- 1College of Pharmacy, Hubei University of Chinese Medicine, Wuhan, China
- 2Hubei Institute for Drug Control, Wuhan, China
- 3Hubei Key Laboratory of Chinese Medicine Resource and Chemistry, Hubei University of Chinese Medicine, Wuhan, China
Introduction: The rhizomes of Coptis plants have been used in traditional Chinese medicine over 2000 years. Due to increasing market demand, the overexploitation of wild populations, habitat degradation and indiscriminate artificial cultivation of Coptis species have severely damaged the native germplasms of species in China.
Methods: Genome-wide simple-sequence repeat (SSR) markers were developed using the genomic data of C. chinensis. Population genetic diversity and structure of 345 Coptis accessions collected from 32 different populations were performed based on these SSRs. The distribution of suitable areas for three taxa in China was predicted and the effects of environmental variables on genetic diversity in relation to different population distributions were further analyzed.
Results: 22 primer pairs were selected as clear, stable, and polymorphic SSR markers. These had an average of 16.41 alleles and an average polymorphism information content (PIC) value of 0.664. In the neighbor-joining (N-J) clustering analysis, the 345 individuals clustered into three groups, with C. chinensis, C. chinensis var. brevisepala and C. teeta being clearly separated. All C. chinensis accessions were further divided into four subgroups in the population structure analysis. The predicted distributions of suitable areas and the environmental variables shaping these distributions varied considerably among the three species.
Discussion: Overall, the amount of solar radiation, precipitation and altitude were the most important environmental variables influencing the distribution and genetic variation of three species. The findings will provide key information to guide the conservation of genetic resources and construction of a core reserve for species.
1 Introduction
The genus Coptis Salisb. of the family Ranunculaceae is important for medicinal herbs and widely used in healthcare (Wang et al., 2019). The rhizomes of Coptis species (Coptidis Rhizome, CR) have been used in traditional Chinese medicine for over 2000 years; the first records of their use in medicines appear in the Divine Farmer’s Materia Medica (Shennong Bencao Jing) during the Han dynasty (202 B.C.–220 A.D.) (Liu et al., 2021). According to the Chinese Pharmacopoeia, CR includes the dry rhizomes of C. chinensis Franch., C. deltoidea C.Y. Cheng et Hsiao, and C. teeta Wall., which are known as ‘Weilian’, ‘Yalian’ and ‘Yunlian’, respectively. Thus far, studies on Coptis plants have primarily focused on the chemical constitution and pharmacological effects of their main active ingredients, which include various protoberberine-type alkaloids such as berberine, coptisine, jatrorrhizine, palmatine, columbamine, and epiberberine (Lv et al., 2016; Yang et al., 2017). These alkaloids are known to have preventative and therapeutic effects on diabetes, cancer, cardiovascular, and nervous system diseases (Wu et al., 2016; Chou et al., 2017).
Coptis plants typically grow in cold, humid, and shady habitats in mountains or valleys at altitudes ranging from 1,200–2,000 m.s.l (Zhao et al., 2021). In China, Coptis species are distributed throughout the southern and southwestern mountain ranges, including the Himalayas (Wang et al., 2020). Owing to their narrow geographical distribution, the natural resources of Coptis are limited (Wang et al., 2022b). The market demand for CR has been increasing, especially in recent years (Ma et al., 2020; Xu et al., 2022). However, overexploitation and habitat destruction over the long-term have severely impacted wild Coptis populations and depleted the natural resources of these plants. At present, almost all CRs available on the market are derived from cultivated germplasms. Most of these CRs are from C. chinensis, with those of C. deltoidea and C. teeta being less common (Wang et al., 2022b). As a variety of C. chinensis, populations of C. chinensis var. brevisepala W.T. Wang et Hsiao are also in decline owing to the habitat degradation and overexploitation (Mamut et al., 2018). The molecular mechanisms modulating the signaling pathway of Coptis pathogenesis have been widely studied (Yamada et al., 2016; He et al., 2018; Chen et al., 2020b). A recent analysis of the C. chinensis genome found that the evolution of a Ranunculales clade-specific gene family, CYP719, greatly contributed to the diversification of protoberberine-type alkaloids in these plants (Liu et al., 2021). However, there is limited knowledge on the genetic relationships between various Coptis species as well as the genetic diversity associated with the adaptation of Coptis species across a range of environments.
The complex interplay between genes and environments shapes the population structures and phenotypes of plant species (Li et al., 2021). Changes in the environment can significantly impact the quality of medicinal herbs (Yang et al., 2018; Perrino et al., 2023). For instance, environmental change can influence the medicinal properties of medicinal herbs by affecting gene expression and consequently altering the abundances of secondary metabolites (Blanquart et al., 2013). At present, the rapid transferring and domestication of wild Coptis into cultivated germplasm as well as the indiscriminate propagation of Coptis are likely to negatively impact the rational layout of CR production and undermine the medicinal qualities of CR in the market (Han et al., 2019). The domestication history of C. chinensis is still largely unknown in the background of mixed germplasm. Whether it is single origin or multi origin domestication, as well as the hypothesis that the specific location of the original domestication center is in which province of Hubei, Sichuan, and Chongqing, the three main production areas of authentic medicinal herbs, has always been controversial. There were also hypothesis dividing cultivated C. chinensis into ‘Nananlian’ and ‘Beianlian’ based on the Yangtze River boundary (Liu et al., 2018). In addition, the impact of changes in environmental variables caused by different terrains on the domestication process of C. chinensis also needs to be explored. There is hence an urgent need to jointly evaluate the genetic variation and ecological suitability of Coptis species. Specifically, to mitigate additional losses of germplasm sources for Coptis, it is necessary to investigate the genetic diversity and architecture of current Coptis germplasms, as this will facilitate the selection and cultivation of superior Coptis germplasms in the future. As environmental conditions strongly influence the distributions of plant species, environmental variables can be employed to simulate plant niches (Poirazidis et al., 2019; Zhang et al., 2019). Knowledge of the distributions of ecologically suitable areas for Coptis species will also help in identifying the geographical and environmental factors influencing the contents of active ingredients in Coptis germplasm, thereby providing crucial data for the conservation and restoration of native Coptis populations.
Simple sequence repeats (SSRs) are effective and economical molecular markers for assessing the levels of genetic variation in plant populations and have been widely used for germplasm identification and breeding of both crops and medicinal herbs (Zhang et al., 2016; Lin et al., 2020; Chen et al., 2020a; Li et al., 2020a). In this study, genome-wide SSR markers for C. chinensis were developed, and 22 pairs of highly polymorphic primers were screened to analyze the genetic diversity and population structures of 345 wild and cultivated accessions derived from three Coptis species in China. Using 68 environmental variables, the potential regional distributions of ecologically suitable areas for the three Coptis taxa were predicted, and the environmental factors that significantly impacted their native distributions and genetic structure were identified. This study provided fundamental data and directions for germplasm conservation and breeding improvement of Coptis resources in future.
2 Materials and methods
2.1 Plant materials and DNA extraction
A total of 307 C. chinensis (including both cultivated and wild plants), 15 wild C. teeta, and 23 wild C. chinensis var. brevisepala were collected from 32 localities in China (Supplementary Table 1). Genomic DNA was extracted from the fresh or dried leaf tissues (using silica gel) using a Plant Genomic DNA Extraction Kit (BioTeke, Wuxi, China) in accordance with the manufacturer’s instructions. The quality and purity of the extracted DNA were evaluated using a NanoDrop 2000 spectrophotometer (Thermo Scientific, USA) and 1% agarose gel electrophoresis (Ouyang et al., 2018). Each DNA sample had a final concentration of approximately 50 ng/μL. The samples were subsequently used for PCR amplification (Liu et al., 2020).
2.2 Identification and experimental validation of SSRs
Based on C. chinensis genome data (Accession number: GCA_15680905.1) downloaded from the NCBI database (Liu et al., 2021), SSRs were searched using the MISA software (http://pgrc.ipk-gatersleben.de/misa/), and the parameters were set to as follows: mono-, di-, tri-, tetra-, penta-, and hexa-nucleotides with minimum repeat numbers of 10, 6, 5, 5, 5, and 5, respectively. Compound SSRs were identified as any two or more SSRs that had a maximum interruption of three bases. SSR primers were designed using Primer Premier 5.0 (http://www.premierbiosoft.com/). Given that dinucleotide and trinucleotide SSRs comprise the majority of highly polymorphic sites in plants (Wang et al., 2014), 180 pairs of dinucleotide and trinucleotide-based primers were generated. The M13 universal connector sequence (GTAAAACGACGGCCAGT) was added to the forward primers 5’ to supplement the generation of the reverse primers. The corresponding M13 connectors were modified by four different fluorescent groups: FAM (blue), HEX (green), ROX (red), and TAMRA (yellow).
