Meta-Analysis Using NGS Data: The Veillonella Species in Dental Caries
- 1Department of Dentistry, School of Health Sciences, University of Brasilia, Brasilia, Brazil
- 2Division of Oral Biology, School of Dentistry, University of Leeds, Leeds, United Kingdom
Objectives: In light of recent technological advances in Next-generation sequencing (NGS) and the accumulation of large, publicly available oral microbiome datasets, the need for meta-analysing data on caries microbiome is becoming feasible and essential. A consensus on the identification of enriched organisms in cariogenic dysbiotic biofilms would be reached. For example, members of the Veillonella genus have been detected in caries biofilms, and may have an underestimated contribution to the dysbiotic process. Hence, we aimed to determine the abundance of Veillonella species in dental caries in studies using NGS data.
Materials and Methods: Analysis was performed according to the Preferred Reporting Items for Systematic Review and Meta-Analysis (registered at PROSPERO: CRD42020204150). Studies investigating microbial composition in saliva, dental biofilm, or carious dentin were included. Six databases and grey literature were searched. Two independent reviewers selected the papers and assessed the methodological quality.
Results: Searches retrieved 1,323 titles, from which 38 studies were included in a qualitative synthesis, comprising a total of 1,374 caries and 745 caries-free individuals. Most studies analysed 16S rRNA amplicons, and only 5 studies used shotgun metagenomics and metatranscriptomics. A geographical bias was observed. The methodological quality was downrated in 81.5% of the studies due to the lack of criteria for defining cases and standard criteria used for measurement of the condition in a reliable way. Six studies on early childhood caries (ECC) were meta-analysed, confirming a significant enrichment of Veillonella spp. in caries-associated biofilms (but not saliva) when compared to caries-free controls [mean difference: 2.22 (0.54–3.90); p = 0.01].
Conclusions: Veillonella spp. is more abundant in individuals suffering with ECC when compared to caries-free controls (very low evidence certainty), and should be considered for further studies to observe their metabolism in dental caries. There is an urgent need for a consensus in methodologies used to allow for more rigorous comparison between NGS studies, particularly including clinical data and details of caries diagnosis, as they are currently scarce. Inconsistent reporting on the NGS data affected the cross-study comparison and the biological connexions of the relative abundances on caries microbiome.
Introduction
Next-generation sequencing (NGS) approaches have been key in revolutionising microbiology, and dental caries microbial communities have been widely explored with these approaches. However, challenges remain when carrying out comparative data analyses due to the differences in methodologies and protocols employed. Recently, literature on carious biofilms investigated by 16S rRNA sequencing has grown considerably, however, much of the research up to now has been descriptive in nature. The existing accounts fail to resolve the contradiction between the abundance of several microorganisms in caries and in health. Studies on either 16S rRNA amplicon or shotgun sequencing allow the investigation of hundreds of taxa in large datasets, but a lack of biological connexions can be observed between those taxa and the disease development. Specific aspects of how some species collaborate to promote caries progression has been unmanageable by describing the microbiota in high abundance enriched in disease-associated biofilms. This is information that varies widely across studies. The enrichment of some species in a dysbiotic biofilm may reflect their unique capacity to exploit their niche, particularly in being favoured by the increase in specific nutrient availability. However, are there virulence factors that promote host damage in enriched taxa? Opportunistic pathogens may contribute to the compositional and/or functional shift towards dysbiosis. To answer that question, first, it would be imperative to identify who they are.
Meta-analyses of NGS data can enable the estimation of the global prevalence of specific members of the oral microbiome in caries for example. By gathering the existing evidence on enriched microorganisms present in caries, other than Streptococcus mutans, dysbiotic signatures may be detected and used as biomarkers or predictors in translational research. After understanding the pattern of microbial distribution, a deeper study of their physiology could infer on health-to-disease mechanisms and have significant clinical benefits. This could facilitate the development of novel treatments by focusing therapeutic strategies on a limited number of bacterial targets which may be able to stabilise the microbial community and revert or prevent its dysbiotic state [1].
Studies exploring the role of Veillonella spp. in oral biofilms are surprisingly infrequent despite their ubiquity and high abundance, and this has been credited to its challenging genetic manipulation [2]. Species from this genus are not among the most studied in dental caries, although their metabolic accountability in the transition of a homeostatic to dysbiotic oral microbial community is arguable as “middle” members of the carbon food chain, meaning that its substrates are derivates of metabolic processes of microbial partners. Veillonella spp. are involved in important inter-species interactions with other organisms such as streptococci through co-aggregation [3–6]. They are also capable of symbiotic relations by using the organic acids produced by several oral streptococci, including S. mutans, which benefits the growth of both species serving as an acid sink. Additional to the interactions in biofilms, a clue of the role of Veillonella in the second stage of root caries development was previously observed, with high expression of genes that code for bacterial collagenolytic proteases [7]. Exploring their abundance in biofilms in health and caries conditions could be relevant to recognise its ecological significance in the complex oral biofilms. Through this systematic review and meta-analyses, we aimed to determine the abundance of Veillonella species in dental caries in studies using NGS methods.
Materials and Methods
Protocol and Registration
This systematic review was performed according to the Preferred Reporting Items for Systematic Review and Meta-Analysis (PRISMA) checklist [8]. The protocol for this study was registered at the International Prospective Register of Systematic Reviews (PROSPERO) database, under the identification number CRD42020204150.
Eligibility Criteria
The acronym PECOs (Population; Exposition; Comparator; Outcomes and Studies) was used to design the search: Participants/population = Humans; Exposure(s) = Dental caries/root caries; Comparator(s)/control = No dental caries / no control; Outcome = Abundance of Veillonella at any taxonomic level (proportion, abundance, average); Studies = Observational and clinical studies applying NGS methods.
Included studies comprised the ones analysing microbial composition by NGS methods in samples (saliva, dental biofilm, or carious dentin) of individuals with dental caries, early childhood caries (ECC), or root caries, compared or not with a control group without caries. Studies eligible for this review were either observational or clinical studies.
