense Technical Information Center: Fort Belvoir, VI.
Meinersmann, R. J., Berrang, M. E., Jackson, C. R., Fedorka-Cray, P., Ladely, S., Little, E., et al. (2008). Salmonella, Campylobacter and Enterococcus spp.: their antimicrobial resistance profiles and their spatial relationships in a synoptic study of the upper Oconee River basin. Microb. Ecol. 55, 444–452. doi: 10.1007/s00248-007-9290-6
Mendes, L. W., Braga, L. P. P., Navarrete, A. A., Souza, D. G., Silva, G. G. Z., and Tsai, S. M. (2017). Using metagenomics to connect microbial community biodiversity and functions. Curr. Issues Mol. Biol. 24, 103–118. doi: 10.21775/cimb.024.103
Merlino, J., Siarakas, S., Robertson, G. J., Funnell, G. R., Gottlieb, T., and Bradbury, R. (1996). Evaluation of CHROMagar orientation for differentiation of GRAM-negative bacilli and Enterococcus species. J. Clin. Microviol. 34, 1788–1793. doi: 10.1128/JCM.34.7.1788-1793.1996
Murray, C. J., Ikuta, K. S., Sharara, F., Swetschinski, L., Aguilar, G. R., Gray, A., et al. (2022). Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655. doi: 10.1016/S0140-6736(21)02724-0
National Academies of Sciences (NAS), Engineering, and Medicine. (2017). Combating antimicrobial resistance: A one health approach to a global threat: Proceedings of a workshop.
NEON. (2011). 2011 science strategy: Enabling continental-scale ecological forecasting. Available at: https://www.neonscience.org/sites/default/files/NEON_Strategy_2011u2_0.pdf (Accessed June 5, 2023)
Niegowska, M., Sanseverino, I., Navarro, A., and Lettieri, T. (2021). Knowledge gaps in the assessment of antimicrobial resistance in surface waters. FEMS Micro. Ecol. 97:viab 140. doi: 10.1093/femsec/fiab140
Nnadozie, C. F., and Odume, O. N. (2019). Freshwater environments as reservoirs of antibiotic resistant Bacteria and their role in the dissemination of antibiotic resistance genes. Environ. Pollut. 254:113067. doi: 10.1016/j.envpol.2019.113067
Nyirabahizi, E., Gregory, G., Dessai, U., Zhao, S., Kabera, C., Crarey, E., et al. (2020). Evaluation of Escherichia coli as an indicator for antimicrobial resistance in Salmonella recovered from the same food or animal ceca samples. Food Control 115:107280. doi: 10.1016/j.foodcont.2020.107280
Ohio EPA. (2021). Ohio Environmental Protection Agency – division of surface water. Loading analysis plan and supporting data acquisition needed for the East fork Little Miami River basin: Total maximum daily load development. Available at: https://epa.ohio.gov/static/Portals/35/tmdl/LAPs/Little%20Miami/EFLMR_LAP.pdf (Accessed May 7, 2023)
O'Neill, J. (2016). Tackling drug-resistant infections globally: Final report and recommendations the review on antimicrobial resistance. 80 pp. Available at: https://wellcomecollection.org/works/thvwsuba (Accessed June 15, 2023)
Ottesen, A., Kocurek, B., Ramachandran, P., Reed, E., Commichaux, S., Engelbach, G., et al. (2022). Advancing antimicrobial resistance monitoring in surface waters with metagenomic and quasimetagenomic methods. PLOS Water 1:e0000067. doi: 10.1371/journal.pwat.0000067
Pal, C., Asiani, K., Arya, S., Rensing, C., Stekel, D. J., Larsson, D. G. K., et al. (2017). Metal resistance and its association with antibiotic resistance. Adv. Microb. Physiol. 70, 261–313. doi: 10.1016/bs.ampbs.2017.02.001
Pal, C., Bengtsson-Palme, J., Kristiansson, E., and Larsson, D. G. (2015). Co-occurrence of resistance genes to antibiotics, biocides, and metals reveals novel insights into their co-selection potential. BMC Genomics 16:964. doi: 10.1186/s12864-015-2153-5