The continually adjusted and optimized reaction system contained 2 µL genomic DNA, 5 µL 2 × Taq PCR MasterMix, 0.05 µL forward primer, 0.24 µL reverse primer, 0.15 µL M13 connector (FAM/HEX/ROX/TAMRA), and 2.56 µL ultrapure water. The mixed PCR amplifications were performed in a BiometraTone 96G PCR cycler (Analytik Jena AG, Jena, Germany) with a PCR amplification cycle of 94 °C pre-denaturation for 5 min, followed by 35 cycles of 94 °C for 30 s, 55 °C annealing for 30 s, 72 °C extension for 30 s, and a 72 °C extension for 10 min. The PCR products were then identified using an ABI 3730xl Genetic Analyzer Sequencer (Sangon Biotech Co., Ltd., Shanghai, China), the outputs were analyzed in the software GeneMarker v2.2.0 (SoftGenetics, State College, Pennsylvania, USA), and the peak information were recorded in diploid form.
2.3 Screening of species occurrence data and environmental variables
In addition to the 32 localities from which Coptis plants had been sampled, 204 occurrence points were identified from previously published articles as well as the public databases of the China National Knowledge Infrastructure (CNKI) (https://www.cnki.net), the China Digital Plant Herbarium (CVH) (https://www.cvh.ac.cn), and the Global Biodiversity Information Facility (GBIF) (https://www.gbif.org). Subsequently, MaxEnt modeling (Steven et al., 2006) was performed using datasets detailing the occurrences of 142 C. chinensis, 30 C. teeta, and 64 C. chinensis var. brevisepala (Supplementary Table 2).
Coptis plants prefer cold, humid, and semi shaded natural environments (Li et al., 2020b). Based on their growth habits, this study focuses on environmental variables such as altitude, temperature, light, rainfall, and water vapor pressure. A suite of bioclimatic variables were retrieved from the WorldClim database (https://www.worldclim.org). These included altitude, 19 ‘bioclimatic’ variables that were measured from 1970 to 2000 (BIO1: Annual Mean Temperature; BIO2: Mean Diurnal Range; BIO3: Isothermality; BIO4: Temperature Seasonality; BIO5: Max Temperature of Warmest Month; BIO6: Min Temperature of Coldest Month; BIO7: Temperature Annual Range; BIO8: Mean Temperature of Wettest Quarter; BIO9: Mean Temperature of Driest Quarter; BIO10: Mean Temperature of Warmest Quarter; BIO11: Mean Temperature of Coldest Quarter; BIO12: Annual Precipitation; BIO13: Precipitation of Wettest Month; BIO14: Precipitation of Driest Month; BIO15: Precipitation Seasonality; BIO16: Precipitation of Wettest Quarter; BIO17: Precipitation of Driest Quarter; BIO18: Precipitation of Warmest Quarter; BIO19: Precipitation of Coldest Quarter), as well as monthly measurements of precipitation (P1-12), solar radiation (SR1-12), water vapor pressure (WVP1-12) and temperature (T1-12). All variables had a spatial resolution of 2.5 arc-minutes. Using data on the occurrences of Coptis species and 68 environmental parameters, the statistical significance of each variable in predicting the occurrences of Coptis species was assessed using the Jackknife approach. Next, Pearson correlation coefficients were calculated between the 68 environmental variables, and statistically significant associations between environmental variables were identified based on a cutoff of |R| ≥ 0.80 (Shcheglovitova and Anderson, 2013). For each pair of substantially correlated environmental variables, only the variable with the larger contribution was retained in the model (Yang et al., 2021; Wei et al., 2021a).
2.4 Model reliability test and classification of suitable regions
Maximum entropy models for C. chinensis, C. teeta, and C. chinensis var. brevisepala were constructed using the MaxEnt v3.4.1 software (Phillips et al., 2017). To ensure that the distributions of the different Coptis species would approximate a normal probability distribution, 75% of the data was used for model training, while the remainder was used for model testing. To reduce the likelihood of model errors, the maximum number of parameters was set to 10,000, and each method was repeated ten times; all other parameters were set to default (Sun et al., 2020; Yan et al., 2020). The accuracies of model predictions were assessed using the area enclosed by the receiver operating characteristic curve (AUC), which had a range of (0, 1). Increasing AUC values indicate higher credibility in distinguishing between appropriate and inappropriate situations, with an AUC value of 0.9 indicating a very accurate model prediction (Guo et al., 2019; Liu et al., 2019).
The result of each MaxEnt model was converted into grid data and imported into ArcGIS 10.5. A reclassification process was then applied to identify ecologically suitable regions for a species. According to previous research results, the maximum test sensitivity plus specificity (MTSPS) criterion is preferable to other threshold choices for the classification of acceptable locations (Tang et al., 2018). Areas with suitability values below the threshold were declared unsuitable in line with the MTSPS criterion. Three equally sized sections of a species’ range of suitability between the MTSPS and 1 were chosen to represent the regions of low, moderate, and high suitability, respectively (Ye et al., 2018; Wei et al., 2021b).
2.5 Statistical analysis
Based on peak position data, GenALEx6.5 software (Peakall and Smouse, 2012) was used to calculate the following parameters of genetic diversity: the number of alleles (Na), the effective number of alleles (Ne), the observed heterozygosity (Ho), the expected heterozygosity (He), the value of Shannon’s information index (I), the inbreeding coefficient (Fis), the values between pairs indicative of population divergence (Fst) and the gene flow between populations (Nm). In addition, GenALEx6.5 was used for the analysis of molecular variance (AMOVA) and a Mantel correlation test between the Nei genetic distances and the geographical distances among separate populations. Polymorphism information content (PIC) was calculated using PowerMarker version 3.25 (Liu and Muse, 2005). To avoid the impact of a small sample size, 19 Coptis populations with a sample size greater than 5 were selected for the calculation of population genetic diversity related parameters. Adjusted genetic information data to “0/1” format, with peaks marked as “1” and no peaks marked as “0”. Based on the “0/1” dataset, Analysis of Phylogenetics and Evolution (APE) package (Paradis and Schliep, 2019) in R software was used to generate a neighbor-joining (N-J) tree of all 345 Coptis samples. A principal coordinates analysis (PCoA) was also performed based on the genetic distance matrix calculated by GenALEx6.5. Population structure was established using STRUCTURE version 2.3.4 (Pritchard et al., 2000). Subsequently, the optimal K value by the method of Evanno (Evanno et al., 2010) was identified by importing the compressed package of the result file into Structure Harvester (https://taylor0.biology.ucla.edu/structureHarvester/).
Geographical populations of Coptis species displaying distinct genetic structures were used in investigations of the relationship between environmental factors and the genetic structures of Coptis plants. The eight environmental factors with the largest contributions to the modeled distribution of each species were chosen. Data for environmental variables were retrieved for each occurrence point using ArcGIS 10.5. The Wekemo Biocloud cloud service (https://bioincloud.tech) was then employed to perform a redundancy analysis (RDA) or a canonical correspondence analysis (CCA) based on the gradient length calculated by a trend correspondence analysis (DCA). To explore the driving effects of high contribution environmental variables on the genetic variation of Coptis species and cultivated C. chinensis, six groups (Group I, II, III-1, III-2, III-3 and III-4) and three subgroups (Group III-2, III-3 and III-4) were analyzed, respectively. Specifically, RDA was used when gradient length < 3.0, CCA was used when gradient length > 4.0, and both methods were used for any gradient length that ranged from 3.0 – 4.0.
3 Result
3.1 Identification and characterization of SSRs
A total of 340,387 SSRs belonging to six classes of microsatellites were detected in the C. chinensis genome, with an average of one SSR per 217.5 bp (Supplementary Tables 3, 4). Of these SSRs, 48,923 were compound SSR loci with motifs that contained two types of repeats (14.37%). The most abundant type of repeat motif was mononucleotide (238,931, 70.20%), followed by dinucleotide (66,049, 19.4%) and trinucleotide (29,001, 8.52%) (Supplementary Tables 3, 4). The repeat number of different SSR motif types varied considerably, from 5 (5.59%) to 12 (6.92%). The three most frequently occurring SSR types had major repeats of 10, 6 and 5 for mononucleotides, dinucleotides, and trinucleotides, respectively (Supplementary Table 5). In addition, the A/T repeat was dominant in mononucleotides (95.16%). In comparison, the AT/AT repeat was more prevalent in dinucleotides (57.18%), while the AAT/ATT repeat type accounted for 44.17% of trinucleotides (Supplementary Table 6).
Primer pairs from 180 randomly selected SSRs were designed and tested for their ability to detect polymorphisms in the Coptis samples. Of these, 22 SSR primer pairs showed high amplification efficiency, good reproducibility, and high polymorphism between samples, while 72 SSRs failed to amplify any product. The 22 SSRs with high polymorphism were thus used for further genetic analysis (Supplementary Table 7). These SSRs resulted in a total of 361 alleles, with the observed number of alleles (Na) varying from 6 (Ccssr49) to 31 (Ccssr3), and an average of 16.41 alleles per locus (Supplementary Table 8). The Ccssr3 locus was associated with the highest level of genetic variation, followed by the Ccssr53 locus. These loci showed high recognition efficiency and will be useful markers for distinguishing closely related Coptis species.