Exclusion criteria were: (1) Animals, in situ or in vitro studies, (2) Studies written in languages not possible to be translated into an electronic translator, (3) Studies including either systemic diseases or syndromes associated with microbiota shift (Sjogren, severe hyposalivation, head and neck cancer, HIV, rheumatoid arthritis, asthma, alcoholism, etc), (4) Non-primary studies (reviews, book chapters, opinions, letters), conference abstracts, study protocols, (5) Numeric data on either prevalence or abundance of Veillonella spp. at any taxonomic level not specifically described, (6) Studies where the protocol for sequencing includes either a cloning step or a previous treatment, (7) A caries group was not defined, and (8) No NGS technique.
Data Sources and Search Strategy
The search process was performed in January 2021. Appendix Table 1 shows the search strategy. “Dental caries, NGS, Oral Microbiota” were used as main search terms that were adapted for each electronic database, namely, MEDLINE via PubMed, LILACS, Web of Science, Scopus, Embase, Cochrane, and Livivo. Grey Literature search was also performed in Google Scholar, ProQuest, and OpenGrey. Moreover, reference lists from included studies were assessed to identify other possibly eligible studies. No language or time restrictions were applied. Duplicates were identified through EndNoteWeb (Clarivate Analytics, Mumbai) and then manually identified at Rayyan QCRI® (Qatar Computer Research Institute, Qatar).
Study Selection
The selection process was performed in two phases. First, two independent and blinded reviewers screened titles and abstracts. A previous inter-reviewer calibration was performed using 20 retrieved studies (unweighted Kappa = 0.8). This phase was carried out in a web application tool designed for systematic reviews (Rayyan QCRI®, Qatar Computing Research Institute). Any disagreement was discussed in a consensus meeting. In a second phase, the same reviewers gathered all the included studies by independently reading full articles. All selected studies had the full-text available online. Once a study was selected in the second phase and the information regarding Veillonella species (at any taxonomic level) was not numerically available in any way through either the full-text or supplementary material, a protocol was performed, in which an email with reminders requesting the data was sent to authors every 3 days for 15 days. Studies with either no answers from the authors or with a negative response were then excluded. Any disagreement in the data extraction was discussed with an expert and the coordinator.
Methodological Quality Assessment
The same reviewers independently assessed the methodological quality of individual studies using the JBI Critical Appraisal Checklist for Analytical Cross-Sectional Studies [9]. For clinical studies, the same tool was applied because only the data from the baseline was considered. Due to the design of included articles, besides all eight questions of the adopted appraisal tool are considered important, two of them were considered highly critical domains to this systematic review, including “Was the exposure measured in a valid and reliable way?” and “Were objective, standard criteria used for measurement of the condition?”. A decision of excluding the studies with “no” answers for at least one of these domains was implemented. Another two were considered critical. These included: “Were the criteria for inclusion in the sample clearly defined”? and “Were the study subjects and the setting described in detail?.” Criteria related to the outcome generated by the NGS methods were considered non-critical (criteria numbers 5–8).
Criteria adopted to this systematic review for considering a low methodological quality were as follows: two “no” or one “no” and one “unclear” or two “unclear” in critical domains, or two “unclear” and one or more “no” in non-critical domains. High methodological quality was considered when an article got a maximum one “no” answer or two “unclear” answers in non-critical domains. Papers with two “no” in highly critical domains were excluded. Decision on critical and non-critical domains and classification system was discussed with the research team before the application of the instrument, as described at JBI Reviewer's Manual [9].
Data Analysis
DerSimonian and Laird Random-Effects Meta-analysis was performed by pooling the mean values of the main outcome (relative abundance) for caries and caries-free groups and calculating the mean difference (MDs) with 95% confidence intervals. Statistical heterogeneity was estimated by the Chi-square test (p < 0.05) and I-squared scores (I2). Review Manager (RevMan; Computer program; Version 5.4) was used to conduct a meta-analysis on the relative abundance (RA) of Veillonella spp. in ECC vs. caries-free (subgroups of saliva, biofilm and carious dentin).
Certainty of the Evidence
The certainty of the evidence was evaluated by the Grading of Recommendations Assessment, Development and Evaluation (GRADE) approach, performed on GRADEpro GDT [GRADEpro Guideline Development Tool (Software). McMaster University, 2015, developed by Evidence Prime, Inc., available from gradepro.org].
Results
Studies Selection and Methodological Quality Assessment
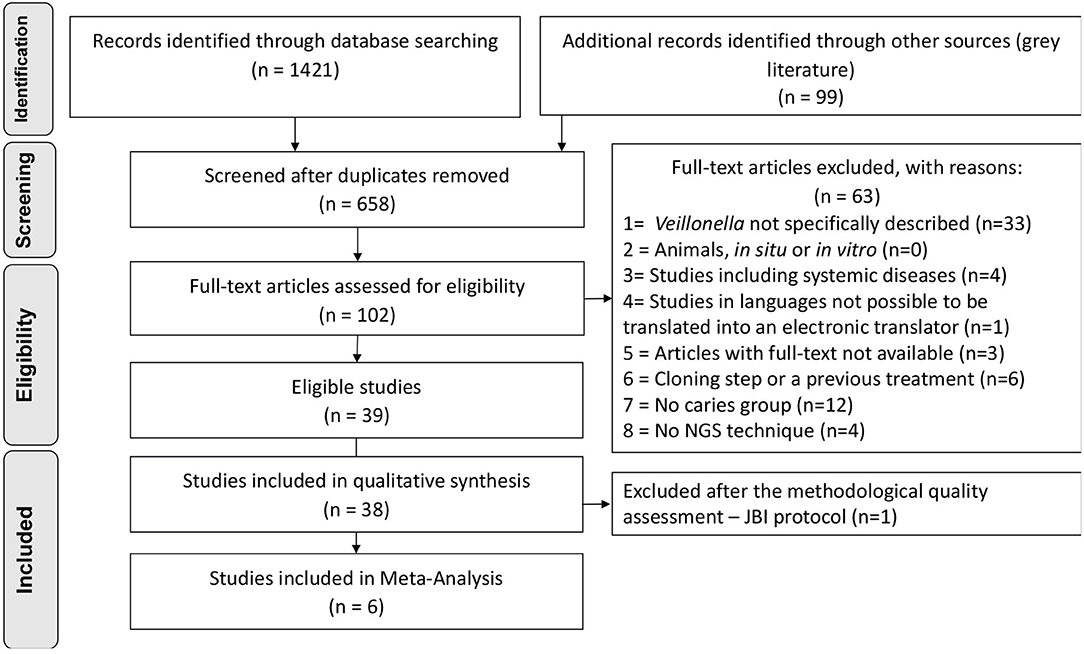
Searches retrieved 1,421 titles through all databases, and 99 titles through grey literature. After duplicates removal, 658 titles remained for screening, and 102 studies continued for full-text reading. The protocol of data request was applied to 37 studies which did not specifically describe numeric data on Veillonella abundance at any taxonomic level. Authors from 13 studies kindly shared their data (the non-respondents were then excluded). After the complete selection process, 39 studies were eligible. The complete selection process, including reasons for exclusions, is described in the PRISMA Flowchart (Figure 1), and the list of the excluded articles can be found as supplementary data (Appendix Table 2).