Panthee, B., Gyawali, S., Panthee, P., and Techato, K. (2022). Environmental and human microbiome for health. Life (Basel) 12:456. doi: 10.3390/life12030456
Pärnänen, K. M., Narciso-da-Rocha, C., Kneis, D., Berendonk, T. U., Cacace, D., Do, T. T., et al. (2019). Antibiotic resistance in European wastewater treatment plants mirrors the pattern of clinical antibiotic resistance prevalence. Sci. Adv. 5:eaau9124. doi: 10.1126/sciadv.aau9124
Paulus, G. K., Hornstra, L. M., Alygizakis, N., Slobodnik, J., Thomaidis, N., and Medema, G. (2019). The impact of on-site hospital wastewater treatment on the downstream communal wastewater system in terms of antibiotics and antibiotic resistance genes. Int. J. Hyg. Environ. Health 222, 635–644. doi: 10.1016/j.ijheh.2019.01.004
Pazda, M., Kumirska, J., Stepnowski, P., and Mulkiewicz, E. (2019). Antibiotic resistance genes identified in wastewater treatment plant systems–a review. Sci. Total Environ. 697:134023. doi: 10.1016/j.scitotenv.2019.134023
Peed, L. A., Nietch, C. T., Kelty, C. A., Meckes, M., Mooney, T., Sivaganesan, M., et al. (2011). Combining land use information and small stream sampling with PCR-based methods for better characterization of diffuse sources of human fecal pollution. Environ. Sci. Technol. 45, 5652–5659. doi: 10.1021/es2003167
Poole, K. (2012). Bacterial stress response as determinants of antimicrobial resistance. J. Antimicrob. Chemo. 67, 2069–2089. doi: 10.1093/jac/dks196
Pruden, A., Vikesland, P. J., Davis, B. C., and deRoda Husman, A. M. (2021). Seizing the moment: now is the time for integrated global surveillance of antimicrobial resistance in wastewater environments. Curr. Opin. Micro. 64, 91–99. doi: 10.1016/j.mib.2021.09.013
Rodriguez-R, L., and Konstantinidis, K. (2014). Estimating coverage in metagenomic data sets and why it matters. ISME J. 8, 2349–2351. doi: 10.1038/ismej.2014.76
Sandegren, L. (2014). Selection of antibiotic resistance at very low antibiotic concentrations. Ups. J. Med. Sci. 119, 103–107. doi: 10.3109/03009734.2014.904457
Sano, D., Wester, A. L., Schmitt, H., Amarasiri, M., Kirby, A., Medlicott, K., et al. (2020). Updated research agenda for water, sanitation, and antimicrobial resistance. J. Water Health 18, 858–866. doi: 10.2166/wh.2020.033
Schenck, K., Rosenblum, L., Ramakrishnan, B., Carson, J. Jr., Macke, D., and Nietch, C. (2015). Correlation of trace contaminants to wastewater management practices in small watersheds. Environ. Sci.: Processes Impacts 17, 956–964. doi: 10.1039/C4EM00583J
Scown, M. W., McManus, M. G., Carson, J. H. Jr., and Nietch, C. T. (2017). Improving predictive medels of in-stream phosphorus concentration based on nationally-available spatial data coverages. JAWRA 53, 944–960. doi: 10.1111/1752-1688.12543
Sharma, M., Handy, E. T., East, C. L., Kim, S., Jiang, C., Callahan, M. T., et al. (2020). Prevalence of Salmonella and Listeria monocytogenes in non-traditional irrigation waters in the mid-Atlantic United States is affected by water type, season, and recovery method. PLoS One 15:e0229365. doi: 10.1371/journal.pone.0229365
Standard Methods Committee of the American Public Health Association, American Water Works Association, and Water Environment Federation (2012). “Enzyme Substrate Coliform Test: 9223B Enzyme Substrate Test” in Standard Methods For the Examination of Water and Wastewater. eds. W. C. Lipps, T. E. Baxter, and E. Braun-Howland (Washington DC: APHA Press)
Stanton, I. C., Bethel, A., Leonard, A. F. C., Gaze, W. H., and Garside, R. (2022). Existing evidence on antibiotic resistance exposure and transmission to humans from the environment: a systematic map. Environ. Evid. 11:8. doi: 10.1186/s13750-022-00262-2