3.2 Genetic diversity and differentiation
The genetic diversity and differentiation of 19 Coptis geographical populations were estimated (Figure 1; Table 1). The average genetic diversity in C. chinensis populations was higher than that in C. teeta populations and C. chinensis var. brevisepala populations, as indicated by both the expected heterozygosity (He) and Shannon’s information index (I). However, wild and cultivated C. chinensis populations did not differ significantly in their levels of genetic diversity. Deviating from expectation of the Hardey-Weinberg principle, the observed heterozygosity (Ho) of the two C. teeta populations TYGSW and TYJPW was considerably higher than expected, while that of the C. chinensis var. brevisepala population BAHSW was considerably lower than expected. This research also compared the level of genetic diversity in samples of cultivated C. chinensis collected from three provinces: Hubei (HB), Sichuan (SC), and Chongqing (CQ). The results showed that the level of genetic diversity in cultivated accessions from HB (I = 1.475, n = 103) was slightly higher than that from SC (I = 1.371, n = 43) and CQ (I = 1.334, n = 43); this was in line with their differences in sample size (Table 2).
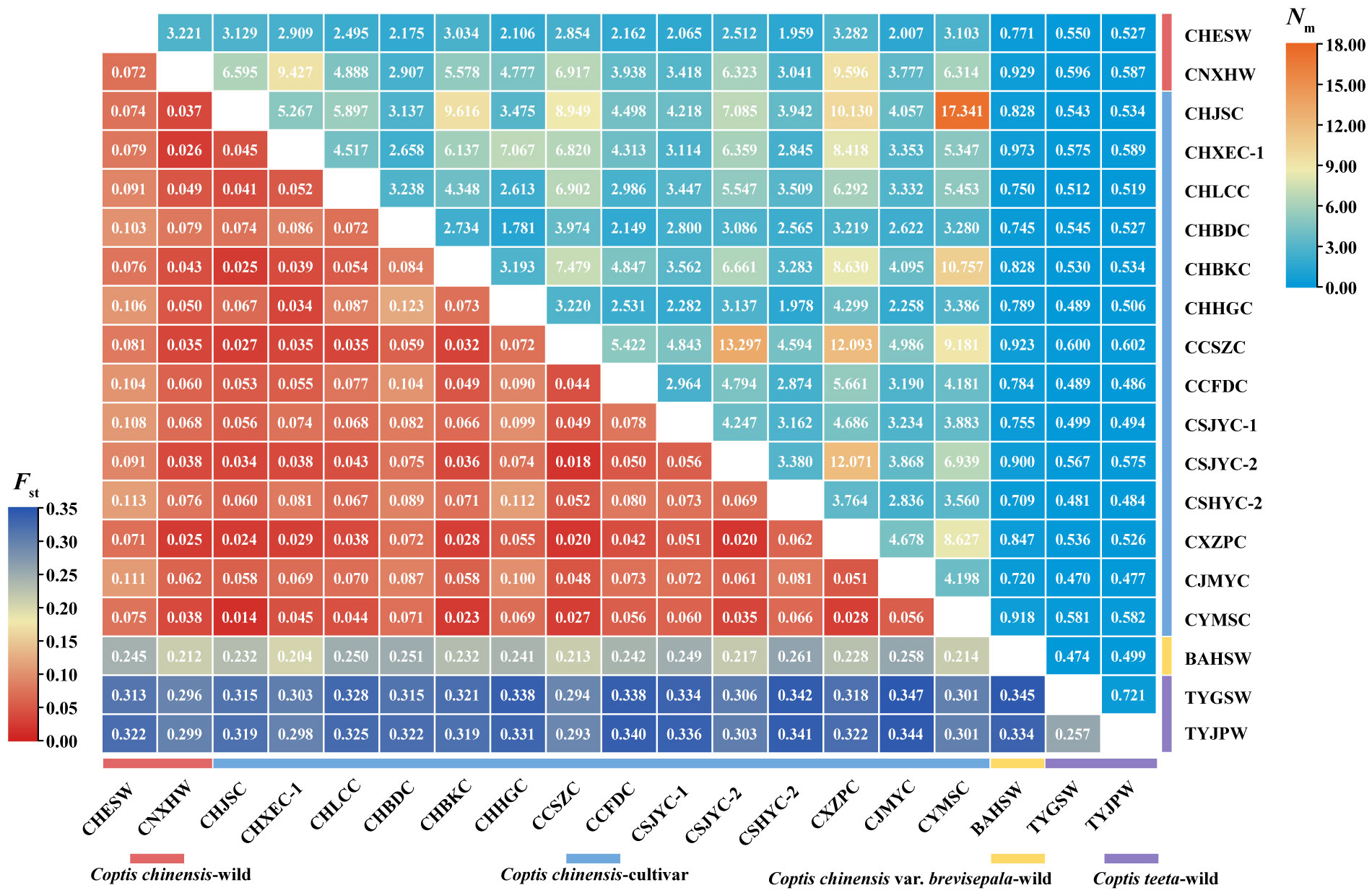
Figure 1 Heatmap of pairwise Fst and Nm values between different geographically Coptis populations. Numbers in the square were the pairwise Fst and Nm values between the populations in horizontal and vertical correspondingly. Colored ranges indicated different germplasm types.
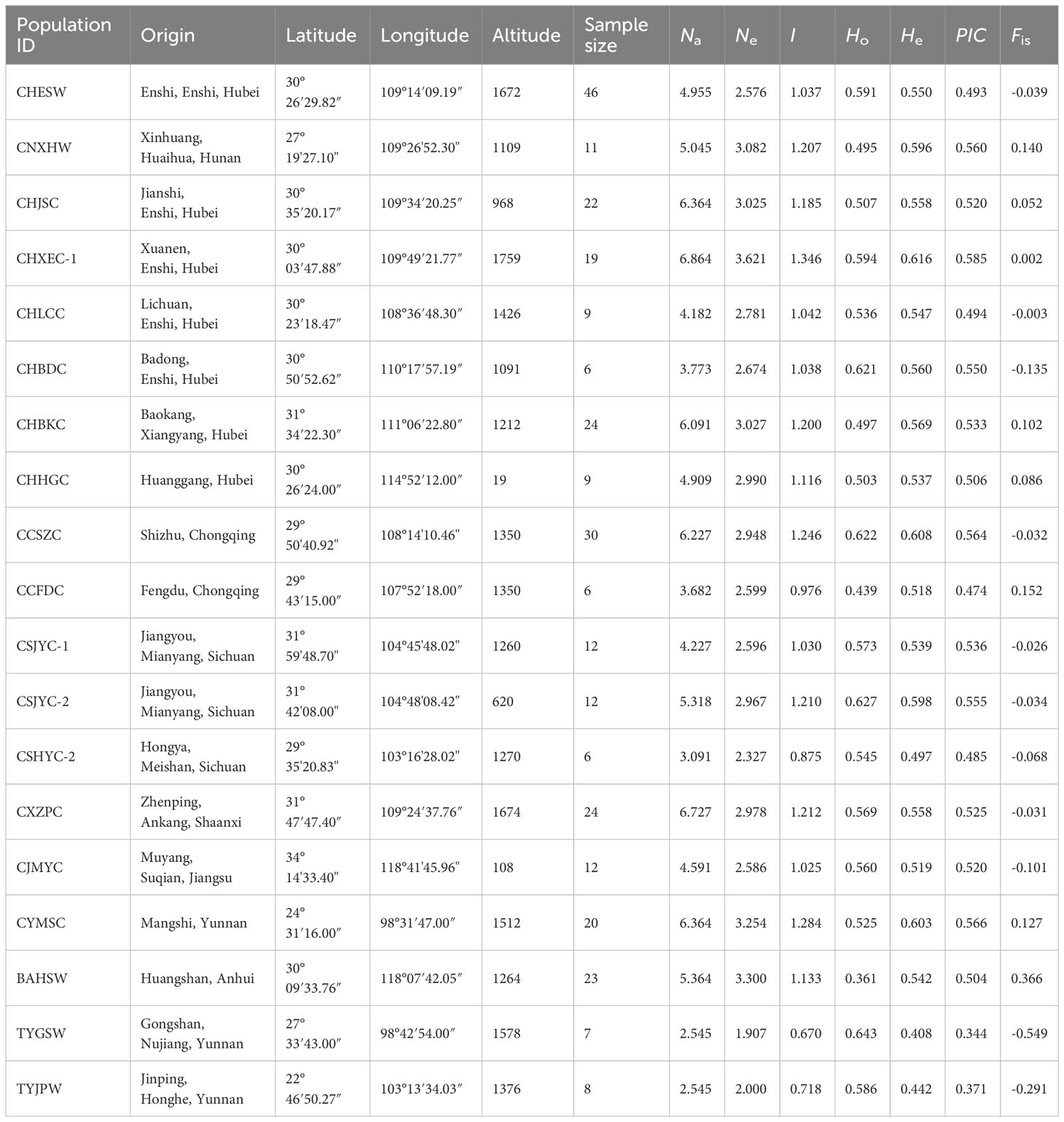
Table 1 Geographical origin and genetic diversity of 19 Coptis populations with sample size more than five.