The quality assessment was applied resulting in 81.6% of the articles with either moderate or low quality (n = 23 studies with low quality) due to the lack of description for defining cases of caries along with standard criteria used for measurement of the caries condition in a reliable way that were considered critical domains in the JBI instrument. The evaluation of caries measurement reliability was based on the description of the number of examiners, their training, and an intra or inter-examiner calibration. One article was then excluded due to “no” answers in one of the highly critical domains [10], reaching the final number of 38 studies included in qualitative synthesis (Figure 1). The overall appraisal for each included study is represented in Table 1, and detailed quality assessment as supplementary data (Appendix Table 3).
Qualitative and Quantitative Synthesis
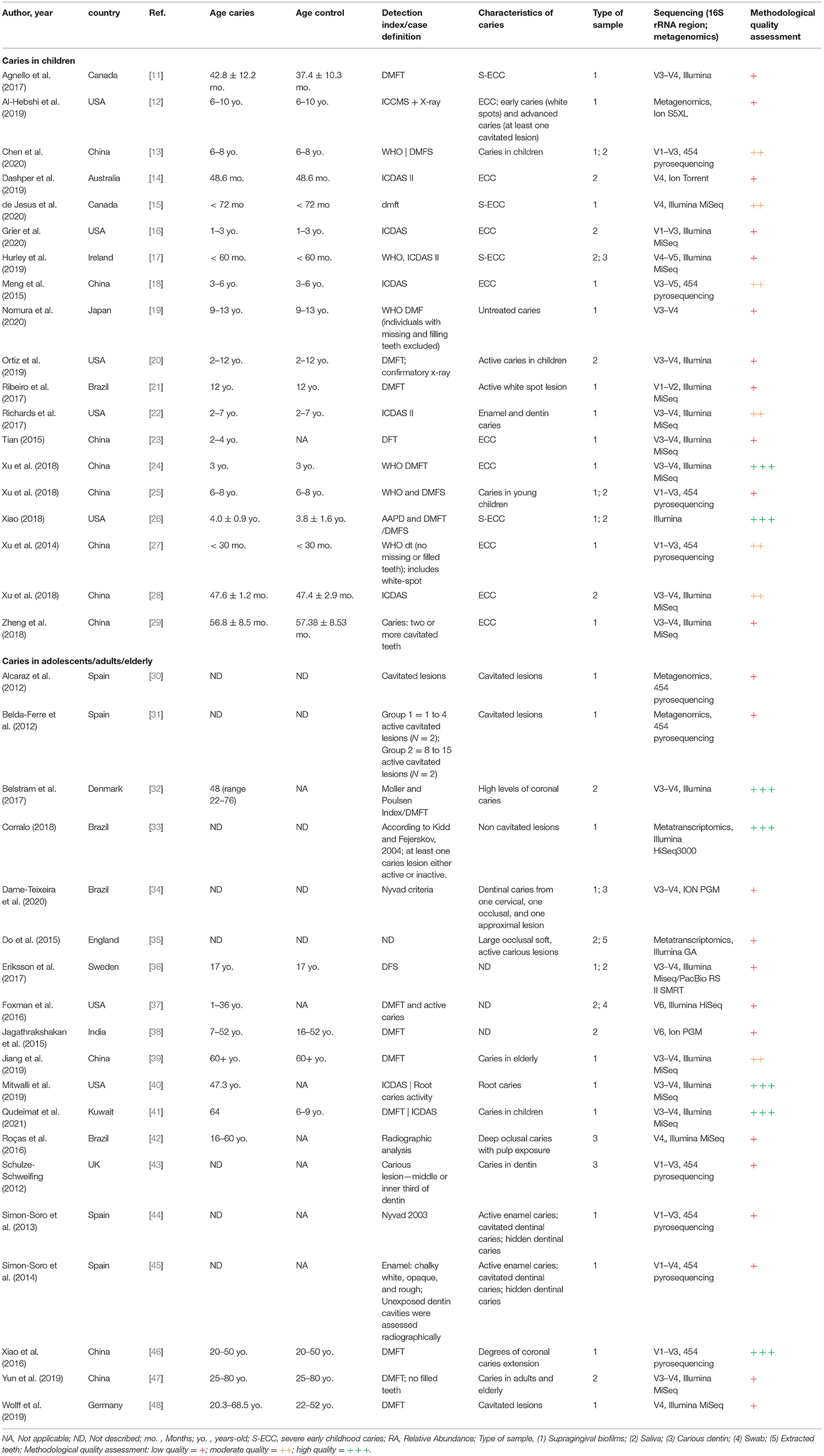
The total of 38 studies comprised a total of 1,374 caries (n = 1,385 samples) and 745 caries-free individuals. Studies from several countries were included as follows: China, United States, Brazil, England, Spain, Canada, Japan, Australia, Denmark, Germany, India, Ireland, Kuwait, and Sweden. Those were published between 2014 and 2021 and are all available in the English language. Geographical patterns of Veillonella distribution in carious biofilms could not be found, although a significantly large number of samples was analysed in only a few countries, which can give rise to a geographical or regional bias. No NGS data on caries from African countries were found, for example (Figure 2). Most studies sampled carious-associated supragingival biofilms (24 studies). Saliva and carious dentin were also frequently sampled, and sometimes combined with analysis of biofilms samples from the same individuals (Table 1). The sample size ranged from 2 to 153 patients per group (median n = 20 for caries and n = 14 for caries-free groups).
A serious weakness across most studies was the lack of description of caries detection criteria and case definition. Only few studies clearly described the caries detection criteria and the conditions of the clinical examination (artificial light, position of the individuals, tooth cleaning and drying conditions, instruments used, etc.). When described, generally, the decayed, missing, and filled teeth (DMFT) criteria from the World Health Organisation (WHO) and the International Caries Detection and Assessment System (ICDAS) were applied (Table 1). Only four studies implemented X-ray as a confirmatory method for caries diagnosis [12, 20, 42, 45] and numerous studies used the DMFT index considering only the presence of cavities to define cases.