Temkin, E., Fallach, N., Almagor, J., Gladstone, B. P., Taconelli, E., and Carmeli, Y. (2018). Estimating the number of infections caused by antibiotic-resistant Escherichia coli and Klebsiella pneumoniae in 2014: a modelling study. Lancet Glob. Health 6, e969–e979. doi: 10.1016/S2214-109X(18)30278-X
Timme, R. E., Wolfgang, W. J., Balkey, M., Venkata, S. L. G., Randolph, R., Allard, M., et al. (2020). Optimizing open data to support one health: best practices to ensure interoperability of genomic data from bacterial pathogens. One Health Outlook 2, 20–11. doi: 10.1186/s42522-020-00026-3
Tollefson, L., Angulo, F. J., and Fedorka-Cray, P. J. (1998). National surveillance for antibiotic resistance in zoonotic enteric pathogens. Vet. Clin. N. Am. Food Anim. Pract. 14, 141–150. doi: 10.1016/s0749-0720(15)30285-1
Topp, E. (2017). Antimicrobial resistance and one health: Agriculture and Agri-Food Canada’s research program on antimicrobial resistance. Can. Commun. Dis. Rep. 43, 224–227. doi: 10.14745/ccdr.v43i11a03
U.S. Centers for Disease Control and Prevention (CDC) (2019). Antibiotic resistance threats in the United States. Atlanta, GA: U.S. Department of Health and Human Services.
U.S. Centers for Disease Control and Prevention (CDC). (2023). “One health basics” Centers for Disease Control and Prevention, National Center for Emerging and Zoonotic Infectious Diseases (NCEZID), 2023. Available at: https://www.cdc.gov/onehealth/basics/index.html
U.S. Environmental Protection Agency (EPA). (2002). Method 1604: Total coliforms and Escherichia coli in water by membrane filtration using a simultaneous detection technique (MI medium). U.S. Environmental Protection Agency (EPA): Washington, DC.
U.S. Environmental Protection Agency (EPA). (2009). Method 1600: enterococci in water by membrane filtration using membrane-Enterococcus Indoxyl- β-D-glucoside agar (mEI). U.S. Environmental Protection Agency (EPA): Washington, DC.
U.S. Environmental Protection Agency (EPA). (2012). Water quality standards handbook: Second edition. Washington, DC: United States Environmental Protection Agency.
U.S. Environmental Protection Agency (EPA). (2014). Method 1603: Escherichia coli (E. coli) in water by membrane filtration using modified membrane-thermotolerant Escherichia coli agar (modified mTEC). Washington, DC: United States Environmental Protection Agency
U.S. Environmental Protection Agency (EPA). (2020). National Rivers and streams assessment 2013–2014: A collaborative survey. Washington, DC: United States Environmental Protection Agency.
U.S. Food and Drug Administration (FDA). (1994a). Summary Minutes, Joint Meeting of the Veterinary Medicine Advisory Committee and Division of Anti-Infective Drugs Advisory Committee, Fluoroquinolones in Food Animal Medicine. Washington, DC: United States Environmental Protection Agency
U.S. Food and Drug Administration (FDA). (1994b). Transcript of the joint meeting of the veterinary medicine advisory committee and anti-infective drugs advisory committee. Washington, DC: United States Environmental Protection Agency
U.S. Food and Drug Administration (FDA) (2000). Enrofloxacin for poultry; opportunity for hearing, notice. Fed. Regist. 65, 64954–64965.
U.S. Food and Drug Administration (FDA). (2018). Summary report on antimicrobials sold or distributed for use in food-producing animals. FDA, Department of Health and Human Services: Washington, DC.
Wellcome Trust. (2020). The global response to AMR momentum, success, and critical gaps. Wellcome Trust: London, UK.