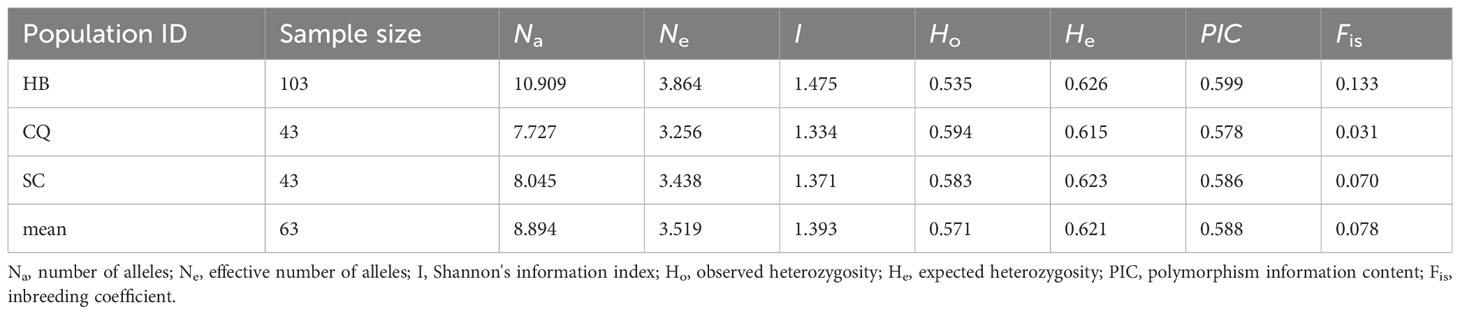
Table 2 Genetic diversity of the cultivated C. chinensis samples in three provinces of Hubei (HB), Sichuan (SC) and Chongqing (CQ).
Very weak genetic differentiation was observed between the cultivated populations of C. chinensis (average pairwise Fst = 0.058), while that between two wild populations of the species (CHESW and CNXHW) was slightly higher (Fst = 0.072). In addition, CNXHW was found to be more genetically related to the cultivated populations of C. chinensis (average pairwise Fst = 0.049) than CHESW (average pairwise Fst = 0.092). A high level of genetic differentiation was observed between the two C. teeta populations, TYGSW and TYJPW (Fst = 0.257) (Figure 1). Importantly, the C. chinensis var. brevisepala population BAHSW showed a high level of genetic differentiation from the C. chinensis (average pairwise Fst = 0.234) and C. teeta populations (average pairwise Fst = 0.340). The level of genetic differentiation between the C. chinensis and C. teeta populations was also high (average pairwise Fst = 0.340) (Figure 1). The pairwise Nm value ranged from 0.470 to 17.341. A high level of gene flow was observed between cultivated C. chinensis populations, such as that between CHJSC and CYMSC (Nm = 17.341) and between CCSZC and CSJYC-2 (Nm = 13.297) (Figure 1). In particular, a very high level of gene flow (Nm > 12) was observed between CCSZC, CSJYC-2, and CXZPC, suggesting these populations were genetically related (Figure 1). The AMOVA analysis further showed that most genetic variation occurred within populations of the respective Coptis species. Nonetheless, relatively high levels of genetic variation among populations of C. teeta were also observed (Supplementary Table 9). A Mantel test showed a significant positive relationship (R2 = 0.2072, P = 0.030) between the genetic and geographical distances of all 19 Coptis populations. However, when the Mantel test was conducted for the C. chinensis populations only, there was no correlation between their genetic and geographical distances (R2 = -6.3777, P = 0.420) (Supplementary Figure 1).
3.3 Genetic relationship and structure of Coptis accessions
An N-J tree grouped all 345 Coptis accessions into three major groups (I, II and III) (Figure 2A). The wild populations of C. teeta and C. chinensis var. brevisepala formed their own groups, namely Group I and Group II, respectively. All C. chinensis individuals were clustered into the larger group, Group III. Within Group III, the wild C. chinensis population CHESW from Hubei was relatively independent, while most individuals of the wild C. chinensis population from Hunan (CNXEW) were mixed with cultivated accessions. These findings were consistent with the lower Fst and higher Nm values observed between CNXEW and the cultivated populations (Figure 1). The results of a PCoA analysis further supported the results of the N-J tree (Figure 2B).
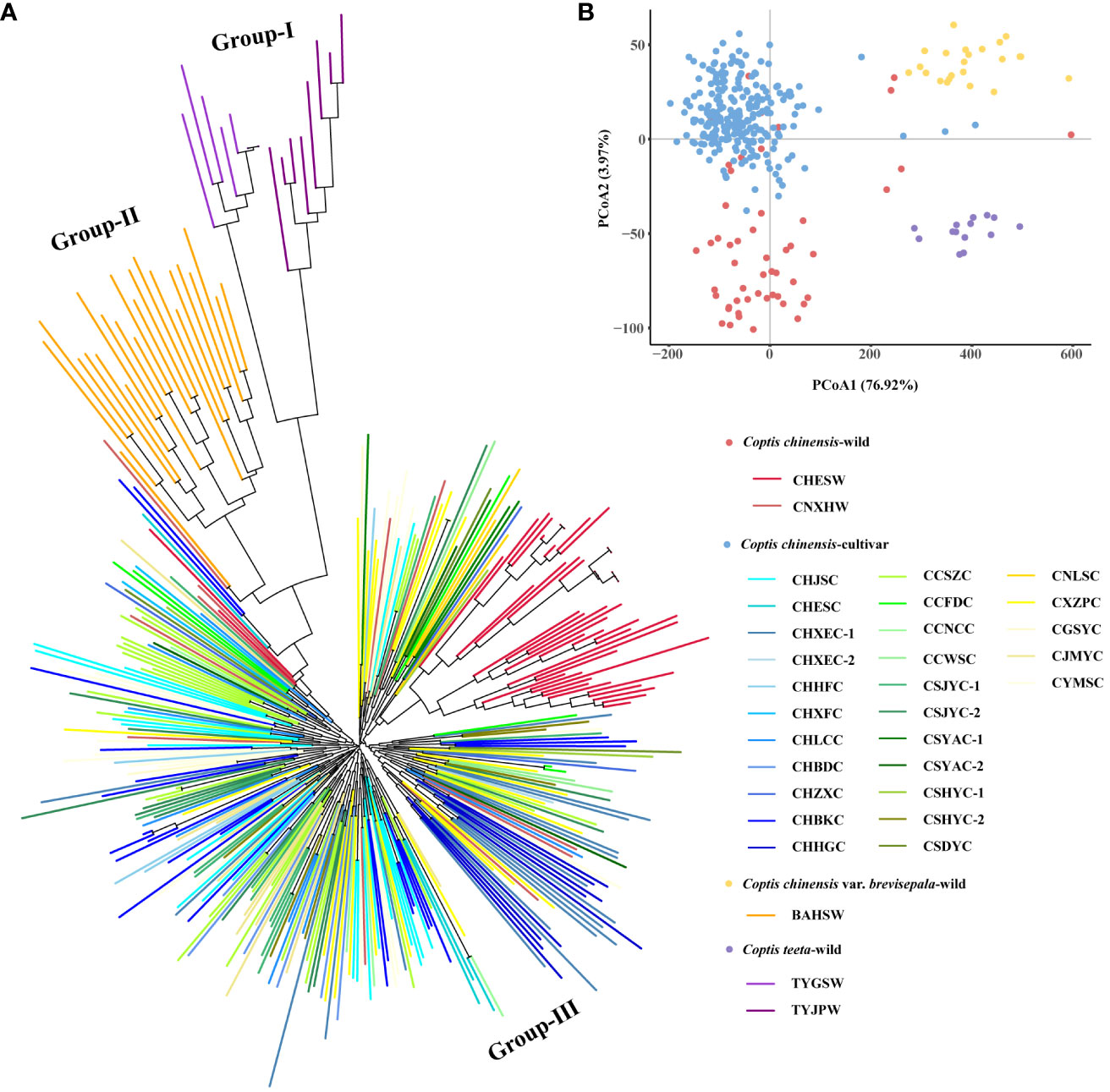
Figure 2 N-J tree (A) and PCoA (B) analysis of 345 accessions. Colored ranges indicated different germplasm types.
In line with the result of the cluster analysis, the STRUCTURE analysis showed that the best population subdivision for all Coptis accessions was achieved when K = 3, corresponding to the three taxa well (Figure 3A). For both wild and cultivated C. chinensis samples in Group III (n = 307), further structural analysis revealed that the most suitable number of subgroups was K = 4 (Figure 3B). Almost all accessions from the wild C. chinensis population in Hubei (CHESW) belonged to Group III-1. The genetic structures of C. chinensis accessions from Sichuan and Chongqing, which were almost unanimous, constituted Group III-4. The wild and cultivated accessions from Hunan constituted Group III-2. In comparison, the cultivated C. chinensis populations from Hubei constituted a genetic admixture, with similar proportions of samples from Groups III-2, 3 and 4 (Figure 3C). This was consistent with the high level of genetic diversity documented in cultivated C. chinensis populations in Hubei (I = 1.475).