Although several differences were observed in both caries diagnosis and sampling, a pattern was observed when molecular methods were used to measure the microbial outcome. The efficacy of DNA/RNA extraction methods could lead to differences in microbial abundances between studies adding some bias to this comparison, however, all but three studies used commercial kits. Illumina sequencers were employed in 22 studies (predominantly MiSeq), and 16S rRNA amplicons of V1-V3 and V3-V4 regions were typically sequenced (Table 1). Only 3 [12, 30, 31] and 2 [33, 35] studies used shotgun metagenomics and metatranscriptomics to analyse the RA of microorganisms and those that are metabolically active, respectively. As seen in Table 2, transcriptomic studies detected higher abundance of Veillonella than the ones sequencing DNA including 16S rRNA amplicons. Regarding the sampling site, higher abundance of Veillonella in supragingival biofilms and carious dentin compared to saliva may indicate its site-specialisation.

Table 2. Relative abundance of Veillonella at different taxonomy levels in caries and caries-free groups.
Although some studies showed data on other species, the most commonly described were Veillonella parvula, Veillonella dispar, and Veillonella atypica. Belstram et al. [32] showed data regarding Veillonella rogosae (RA = 0.6%) and Veillonella denticariosi (RA = 0.11%) in saliva of individuals with high levels of coronal caries. Ortiz et al. [20] analysed saliva of children with active caries lesions and a control group without caries and showed an RA of V. rogosae of 0.21% in caries and 0.54% in caries-free. When data on species were available, V. parvula and V. dispar had higher abundance than V. atypica and other species. By analysing those species altogether, Dashper et al. [14] showed a difference of RA = 0.15 between caries and caries-free groups. Furthermore, it seems that V. dispar and V. atypica RA drop sharply in caries environment, while V. parvula was enriched in dental caries in dentinal caries lesions analysed by RNA-seq (16.62%). It is possible to qualitatively compare these data with the study by Roças et al. [42], which also evaluated carious dentin and presented a much lower proportion of V. parvula (1.30%) in carious dentin.
The numerical range of the RA is in the magnitude of 10−2 to 102 (Table 2), and it might be related either to methodological or differences in data analysis and reporting. However, the meta-analysis of six studies on ECC confirmed a significant enrichment of Veillonella spp. in children with caries when compared to caries-free controls [mean RA difference: 2.22 (0.54–3.90); p = 0.01] with considerable statistical heterogeneity (I2 81%) despite one study which found conflicting results by analysing the salivary microbiome [28]. In the analysis of subgroups, the difference was clearly defined by the studies on supragingival biofilms, and the salivary enrichment of Veillonella in caries could not be observed (Figure 3).
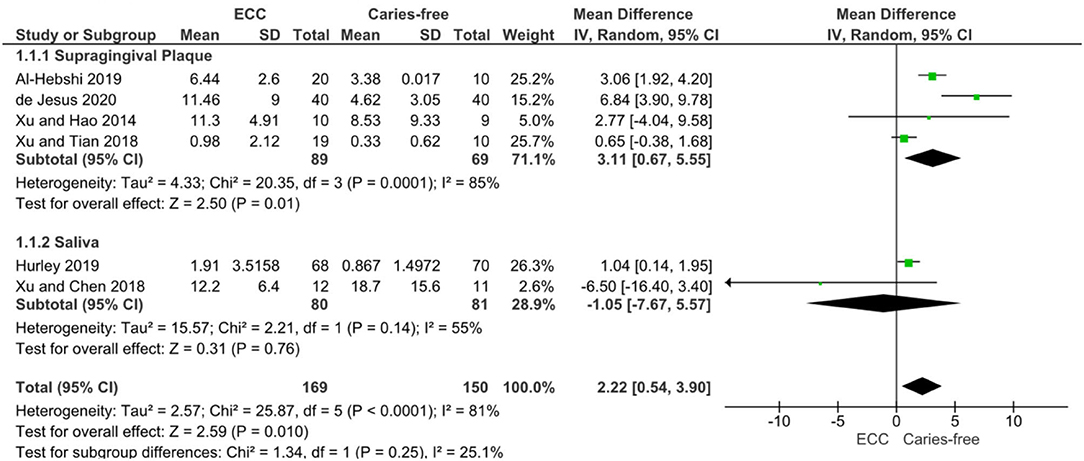
Figure 3. DerSimonian and Laird Random-Effects Meta-analysis of Veillonella spp. abundance in Early Childhood Caries (ECC) individuals compared to caries-free individuals. A subgroup analysis was performed for studies on supragingival plaque (1.1.1) and saliva (1.1.2), as well as the pooled results.
Certainty of evidence evaluated through GRADE approach was rated as “very low” for Veillonella RA in ECC individuals compared with caries-free individuals. Considering that observational studies initiate the GRADE assessment as “low,” the certainty was further downgraded by one additional point due to low-quality of two included studies [12, 17] after sensitivity analysis (since the removal of these two low-quality studies from meta-analysis resulted in summary effect size modification—MD 2.07 CI 95% −2.44 to 6.58, p > 0.05). The other GRADE approach domains were not downgraded (inconsistency, indirectness, imprecision, and publication bias), nor upgraded (great magnitude, dose-response, residual confounders). Appendix Table 4 shows the summary of GRADE of finding table with detailed decisions.
Discussion
In light of recent technological advances in NGS and the accumulation of large and publicly available oral microbiome datasets, the need for meta-analysing data on caries microbiome is becoming essential. Developing a versatile analysis pipeline for the raw sequencing data of a wide range of studies would be particularly complex due to the heterogeneity of the datasets (differences in nucleic acid isolation, library preparation, sequencing platforms used, etc.) and their associated information on clinical data. Systematic reviews can, however, be a worthy strategy to achieve a cross-study comparison of the microbiome using NGS data, as we validated here. Although a meta-analysis approach is still subject to heterogeneity, it can nevertheless be feasible. As far as we know, this is the first meta-analysis of the NGS data on dental caries. After a comprehensive literature search, 39 studies presenting Veillonella data from caries samples were eligible, of which 38 and 6 were qualitatively and quantitatively analysed, respectively. We demonstrated a significant enrichment of Veillonella spp. in ECC samples when compared to samples from caries-free samples, yet with very low certainty. However, the most striking result here is lack of clarity in defining cases and controlling confounding factors, and the need for changes in the reports on the clinical data from the donors.