Wilkinson, M. D., Dumontier, M., Aalbersberg, I. J., Appleton, G., Axton, M., Baak, A., et al. (2016). The FAIR guiding principles for scientific data management and stewardship. Sci. Data 3:160018. doi: 10.1038/sdata.2016.18
World Health Organization. (2000). Overcoming antimicrobial resistance. Geneva: World Health Organization.
World Health Organization. (2015). Global action plan on antimicrobial resistance. Geneva: World Health Organization
World Health Organization. (2018). Critically important antimicrobials for human medicine. Geneva: World Health Organization
World Health Organization. (2021). WHO integrated global surveillance on ESBL-producing E. coli using a “one health” approach: Implementation and opportunities. Geneva: World Health Organization.
Yilmaz, P., Kottmann, R., Field, D., Knight, R., Cole, J. R., Amaral-Zettler, L., et al. (2011). Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MIxS) specifications. Nat. Biotechnol. 29, 415–420. doi: 10.1038/nbt.1823
Yin, X., M’ikanatha, N. M., Nyirabahizi, E., McDermott, P., and Tate, H. (2021). Antimicrobial resistance in non-Typhoidal Salmonella from retail poultry meat by antibiotic usage-related production claims –United States, 2008-2017. Int. J. Food Microbiol. 342:109044. doi: 10.1016/j.ijfoodmicro.2021.109044
Yoshizawa, N., Usui, M., Fukuda, A., Asai, T., Higuchi, H., Okamoto, E., et al. (2020). Manure compost is a potential source of tetracycline-resistant Escherichia coli and tetracycline resistance genes in Japanese farms. Antibiotics 9:76. doi: 10.3390/antibiotics9020076
Zhang, Z., Zhang, Q., Wang, T., Xu, N., Lu, T., Hong, W., et al. (2022). Assessment of global health risk of antibiotic resistance genes. Nat. Commun. 13:1553. doi: 10.1038/s41467-022-29283-8
Keywords: antimicrobial resistance, surface waters, monitoring, one health, freshwater, environment, human health
Citation: Franklin AM, Weller DL, Durso LM, Bagley M, Davis BC, Frye JG, Grim CJ, Ibekwe AM, Jahne MA, Keely SP, Kraft AL, McConn BR, Mitchell RM, Ottesen AR, Sharma M, Strain EA, Tadesse DA, Tate H, Wells JE, Williams CF, Cook KL, Kabera C, McDermott PF and Garland JL (2024) A one health approach for monitoring antimicrobial resistance: developing a national freshwater pilot effort. Front. Water. 6:1359109. doi: 10.3389/frwa.2024.1359109
Edited by:
Santanu Hazra, Government of West Bengal, IndiaReviewed by:
Liang Zhao, Michigan State University, United StatesSihem Jebri, National Center for Nuclear Science and Technology, Tunisia
Copyright © 2024 Franklin, Weller, Durso, Bagley, Davis, Frye, Grim, Ibekwe, Jahne, Keely, Kraft, McConn, Mitchell, Ottesen, Sharma, Strain, Tadesse, Tate, Wells, Williams, Cook, Kabera, McDermott and Garland. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alison M. Franklin, Franklin.alison@epa.gov
 Alison M. Franklin
Alison M. Franklin Daniel L. Weller
Daniel L. Weller Lisa M. Durso3
Lisa M. Durso3 Mark Bagley
Mark Bagley Benjamin C. Davis
Benjamin C. Davis Jonathan G. Frye
Jonathan G. Frye Christopher J. Grim
Christopher J. Grim Scott P. Keely
Scott P. Keely Autumn L. Kraft
Autumn L. Kraft Richard M. Mitchell
Richard M. Mitchell Manan Sharma
Manan Sharma Errol A. Strain
Errol A. Strain Daniel A. Tadesse
Daniel A. Tadesse Heather Tate
Heather Tate Jim E. Wells
Jim E. Wells Clinton F. Williams
Clinton F. Williams Kim L. Cook
Kim L. Cook Claudine Kabera
Claudine Kabera Patrick F. McDermott
Patrick F. McDermott Jay L. Garland
Jay L. Garland