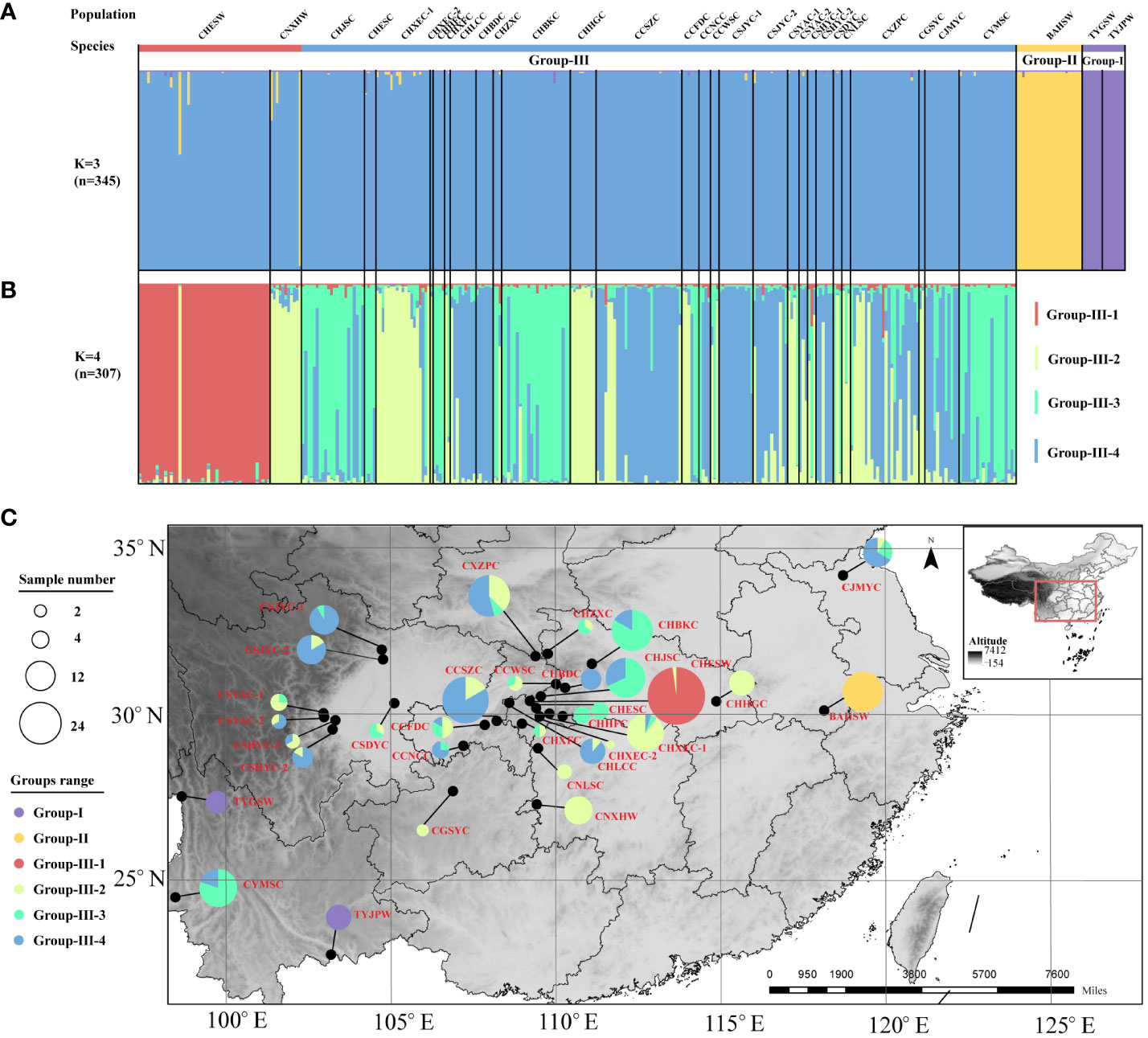
Figure 3 STRUCTURE analysis based on the entire SSR dataset. (A) The population structure of Coptis accessions (n = 345) when K = 3. (B) The population structure of C. chinensis accessions (n = 307) when K = 4. (C) The geographic distribution of the 32 Coptis populations inferred with STRUCTURE analysis. The different colors of the pie charts represented the proportions of the populations in the 6 Groups. Different circle sizes represented sample sizes.
3.4 Potentially distribution of ecologically suitable areas and functional environment variables
Under current climatic conditions, the distributions of suitable areas for the three Coptis species show little overlap (Figure 4). Areas that are predicted to be suitable for C. chinensis primarily occur in Hubei, Sichuan, Shaanxi, Hunan, Guizhou and Chongqing. Among these, highly suitable areas for C. chinensis are concentrated in two regions. The first region corresponds to the intersection of western Hubei, eastern Chongqing, northwestern Hunan, southern Shaanxi and northeastern Guizhou, while the second region is eastern Sichuan (Figure 4A). In comparison, the areas that are predicted to be suitable for C. teeta and C. chinensis var. brevisepala are considerably smaller and independent from those that are suitable for C. chinensis (Figures 4B, C). The distribution of suitable areas for C. chinensis var. brevisepala is relatively scattered, with highly suitable areas sporadically distributed across the provinces of Guizhou, Guangxi, Hubei, Hunan, Anhui, Zhejiang and Fujian (Figure 4B). Areas suitable for C. teeta are mainly distributed in Yunnan and eastern Tibet, and highly suitable areas of this species are restricted to the Gaoligong Mountains (Figure 4C).
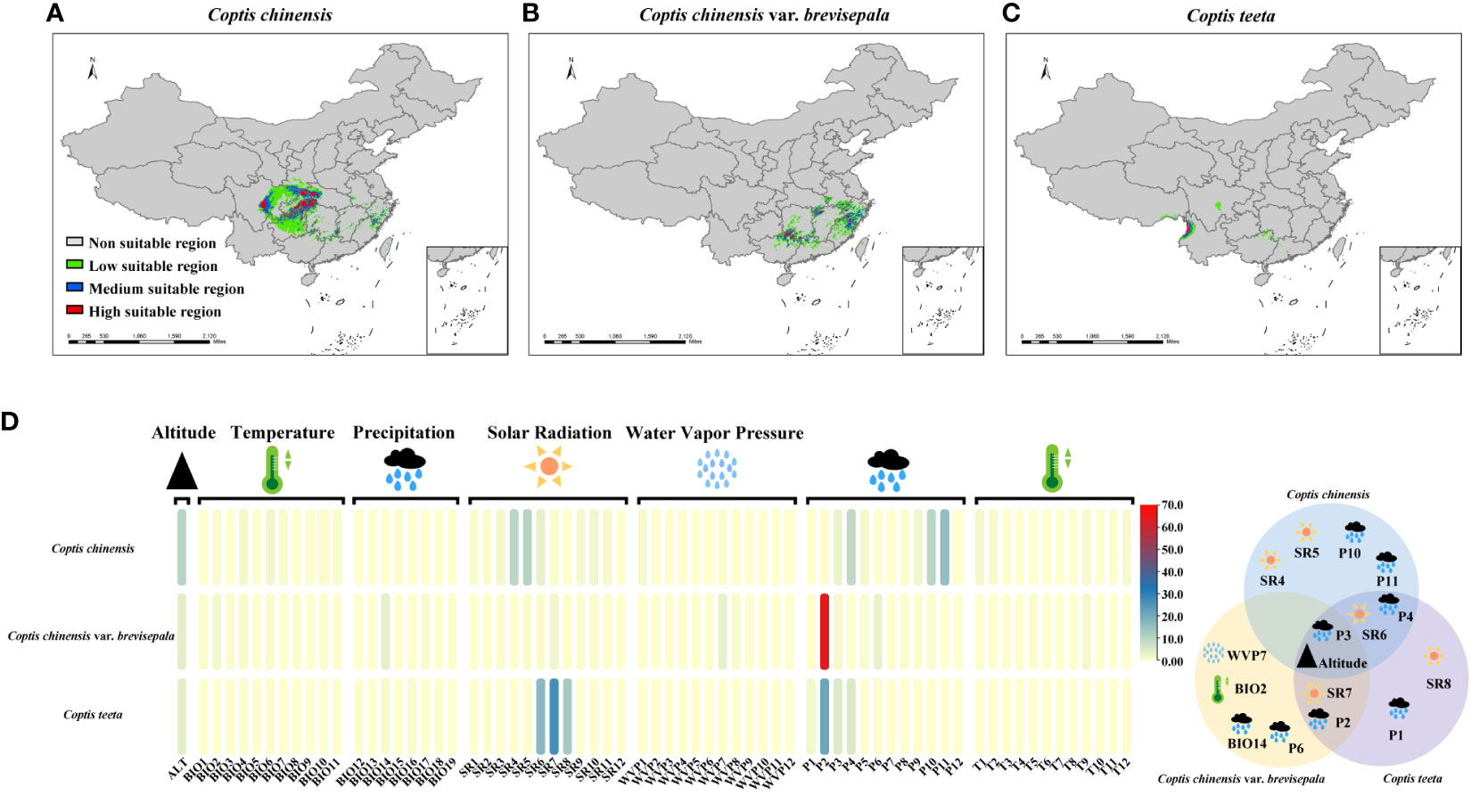
Figure 4 Analysis of ecological suitability of three Coptis species. The colored ranges indicated different species and suitable areas at different levels. The potential suitable distribution of C. chinensis (A), C. chinensis var. brevisepala (B) and C. teeta (C) in China. (D) Contribution of 68 environmental variables to the distribution of three Coptis species. (E) Venn diagram of 16 contributing environmental variables affecting the distribution of three Coptis species. Effects of environmental variables classified into five categories (temperature, precipitation, solar radiation, water vapor pressure and altitude) on the distribution of Coptis species.