Here we showed differences in Veillonella proportion in different lesions and type of sampling, being the supragingival biofilms more prone to demonstrate differences in RA in health-to-disease. This higher abundance of Veillonella in supragingival biofilms and carious dentin compared to saliva may indicate its site-specialisation. They are evidently likely to benefit from the ecological conditions of low pH environment commonly observed in carious lesions. However, Veillonella spp. enrichment can also suggest their non-negligible role in caries dysbiosis. A relevant research question could perhaps consider Veillonella spp. to have virulence potential in caries, particularly in ECC. Veillonella spp. are considered commensal bacteria present in the oral cavity and the gastrointestinal tract of humans, due to being part of the core microbiome [49, 50]. They are known to be involved in cross-feeding and co-aggregation with acidogenic species in other environments [51], using their metabolic intermediates, which suggest that they act as an acid sink. However, in an in vitro study, it was shown that Veillonella, when associated with S. mutans, did not increase the biofilm pH as expected, as well as promote the growth and extracellular polysaccharide synthesis [49]. We believe that the identification of these virulence factors and a more detailed molecular and functional characterisation of Veillonella in caries-associated biofilms, in combination with a more thorough characterisation of other microorganisms, could be the key for the development of strategies to modulate the microbiota by using pre or probiotics. A more recent molecular characterisation of oral Veillonella revealed that the genomes of oral Veillonella species were remarkably diverse, and that these Veillonella have conserved pathways that utilise carbohydrates other than lactate as an energy source [50], which might be an interesting finding of their potential pathogenic traits as oral species. Furthermore, RNA sequencing seems to show higher abundances of Veillonella than DNA sequencing. Although the result should be interpreted with caution as no statistical analysis was achievable. In addition, it could also suggest some inactive or dead cells in the DNA studies. Benítez-Páez showed important differences in the RA of bacterial genera from metagenomic and metatranscriptomic data by analysing dental biofilms, and Veillonella was one of the most commonly found in the total DNA-based metagenome, however, with a slightly lower value in RNA-based metatranscriptomics [52].
Important limitations on the heterogeneity of the primary studies should be considered. There is a concerning pattern of lack of thorough definition and description of caries cases, even for the same condition, occurring in ECC which is defined differently throughout studies. Most studies used the standard criterion of the World Health Organisation (the classic DMFT), or solely the presence of cavities to define cases. However, this can add an important bias due to the overlooking of non-cavitated caries and caries activity. Furthermore, several studies did not describe whether the “caries-free” group had past caries experience, such as the presence of surfaces with inactive lesions, fillings, or missing teeth, making unclear if this could impact the microbial composition in the microbiome after caries management and arrestment. We hypothesise the presence of a “scarf of dysbiosis” in biofilms from individuals with past caries experience by observing findings from Corralo where microbial profiles of inactive caries-associated biofilms were unlike those of caries-free biofilms [33]. Different detection criteria should be used to assess dental caries in a more accurate way, such as the Nyvad Criterion [53] and the International Caries Detection and Assessment System (ICDAS) [54]. The criterion to determine cases plays an important role in investigating caries-associated microbiota. Furthermore, most studies did not evaluate disease in the same way and did not address the calibration of dental examiners, which would have improved the certainty of the evidence in the field. New studies on caries microbiome should have plans and strategies for a better definition of caries.
Despite advances in the 16S rRNA sequencing and the shotgun whole-genome metagenomic methods, there is an urgent need to define patterns in methodological data from studies on caries microbiology to make them comparable. Although our search found a considerable number of primary studies, it was extremely complex and difficult to make a synthesis within the current available data from the literature. The microbial composition was shown in different perspectives and taxonomic levels. Other sources of variation, such as the complexity of the related clinical elements in each study, may have a great impact on the overall microbiome investigation and is often omitted in many studies. Examples include the lack of the clarity at the selection criteria and the control of confounding factors. In the present study, 20 out of 39 studies were downrated at the JBI domain: “Were confounding factors identified?”. Those confounding factors could include a large range of age, gender, patient habits (including diet, smoking, oral hygiene practises, etc.), caries extent, caries activity, salivary or biofilm pH, salivary flow, other oral conditions (gingivitis, periodontitis, tooth loss, use of prosthesis), systemic diseases, etc. We believe that further study should provide those details as they will favour more rigorous and accurate analyses of the oral microbiome, particularly when saliva is sampled. Zaura et al. already described the need for improving the clinical characterisation in future NGS studies on the oral microbiome so that the quality of the scientific literature would be improved throughout more carefully designed papers [55].
We also identified a lack of standardisation in reporting this type of study, suggesting the need of a comprehensive guideline checklist development. Reports on NGS data following guideline checklists, such as the STROBE for observational studies [56] or the CONSORT for randomised clinical trials [57] promote research reproducibility, better-quality study designs, and help the development of meta-analyses. There is a need for a specific checklist for microbiological data as we faced difficulties in obtaining accurate Veillonella spp. RA data from studies. Authors were requested to share their data on Veillonella RA and the response rate was 34%, resulting in information losses that add a selective reporting bias. Although it would be more reasonable to meta-analyse several microorganisms as caries is a polymicrobial disease, it is not logistically easy due to the differences in these reports. In order to overcome this problem, additional tables containing the average RA of all species might be uploaded as a supplementary material in further studies. Besides, not all studies that presented data on Veillonella clearly defined the unit used for the RA. On a positive note, the laboratory experiments and bioinformatic pipelines were adequately described in all studies.
Zhou et al. lay emphasis on the need to optimise and standardise metagenomic studies [58] as technical or methodologic heterogeneity produces systematic biases that could obscure biologically meaningful information on the compositional differences. More homogeneous studies regarding the DNA/RNA extraction methods, amplicons region sequencing, pipeline (ASV or OTUs), etc. [59] ought to lead to additional meta-analyses. Another important bias control is to develop a sample size calculation, since none of the studies retrieved here presented, thus reducing their external validity (generalisation). Low sample sizes (median of 20 individuals sampled) can be explained due to the high costs of the NGS studies. However, power calculation methods for microbiome studies are now available [60], and specific sample sizes based on the prevalence of microorganisms can also be estimated.