To investigate environmental factors influencing the distributions of the three Coptis taxa, 68 environmental variables indicative of temperature (BIO1-11, T1-12), precipitation (BIO12-19, P1-12), solar radiation (SR1-12), water vapor pressure (WVP1-12) and altitude were analyzed. In general, variables relating to solar radiation and precipitation were found to be the main factors determining the current distribution of Coptis species; these were followed by variables relating to altitude and temperature (Figure 4D; Supplementary Table 10). In terms of seasonality, the most important variable affecting the distribution of C. chinensis in spring and early summer was the amount of solar radiation (25.2% in March, April, May, and June), while that for spring and autumn was precipitation (13.4% in March, April, and May and 28% in September, October, and November). Likewise, the dominant variable determining the distribution of C. teeta in summer was solar radiation (56.3% in June, July, and August), while that for late winter and early spring was precipitation (33% in December, January, February, and March). In comparison, the distribution of C. chinensis var. brevisepala was found to be influenced most by the level of precipitation in February, which accounted for 64.6% of variation in the species’ distribution.
3.5 Environmental variables related to population structure
Based on the results of the STRUCTURE analysis, 22 geographical populations (in each population ≥ 75% samples had a similar genetic background) of Coptis species belonging to six distinct genetic groups were selected for a study on the relationship between environmental and genetic variation in Coptis plants. Sixteen environmental variables, corresponding to the top eight environmental variables influencing the distribution of each of the three Coptis species, were selected for the study (Figure 4E). Of these, six variables had a large effect on the distribution of at least two species, while altitude and the level of precipitation in March significantly affected the distributions of all three Coptis species.
A CCA analysis of the sample allele frequencies from the six genetic groups and the 16 environmental variables showed that variables relating to precipitation (P1, P2, P3, P6) had longer arrows that were directed towards Group II, suggesting a strong influence (explanatory value) of these environmental variables on the genetic differentiation of C. chinensis var. brevisepala from the other Coptis species (Figure 5A). In addition, the variables SR4, SR5, P2 and altitude displayed long arrows at a small angle from Group I, indicating that these had a strong influence on the genetic differentiation of C. teeta (Figure 5A). The variable P2 had a small angle with both C. chinensis var. brevisepala and C. teeta, and was more inclined towards the former. This was consistent with the results of the ecologically suitable analysis, which showed that P2 was an important variable shaping the distributions of all three species, especially C. chinensis var. brevisepala (Figure 4D).
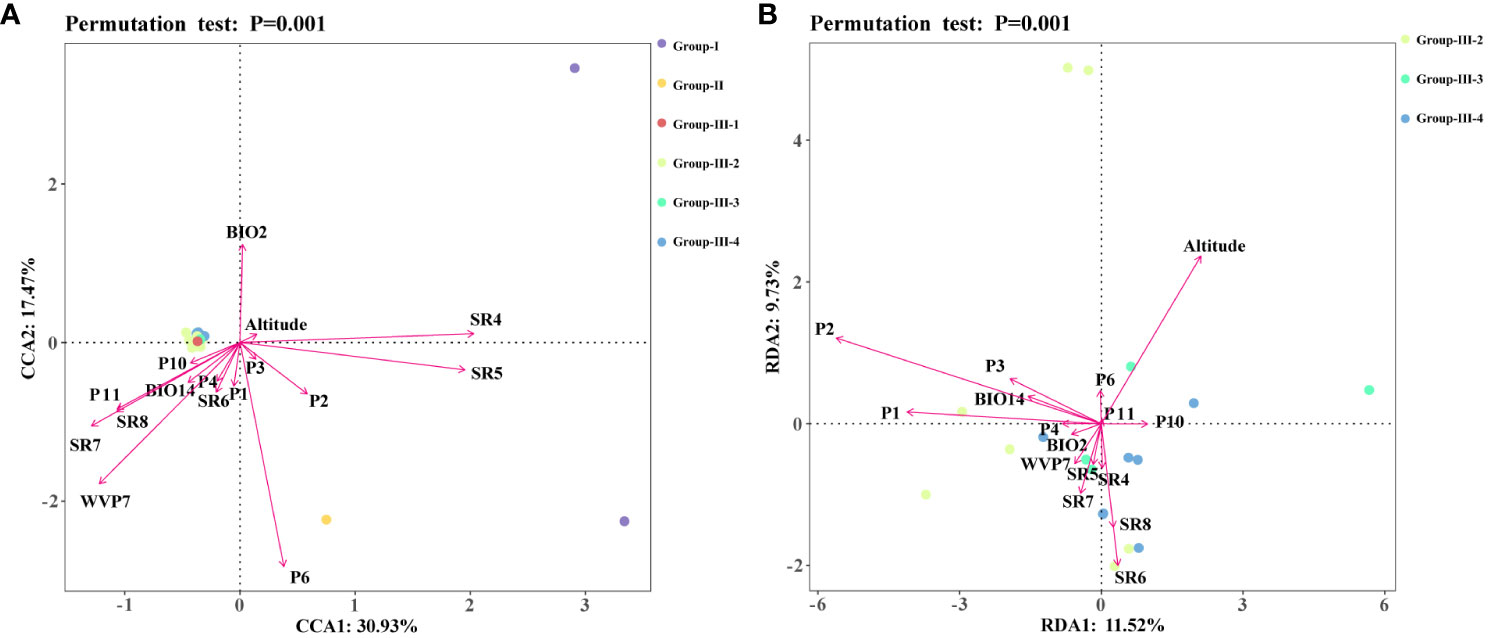
Figure 5 Redundancy analysis (RDA) or canonical correlation analysis (CCA) of 16 high contribution environmental variables and genetic structures. Different colored dots represented different Groups, and different arrows represented environmental variables. (A) The results of CCA analysis. (B) The results of RDA analysis.
The RDA results for three cultivation subgroups showed that the level of genetic variation in Group-III-4 was positively correlated with the amount of solar radiation in spring and summer (Figure 5B). In comparison, nearly all of the 16 environmental variables were found to influence the genetic differentiation in Group III-2 and Group 3 to different degrees. Overall, the results showed that the main environmental factors influencing the genetic variation and domestication history of C. chinensis are altitude, solar radiation and precipitation (Figure 4E).
4 DISCUSSION
4.1 Population genetic diversity and structure
The results revealed relatively high levels of genetic diversity in both wild and cultivated Coptis germplasms. In particular, the observed heterozygosity of the C. chinensis var. brevisepala population (BAHSW) was considerably lower than expected (Table 1). Moreover, this population had a high inbreeding coefficient (Fis = 0.366) (Table 1). Indeed, inbreeding could have occurred among the small natural populations of C. chinensis var. brevisepala, which have a restricted distribution in the Middle and Lower Yangtze Valley Plain. In contrast, the observed heterozygosity of the two wild C. teeta populations was higher than expected. Given that C. teeta is a species adapted to the environments of high mountains and deep valleys in the Hengduan Mountains and the Yunnan-Guizhou Plateau, geographical isolation and limited gene flow (Figure 1) could have accelerated the genetic differentiation among C. teeta individuals. This result was further supported by the high genetic differentiation (Fst = 0.257) observed between the two wild C. teeta populations, TYGSW and TYJPW.
Collectively, the results of the N-J tree, PCoA and STRUCTURE analyses supported the existence of independent genetic groups for C. teeta, C. chinensis var. brevisepala and C. chinensis, suggesting that substantial genetic differentiation has occurred between these Coptis taxa. A further delineation of the subgroups of C. chinensis by structure analysis yielded four subgroups. Among these, Group III-1 comprised the wild C. chinensis samples (CHESW) collected from Enshi, Hubei. The CHESW population germplasm may represent the early main domestication source in the authentic region of Hubei. Under the optimal number of subgroups (K = 4), cultivated C. chinensis were divided into three subgroups, indicating that there may be other domestication centers besides the CHESW population, and domestication in different regions may not be completed simultaneously. For example, another wild population, CNXHW, was found to be genetically similar to the cultivated C. chinensis samples, as evidenced by both the N-J tree and the STRUCTURE analysis. It can be speculated that the CNXHW population comprises individuals that were accidentally established in the wild from propagules of cultivated individuals domesticated in early stages rather than genuine primitive wild C. chinensis. Alternatively, CNXHW population may be a relatively late-stage domestication center. This research also found that the levels of genetic diversity among wild and cultivated C. chinensis populations did not differ significantly. This is consistent with previous claims that the genetic diversity of cultivated C. chinensis populations have not been eroded by domestication (Shi et al., 2008).
The cultivated C. chinensis germplasm was divided into three subgroups (Group III-2, 3 and 4), with each group showing a distinct geographical distribution. This indicated that various climatic, habitat and geomorphological conditions have influenced the genetic constituents of cultivated C. chinensis germplasms to some degree. However, that no particularly clear boundary was apparent between the clustering patterns of cultivated germplasm and different habitats suggested that long-term domestication and cultivation had led to gene transfer and germplasm exchange among groups in different regions (Wang et al., 2020; Wang et al., 2022b). For example, several cultivated populations that were geographically separated (CYMSC from Yunnan and CHJSC from Hubei; CSJYC-2 from Sichuan, CCSZC from Chongqing and CXZPC from Shaanxi) from one another showed a lower Fst value and a higher Nm value, suggesting that artificial introduction and cultivation had occurred. This was also fully consistent with the results of the analysis of their population structure at K = 4 (n=307).
Owing to rampant overexploitation over the long term, most wild Coptis populations are currently at risk of extinction. C. chinensis has been cultivated and domesticated over a long period of time through mutual introduction and commercial trade in different regions. The Mantel test showed a lack of genetic variation among populations of C. chinensis cultivated in different regions (Supplementary Figure 1). This suggests that a small genetic bottleneck event possibly occurred during the domestication of C. chinensis. This analyses further revealed that the level of genetic variation within populations was higher than that among populations, as was reported for Angelica sinensis (Liu et al., 2020).
4.2 Ecological requirements of three Coptis species
Previously, based on an analysis incorporating BIO1-19, Zhao et al. reported that the potentially suitable areas for C. chinensis were concentrated in the region extending from the Sichuan Basin to the middle and lower reaches of the Yangtze River (Zhao et al., 2021), and the distributions of potentially suitable areas for three Coptis species as well as the potential effects of climate change were also modelled in recent studies (Li et al., 2020b; Wang et al., 2022a).
In the present study, a comprehensive suite of 68 environmental variables (including BIO1-19) was used to predict the distribution of suitable areas for three Coptis species. Based on the present findings, the distribution of areas suitable for C. chinensis is generally consistent with those reported previously (Zhao et al., 2021). Furthermore, the results showed that highly suitable areas for the species are mainly distributed across the Sichuan Basin and in several plains located in the middle and lower reaches of the Yangtze River. The study found that there was a high degree of geographical correspondence between the three red high suitability regions and the three cultivated C. chinensis subpopulations, which further supported the domestication of C. chinensis in different suitable regions and the generation of germplasm with different genetic backgrounds through multiple domestication centers. In addition, the areas suitable for C. chinensis var. brevisepala are concentrated in the middle and lower reaches of the Yangtze River, while most areas suitable for C. teeta are located in Yunnan. The distinct distributions of the three Coptis species further support their independent genetic relationships. Therefore, based on the extensive domestication history of Coptis, C. teeta, which occupies a more basal position in the Coptis phylogeny, has had a longer population history than the other two species, and derived sister species such as C. chinensis, C. omeiensis (Chen) C. Y. Cheng and C. deltoidea as it reached the Sichuan Basin in the northeast, before deriving the variety C. chinensis var. brevisepala in the southeast.
According to experienced breeders, Coptis plants require environments with low temperatures, high levels of humidity and little solar radiation. Therefore, in this study, in addition to BIO1-19 (mainly temperature and precipitation), other environmental variables such as solar radiation, altitude, and water vapor pressure, as well as monthly data, were added as environmental factors. In particular, solar radiation and altitude have high contribution values in the ecological suitability analysis of Coptis plants, indicating their environmental driving role in the adaptive evolution process of Coptis plants to a certain extent. These previous observations are further confirmed by the findings of the present study. Separate environmental variables had different effects on the distributions of the three Coptis species. The environmental variables that most influenced their distributions were altitude, as well as the amount of solar radiation and precipitation, which both varied seasonally. This finding also reflected the extensive effects of environmental change on the evolution of Coptis species. From C. teeta to C. chinensis, solar radiation and precipitation were developed in the directions of summer to spring and spring to autumn, respectively; that is, Coptis species have been continuously adapting to novel seasonal changes in environmental conditions. The differences in monthly environmental variable data in the ecological suitability analysis of three species also reflected that Coptis plants not only adapt to changes in geographical environment, but also include seasonal changes. Such information can help guide the conservation and cultivation of different Coptis species.
4.3 Impacts of environment on genetic structure of three Coptis species
The results demonstrated that genetic variation, the foundation of species survival and biodiversity (Blanquart et al., 2013; Li et al., 2021), is largely influenced by changes in the environment. In the results of the CCA analysis, almost all environmental variables pointed to Groups I, II and III, which were represented by different Coptis species. The difference of solar radiation and precipitation caused by the environment showed key driving forces for the adaptive evolution of Coptis species. This was also consistent with the results of the Mantel test. Similarly, in the RDA analysis of the cultivated C. chinensis populations, no particularly significant correlation between the subgroups and environmental variables. However, considering that C. chinensis has a low domestication bottleneck and has been in introduction and cultivation for a long time, different environmental variables have shown a high driving force on the overall genetic variation of the cultivated C. chinensis populations. Especially the altitude, precipitation in spring, and solar radiation in autumn have formed three different directional clusters guiding the domestication direction of C. chinensis. In particular, the relatively concentrated Group III-4 from the high mountains around the Sichuan Basin was strongly associated with the amount of solar radiation in spring and summer. It can be inferred that the mountains around the Sichuan Basin reach high altitudes and are therefore exposed to different levels of solar radiation. This may have caused the variation in the levels of genetic diversity and in the contents of the active components of different Coptis accessions. Additional investigations on the metabolic components of different Coptis populations will further benefit efforts to identify ideal germplasms for cultivating Coptis species.
5 Conclusion
In this study, a set of highly polymorphic genomic-SSR markers was developed based on the Coptis chinensis genome. A genetic analysis of 345 Coptis accessions from 32 geographical populations based on 22 SSR markers was conducted. A high level of genetic diversity was observed in both wild and cultivated Coptis populations. Most wild populations showed clear levels of genetic differentiation. Additionally, there was evidence of frequent gene flow between various cultivated C. chinensis populations, with signs of multiple artificial introductions. The genetic structure of Coptis was found to be characterized by three major groups, while the C. chinensis germplasm could be further divided into four subgroups. The wild C. chinensis population in Hubei (CHESW) may represent the major domestication center of the species. The distributions of suitable areas for the three Coptis species were predicted, and environmental variables shaping these distributions and the genetic structures of the species were identified. Coptis plants have had enormous medicinal and economic value from ancient times to the present day, but currently face the problem of endangered wild resources and chaotic introduction sources. Our research will provide theoretical support for subsequent resource protection and healthy development patterns tailored to local conditions.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
YC: Formal Analysis, Investigation, Methodology, Resources, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. CL: Data curation, Formal Analysis, Writing – review & editing. WL: Investigation, Supervision, Writing – review & editing. XT: Resources, Validation, Writing – review & editing. JH: Data curation, Software, Writing – review & editing. BW: Funding acquisition, Writing – review & editing. DL: Funding acquisition, Methodology, Validation, Writing – review & editing. YL: Funding acquisition, Investigation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China: 32100299, 81973429; the Funds for Key Program for Traditional Chinese Medicine of Hubei University of Chinese Medicine: 2022ZZXZ002; and the National Key Research and Development Program of China: 2019YFC1711100.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1341996/full#supplementary-material
Supplementary Figure 1 | The mantel test of all populations and C. chinensis populations.