In order to overcome the apparent geographical bias, with data originating from selected few countries (only 14), those with perhaps greater facilities and funds for conducting NGS research, new multicentric studies, in particular, should be developed with a wider interest in the microbiomes of all human populations and ethnicities. More meta-analyses of NGS data investigating other microorganisms (or a group of) in dental caries would help gather and confirm invaluable evidence of enriched microorganisms, such as Veillonella spp. Understanding the pattern of their distribution and physiology could help elucidate the mechanism of the health-to-disease transition, with crucial information on colonisation, survival, and interactions, which would help define research priorities in the field of caries microbiology.
Conclusions
Veillonella spp. are more abundant in individuals suffering with ECC when compared to caries-free controls (very low evidence certainty), and should be considered in further studies to better understand their metabolism and contribution to dental caries. There is an urgent need for a consensus in methodologies used to allow for more rigorous comparison, particularly including clinical data and details of caries diagnosis, which are currently scarce. Inconsistent reporting on the NGS data affected the cross-study comparison and the biological connexions of the relative abundances on caries microbiome.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
ND-T contributed to conception, design, data acquisition and interpretation, drafted, and critically revised the manuscript. AL contributed to design, data acquisition and interpretation, drafted, and critically revised the manuscript. TD contributed to conception, design, and critically revised the manuscript. CS contributed to conception, design, data acquisition and interpretation, performed statistical analysis, drafted, and critically revised the manuscript. All authors gave their final approval and agree to be accountable for all aspects of the work.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We gratefully acknowledge the UK's Academy of Medical Sciences Newton International Fellowship (NIF\R5\242) for their support. We also thank the authors who kindly responded to our inquiry to share their data on Veillonella RA.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/froh.2021.770917/full#supplementary-material
References
1. Hajishengallis G, Darveau RP, Curtis MA. The keystone-pathogen hypothesis. Nat Rev Microbiol. (2012) 10:717–25. doi: 10.1038/nrmicro2873
2. Knapp S, Brodal C, Peterson J, Qi F, Kreth J, Merritt J. Natural competence is common among clinical isolates of Veillonella parvula and is useful for genetic manipulation of this key member of the oral microbiome. Front Cell Infect Microbiol. (2017) 7:139. doi: 10.3389/fcimb.2017.00139
3. Kolenbrander PE. Intergeneric coaggregation among human oral bacteria and ecology of dental plaque. Annu Rev Microbiol. (1988) 42:627–56. doi: 10.1146/annurev.mi.42.100188.003211
4. Mutha NVR, Mohammed WK, Krasnogor N, Tan GYA, Wee WY, Li Y, et al. Transcriptional profiling of coaggregation interactions between Streptococcus gordonii and Veillonella parvula by Dual RNA-Seq. Sci Rep. (2019) 9:1–14. doi: 10.1038/s41598-019-43979-w
5. Periasamy S, Kolenbrander PE. Central role of the early colonizer Veillonella sp. in establishing multispecies biofilm communities with initial, middle, and late colonizers. Enamel J Bacteriol. (2010) 192:2965–72. doi: 10.1128/JB.01631-09
6. Zhou P, Liu J, Li X, Takahashi Y, Qi F. The sialic acid binding protein, hsa, in Streptococcus gordonii DL1 also mediates intergeneric coaggregation with Veillonella species. PLoS ONE. (2015) 10:e0143898. doi: 10.1371/journal.pone.0143898
7. Damé-Teixeira N, Parolo C, Maltz M, Rup A, Devine D, Do T. Gene expression of bacterial collagenolytic proteases in root caries. J Oral Microbiol. (2018) 10:1424475. doi: 10.1080/20002297.2018.1424475
8. Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ. (2009) 339:b2535. doi: 10.1136/bmj.b2535
9. Moola S, Munn Z, Tufanaru C, Aromataris E, Sears K, Sfetcu R, et al. Chapter 7: systematic reviews of etiology and risk. In: Aromataris E, Munn Z, editors. JBI Manual for Evidence Synthesis. JBI; 2020.
10. Obata J, Takeshita T, Shibata Y, Yamanaka W, Unemori M, Akamine A, et al. Identification of the microbiota in carious dentin lesions using 165 rRNA gene sequencing. PloS ONE. (2014) 9:3712. doi: 10.1371/journal.pone.0103712
11. Agnello M, Marques J, Cen L, Mittermuller B, Huang A, Tran NC, et al. Microbiome associated with severe caries in canadian first nations children. J Dent Res. (2017) 96:1378–85. doi: 10.1177/0022034517718819
12. Al-Hebshi NN, Baraniya D, Chen T, Hill J, Puri S, Tellez M, et al. Metagenome sequencing-based strain-level and functional characterization of supragingival microbiome associated with dental caries in children. J Oral Microbiol. (2019) 11:1557986. doi: 10.1080/20002297.2018.1557986
13. Chen W, Jiang Q, Yan G, Yang D. The oral microbiome and salivary proteins influence caries in children aged 6 to 8 years. BMC Oral Health. (2020) 20:295. doi: 10.1186/s12903-020-01262-9
14. Dashper SG, Mitchell HL, Ka LC, Carpenter L, Gussy MG, Calache H, et al. Temporal development of the oral microbiome and prediction of early childhood caries. Sci Rep. (2019) 9:19732. doi: 10.1038/s41598-019-56233-0
15. de Jesus VC, Shikder R, Oryniak D, Mann K, Alamri A, Mittermuller B, et al. Sex-based diverse plaque microbiota in children with severe caries. J Dent Res. (2020) 99:703–12. doi: 10.1177/0022034520908595
16. Grier A, Myers JA, O'Connor TG, Quivey RG, Gill SR, Kopycka-Kedzierawski DT. Oral microbiota composition predicts early childhood caries onset. J Dent Res. (2020) 220:34520979926. doi: 10.1177/0022034520979926
17. Hurley E, Barrett MPJ, Kinirons M, Whelton H, Ryan CA, Stanton C, et al. Comparison of the salivary and dentinal microbiome of children with severe-early childhood caries to the salivary microbiome of caries-free children. BMC Oral Health. (2019)19:13. doi: 10.1186/s12903-018-0693-1
18. Meng Z, Yongxing C, Lingzhi X, Yuhong L, Han J, Minquan D. Pyrosequencing of plaque microflora in twin children with discordant caries phenotypes. PLoS ONE 10:141310. doi: 10.1371/journal.pone.0141310
19. Nomura Y, Otsuka R, Hasegawa R, Hanada N. Oral microbiome of children living in an isolated area in Myanmar. Int J Environ Res Public Health. (2020) 17:114033. doi: 10.3390/ijerph17114033
20. Ortiz S, Herrman E, Lyashenko C, Purcell A, Raslan K, Khor B, et al. Sex-specific differences in the salivary microbiome of caries-active children. J Oral Microbiol. (2019) 11:1653124. doi: 10.1080/20002297.2019.1653124
21. Ribeiro AA, Azcarate-Peril MA, Cadenas MB, Butz N, Paster BJ, Chen T, et al. The oral bacterial microbiome of occlusal surfaces in children and its association with diet and caries. PLoS ONE. (2017) 12:e0180621. doi: 10.1371/journal.pone.0180621
22. Richards VP, Alvarez AJ, Luce AR, Bedenbaugh M, Mitchell ML, Burne RA, et al. Microbiomes of sitespecific dental plaques from children with different caries status. Infect Immun. (2017) 85:17. doi: 10.1128/IAI.00106-17
23. Tian J, Qin M, Ma WL, Xia B, Xu H, Zhang Q, et al. Microbiome interaction with sugar plays an important role in relapse of childhood caries. Biochem Biophys Res Commun. (2015) 468:294–9. doi: 10.1016/j.bbrc.2015.10.110
24. Xu H, Tian J, Hao W, Zhang Q, Zhou Q, Shi W, et al. Oral microbiome shifts from caries-free to caries-affected status in 3-year-old chinese children: a longitudinal study. Front Microbiol. (2018) 9:2009. doi: 10.3389/fmicb.2018.02009
25. Xu Y, Jia YH, Chen L, Huang WM, Yang DQ. Metagenomic analysis of oral microbiome in young children aged 6-8 years living in a rural isolated Chinese province. Oral Dis. (2018) 24:1115–25. doi: 10.1111/odi.12871
26. Xiao J, Grier A, Faustoferri RC, Alzoubi S, Gill AL, Feng C, et al. Association between oral candida and bacteriome in children with severe ECC. J Dent Res. (2018) 97:1468–76. doi: 10.1177/0022034518790941
27. Xu H, Hao W, Zhou Q, Wang W, Xia Z, Liu C, et al. Plaque bacterial microbiome diversity in children younger than 30 months with or without caries prior to eruption of second primary molars. PLoS ONE. (2014) 9:e89269. doi: 10.1371/journal.pone.0089269
28. Xu L, Chen X, Wang Y, Jiang W, Wang S, Ling ZX, et al. Dynamic alterations in salivary microbiota related to dental caries and age in preschool children with deciduous dentition: a 2-year follow-up study. Front Physiol. (2018) 9:342. doi: 10.3389/fphys.2018.00342
29. Zheng YQ, Zhang M, Li J, Li YH, Teng F, Jiang H, et al. Comparative analysis of the microbial profiles in supragingival plaque samples obtained from twins with discordant caries phenotypes and their mothers. Front Cell Infect Microbiol. (2018) 8:361. doi: 10.3389/fcimb.2018.00361
30. Alcaraz LD, Belda-Ferre P, Cabrera-Rubio R, Romero H, Simón-Soro Á, Pignatelli M, et al. Identifying a healthy oral microbiome through metagenomics. Clin Microbiol Infect. (2012) 18:54–7. doi: 10.1111/j.1469-0691.2012.03857.x
31. Belda-Ferre P, Alcaraz LD, Cabrera-Rubio R, Romero H, Simón-Soro A, Pignatelli M, et al. The oral metagenome in health and disease. ISME J. (2012) 6:46–56. doi: 10.1038/ismej.2011.85
32. Belstrøm D, Holmstrup P, Fiehn NE, Kirkby N, Kokaras A, Paster BJ, et al. Salivary microbiota in individuals with different levels of caries experience. J Oral Microbiol. (2017) 9:1270614. doi: 10.1080/20002297.2016.1270614
33. Corralo DJ. Bacterial composition of microbiome associated to supragingival biofilm in health and caries disease (Thesis). Porto Alegre: Federal University of Rio Grande do Sul Brazil (2018). Available online at: https://lume.ufrgs.br/handle/10183/199180
34. Dame-Teixeira N, de Cena JA, Côrtes DA, Belmok A, Dos Anjos Borges LG, Marconatto L, et al. Presence of archaea in dental caries biofilms. Arch Oral Biol. (2020) 110:104606. doi: 10.1016/j.archoralbio.2019.104606
35. Do T, Sheehy EC, Mulli T, Hughes F, Beighton D. Transcriptomic analysis of three veillonella spp. present in carious dentine and in the saliva of caries-free individuals. Front Cell Infect Microbiol. (2015) 5:25. doi: 10.3389/fcimb.2015.00025
36. Eriksson L, Lif Holgerson P, Johansson I. Saliva and tooth biofilm bacterial microbiota in adolescents in a low caries community. Sci Rep. (2017) 7:5861. doi: 10.1038/s41598-017-06221-z
37. Foxman B, Luo T, Srinivasan U, Ramadugu K, Wen A, Goldberg D, et al. The effects of family, dentition, and dental caries on the salivary microbiome. Ann Epidemiol. (2016) 26:348–54. doi: 10.1016/j.annepidem.2016.03.006
38. Jagathrakshakan SN, Sethumadhava RJ, Mehta DT, Ramanathan A. 16S rRNA gene-based metagenomic analysis identifies a novel bacterial co-prevalence pattern in dental caries. Eur J Dent. (2015) 9:127–32. doi: 10.4103/1305-7456.149661
39. Jiang Q, Liu J, Chen L, Gan N, Yang D. The oral microbiome in the elderly with dental caries and health. Front Cell Infect Microbiol. (2019) 9:442. doi: 10.3389/fcimb.2018.00442
40. Mitwalli H, Mourao MDA, Dennison J, Yaman P, Paster BJ, Fontana M. Effect of silver diamine fluoride treatment on microbial profiles of plaque biofilms from root/cervical caries lesions. Caries Res. (2019) 53:555–66. doi: 10.1159/000499578
41. Qudeimat MA, Alyahya A, Karched M, Behbehani J, Salako NO. Dental plaque microbiota profiles of children with caries-free and caries-active dentition. J Dent. (2021)104:103539. doi: 10.1016/j.jdent.2020.103539
42. Rôças IN, Alves FRF, Rachid CTCC, Lima KC, Assunção IV, Gomes PN, et al. Microbiome of deep dentinal caries lesions in teeth with symptomatic irreversible pulpitis. PLoS ONE. (2016) 11:e0154653. doi: 10.1371/journal.pone.0154653
43. Schulze-Schweifing K. Molecular characterisation of the bacterial community in dentinal caries (Thesis). United Kingdom: University of London, King's College (2012). Available online at: https://kclpure.kcl.ac.uk/portal/en/theses/molecular-characterisation-of-the-bacterial-community-in-dentinal-caries(a71b7ef7-e5f2-4b90-8545-b120b52d9b1f).html
44. Simón-Soro Á, Belda-Ferre P, Cabrera-Rubio R, Alcaraz LD, Mira A. A tissue-dependent hypothesis of dental caries. Caries Res. (2013) 47:591–600. doi: 10.1159/000351663
45. Simón-Soro A, Guillen-Navarro M, Mira A. Metatranscriptomics reveals overall active bacterial composition in caries lesions. J Oral Microbiol. (2014) 6:25443. doi: 10.3402/jom.v6.25443
46. Xiao CC, Ran SJ, Hunag ZW, Liang JP. Bacterial diversity and community structure of supragingival plaques in adults with dental health or caries revealed by 16S pyrosequencing. Front Microbiol. (2016) 7:1145. doi: 10.3389/fmicb.2016.01145
47. Yun C, Zhiyan L, Chong Z, Jing L, Xin Z, Derui Z. Illumina-based sequencing analysis of pathogenic microorganisms in dental caries patients of different chinese ethnic groups. J Int Med Res. (2019) 47:5037–47. doi: 10.1177/0300060519866939
48. Wolff D, Frese C, Schoilew K, Dalpke A, Wolff B, Boutin S. Amplicon-based microbiome study highlights the loss of diversity and the establishment of a set of species in patients with dentin caries. PLoS ONE. (2019) 219714:219714. doi: 10.1371/journal.pone.0219714
49. Liu S, Chen M, Wang Y, Zhou X, Peng X, Ren B, et al. Effect of Veillonella parvula on the physiological activity of Streptococcus mutans. Arch Oral Biol. (2020) 109:104578. doi: 10.1016/j.archoralbio.2019.104578
50. Mashima I, Liao Y-C, Lin C-H, Nakazawa F, Haase EM, Kiyoura Y, et al. Comparative pan-genome analysis of oral veillonella species. Microorganisms. (2021) 9:1775. doi: 10.3390/microorganisms9081775
51. Hinton A, Hume ME, Deloach JR. Role of metabolic intermediates in the inhibition of Salmonella typhimurium and Salmonella enteritidis by Veillonella. J Food Prot. (1993) 56:932–7. doi: 10.4315/0362-028X-56.11.932
52. Benítez-Páez A, Belda-Ferre P, Simón-Soro A, Mira A. Microbiota diversity and gene expression dynamics in human oral biofilms. BMC Genomics. (2014) 15:311. doi: 10.1186/1471-2164-15-311
53. Nyvad B, Baelum V. Nyvad criteria for caries lesion activity and severity assessment: a validated approach for clinical management and research. Caries Res. (2018) 52:397–405. doi: 10.1159/000480522
54. Ismail AI, Sohn W, Tellez M, Amaya A, Sen A, Hasson H, et al. The international caries detection and assessment system (icdas): an integrated system for measuring dental caries. Commun Dent Oral Epidemiol. (2007) 35:170–8. doi: 10.1111/j.1600-0528.2007.00347.x
55. Zaura E, Pappalardo VY, Buijs MJ, Volgenant CMC, Brandt BW. Optimizing the quality of clinical studies on oral microbiome: a practical guide for planning, performing, and reporting. Periodontol. (2000) 85:210–36. doi: 10.1111/prd.12359
56. Vandenbroucke JP, von Elm E, Altman DG, Gøtzsche PC, Mulrow CD, Pocock SJ, et al. Strengthening the reporting of observational studies in epidemiology (STROBE): explanation and elaboration. Int J Surg. (2014) 12:1500–24. doi: 10.1016/j.ijsu.2014.07.014
57. Moher D, Hopewell S, Schulz KF, Montori V, Gøtzsche PC, Devereaux PJ, et al. CONSORT 2010 explanation and elaboration: updated guidelines for reporting parallel group randomised trials. J Clin Epidemiol. (2010) 63:e1–37. doi: 10.1016/j.jclinepi.2010.03.004
58. Zhou Y, Gao H, Mihindukulasuriya KA, La Rosa PS, Wylie KM, Vishnivetskaya T, et al. Biogeography of the ecosystems of the healthy human body. Genome Biol. (2013) 14:R1. doi: 10.1186/gb-2013-14-1-r1
59. Wade WG, Prosdocimi EM. Profiling of oral bacterial communities. J Dent Res. (2020) 99:621–9. doi: 10.1177/0022034520914594
Keywords: next-generation sequencing, 16S rRNA amplicon sequencing, oral microbiology, systematic reviews, meta-analysis
Citation: Dame-Teixeira N, de Lima AKA, Do T and Stefani CM (2021) Meta-Analysis Using NGS Data: The Veillonella Species in Dental Caries. Front. Oral. Health 2:770917. doi: 10.3389/froh.2021.770917
Received: 05 September 2021; Accepted: 22 September 2021;
Published: 22 October 2021.
Edited by:
Georgios N. Belibasakis, Karolinska Institutet (KI), SwedenReviewed by:
Takuichi Sato, Niigata University, JapanPeng Zhou, The University of Texas Health Science Centre at San Antonio, United States
Copyright © 2021 Dame-Teixeira, de Lima, Do and Stefani. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Naile Dame-Teixeira, nailedame@unb.br; nailedame@hotmail.com
 Naile Dame-Teixeira
Naile Dame-Teixeira Ana Karolina Almeida de Lima1
Ana Karolina Almeida de Lima1  Thuy Do
Thuy Do