References
Blanquart, F., Kaltz, O., Nuismer, S. L., Gandon, S. (2013). A practical guide to measuring local adaptation. Ecol. Lett. 16, 1195–1205. doi: 10.1111/ele.12150
Chen, C., Chu, Y. G., Ding, C. J., Su, X. H., Huang, Q. J. (2020a). Genetic diversity and population structure of black cottonwood (Populus deltoides) revealed using simple sequence repeat markers. BMC Genet. 21, 2. doi: 10.1186/s12863-019-0805-1
Chen, H., Li, J., He, Y. (2020b). Overexpression of a novel E3 ubiquitin ligase gene from Coptis chinensis Franch enhances drought tolerance in transgenic tobacco. Z Naturforsch. C J. Biosci. 75, 417–424. doi: 10.1515/znc-2019-0211
Chou, S. T., Hsiang, C. Y., Lo, H. Y., Huang, H. F., Lai, M. T., Hsieh, C. L., et al. (2017). Exploration of anti-cancer effects and mechanisms of Zuo-Jin-Wan and its alkaloid components in vitro and in orthotopic HepG2 xenograft immunocompetent mice. BMC Complement Altern. Med. 17, 121. doi: 10.1186/s12906-017-1586-6
Evanno, G., Regnaut, S., Goudet, J. (2010). Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294x.2005.02553.x
Guo, Y. L., Li, X., Zhao, Z. F., Nawaz, Z. (2019). Predicting the impacts of climate change, soils and vegetation types on the geographic distribution of Polyporus umbellatus in China. Sci. Total Environ. 648, 1–11. doi: 10.1016/j.scitotenv.2018.07.465
Han, L., Wang, R. Z., Zhang, X., Yu, X., Zhou, L., Song, T., et al. (2019). Advances in processing and quality control of traditional chinese medicine coptidis rhizoma (Huanglian): A review. J. Aoac Int. 102, 699–707. doi: 10.5740/jaoacint.18-0303
He, S. M., Liang, Y. L., Cong, K., Chen, G., Zhao, X., Zhao, Q. M., et al. (2018). Identification and characterization of genes involved in benzylisoquinoline alkaloid biosynthesis in coptis species. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00731
Li, D. Y., Long, C., Pang, X. M., Ning, D. L., Wu, T., Dong, M. L., et al. (2020a). The newly developed genomic-SSR markers uncover the genetic characteristics and relationships of olive accessions. Peerj 8, e8573. doi: 10.7717/peerj.8573
Li, J., Fan, G., He, Y. (2020b). Predicting the current and future distribution of three Coptis herbs in China under climate change conditions, using the MaxEnt model and chemical analysis. Sci. Total Environ. 698, 134141. doi: 10.1016/j.scitotenv.2019.134141
Li, Y., Cao, K., Li, N., Zhu, G. R., Fang, W. C., Chen, C. W., et al. (2021). Genomic analyses provide insights into peach local adaptation and responses to climate change. Genome Res. 31, 592–606. doi: 10.1101/gr.261032.120
Lin, E. P., Zhuang, H. B., Yu, J. J., Liu, X. Y., Huang, H. H., Zhu, M. Y., et al. (2020). Genome survey of Chinese fir (Cunninghamia lanceolata): Identification of genomic SSRs and demonstration of their utility in genetic diversity analysis. Sci. Rep-UK 10, 4698. doi: 10.1038/s41598-020-61611-0
Liu, B. Y., Gao, X., Ma, J., Jiao, Z. H., Xiao, J. H., Hayat, M. A., et al. (2019). Modeling the present and future distribution of arbovirus vectors Aedes aEgypti and Aedes albopictus under climate change scenarios in Mainland China. Sci. Total Environ. 664, 203–214. doi: 10.1016/j.scitotenv.2019.01.301
Liu, M., Hu, X., Wang, X., Zhang, J. J., Peng, X. B., Hu, Z. G., et al. (2020). Constructing a core collection of the medicinal plant angelica biserrata using genetic and metabolic data. Front. Plant Sci. 11, 600249. doi: 10.3389/fpls.2020.600249
Liu, K., Muse, S. V. (2005). PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21, 2128–2129. doi: 10.1093/bioinformatics/bti282
Liu, Y., Wang, B., Shu, S., Li, Z., Song, C., Liu, D., et al. (2021). Analysis of the Coptis chinensis genome reveals the diversification of protoberberine-type alkaloids. Nat. Commun. 12, 3276. doi: 10.1038/s41467-021-23611-0
Liu, D., Wu, T., Meng, X., Cui, N., Wang, B., Liu, X., et al. (2018). Research on technology regulation of non-polluted cultivation and strategies of coptis herba cultivation. Modern Chin. Med. 20, 1396–1403. doi: 10.13313/j.issn.1673-4890.20180910008
Lv, X., Li, Y., Tang, C., Zhang, Y., Zhang, J., Fan, G. (2016). Integration of HPLC-based fingerprint and quantitative analyses for differentiating botanical species and geographical growing origins of Rhizoma coptidis. Pharm. Biol. 54, 3264–3271. doi: 10.1080/13880209.2016.1223699
Ma, K., Wang, X., Feng, S., Xia, X., Zhang, H., Rahaman, A., et al. (2020). From the perspective of traditional Chinese medicine: treatment of mental disorders in COVID-19 survivors. Biomed. Pharmacother. 132, 110810. doi: 10.1016/j.biopha.2020.110810
Mamut, R., Yisilam, G., Zhang, H. W., Hina, F., Yang, Z. P. (2018). The first complete chloroplast genome of Coptis chinensis var. brevisepala, with implication for the phylogeny of Ranunculaccae. Mitochondrial DNA B 3, 953–954. doi: 10.1080/23802359.2018.1501308
Ouyang, P. Y., Kang, D. L., Mo, X. L., Tian, E. W., Hu, Y. Y., Huang, R. S., et al. (2018). Development and characterization of high-throughput EST-based SSR markers for pogostemon cablin using transcriptome sequencing. Molecules 23, 2014. doi: 10.3390/molecules23082014
Paradis, E., Schliep, K. (2019). Ape 50: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35, 526–528. doi: 10.1093/bioinformatics/bty633
Peakall, R., Smouse, P. E. (2012). GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research–an update. Bioinformatics 28, 2537–2539. doi: 10.1093/bioinformatics/bts460
Perrino, E. V., Mahmoud, Z. N. A., Valerio, F., Tomaselli, V., Wagensommer, R. P., Trani, A. (2023). Synecology of Lagoecia cuminoides L. @ in Italy and evaluation of functional compounds presence in its water or hydroalcoholic extracts. Sci. Rep. 13, 20906. doi: 10.1038/s41598-023-48065-w
Phillips, S. J., Anderson, R. P., Dudik, M., Schapire, R. E., Blair, M. E. (2017). Opening the black box: an open-source release of Maxent. Ecography 40, 887–893. doi: 10.1111/ecog.03049
Poirazidis, K., Bontzorlos, V., Xofis, P., Zakkak, S., Xirouchakis, S., Grigoriadou, E., et al. (2019). Bioclimatic and environmental suitability models for capercaillie (Tetrao urogallus) conservation: Identification of optimal and marginal areas in Rodopi Mountain-Range National Park (Northern Greece). Glob Ecol. Conserv. 17, e00526. doi: 10.1016/j.gecco.2019.e00526
Pritchard, J. K., Stephens, M., Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959. doi: 10.1093/genetics/155.2.945
Shcheglovitova, M., Anderson, R. P. (2013). Estimating optimal complexity for ecological niche models: A jackknife approach for species with small sample sizes. Ecol. Model. 269, 9–17. doi: 10.1016/j.ecolmodel.2013.08.011
Shi, W., Yang, C. F., Chen, J. M., Guo, Y. H. (2008). Genetic variation among wild and cultivated populations of the Chinese medicinal plant Coptis chinensis (Ranunculaceae). Plant Biol. (Stuttg) 10, 485–491. doi: 10.1111/j.1438-8677.2008.00035.x
Steven, J. P., Robert, P. A., Robert, E. S. (2006). Maximum entropy modeling of species geographic distributions. Ecol. Model. 190, 231–259. doi: 10.1016/j.ecolmodel.2005.03.026
Sun, S. X., Zhang, Y., Huang, D. Z., Wang, H., Cao, Q., Fan, P. X., et al. (2020). The effect of climate change on the richness distribution pattern of oaks (Quercus L.) in China. Sci. Total Environ. 744. doi: 10.1016/j.scitotenv.2020.140786
Tang, C. Q., Matsui, T., Ohashi, H., Dong, Y. F., Momohara, A., Herrando-Moraira, S., et al. (2018). Identifying long-term stable refugia for relict plant species in East Asia. Nat. Commun. 9. doi: 10.1038/s41467-018-06837-3
Wang, X., Liu, X. Q., Ko, Y. Z., Jin, X. L., Sun, J. H., Huang, Q. W., et al. (2020). Genetic diversity and phylogeography of the important medical herb, cultivated huang-lian populations, and the wild relatives coptis species in China. Front. Genet. 11. doi: 10.3389/fgene.2020.00708
Wang, Y., Sun, J., Qiao, P., Wang, J., Wang, M., Zhao, Z. Y., et al. (2022a). Evolutionary history of genus Coptis and its dynamic changes in the potential suitable distribution area. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1003368
Wang, Y. H., Sun, J. H., Zhao, Z. Y., Xu, C., Qiao, P., Du, Y. X., et al. (2022b). Multiplexed massively parallel sequencing of plastomes provides insights into the genetic diversity, population structure, and phylogeography of wild and cultivated coptis chinensis. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.923600
Wang, J., Wang, L., Lou, G. H., Zeng, H. R., Hu, J., Wang, S., et al. (2019). Coptidis Rhizoma: a comprehensive review of its traditional uses, botany, phytochemistry, pharmacology and toxicology. Pharm. Biol. 57, 193–225. doi: 10.1080/13880209.2019.1577466
Wang, Z., Yu, G., Shi, B., Wang, X., Qiang, H. (2014). Development and Characterization of Simple Sequence Repeat (SSR) Markers Based on RNA-Sequencing of Medicago sativa and In silico Mapping onto the M. truncatula Genome. PloS One 9, e92029. doi: 10.1371/journal.pone.0092029
Wei, T., Van Treuren, R., Liu, X., Zhang, Z., Chen, J., Liu, Y., et al. (2021a). Whole-genome resequencing of 445 Lactuca accessions reveals the domestication history of cultivated lettuce. Nat. Genet. 53, 752–760. doi: 10.1038/s41588-021-00831-0
Wei, Y. Q., Zhang, L., Wang, J. N., Wang, W. W., Niyati, N., Guo, Y. L., et al. (2021b). Chinese caterpillar fungus (Ophiocordyceps sinensis) in China: Current distribution, trading, and futures under climate change and overexploitation. Sci. Total Environ. 755, 142548. doi: 10.1016/j.scitotenv.2020.142548
Wu, J., Zhang, H., Hu, B., Yang, L., Wang, P., Wang, F., et al. (2016). Coptisine from Coptis chinensis inhibits production of inflammatory mediators in lipopolysaccharide-stimulated RAW 264.7 murine macrophage cells. Eur. J. Pharmacol. 780, 106–114. doi: 10.1016/j.ejphar.2016.03.037
Xu, X., Liu, L., Cao, X., Long, X., Peng, S., Zhang, G. (2022). Network pharmacology and molecular docking analysis on molecular targets and mechanism prediction of huanglian jiedu decoction in the treatment of COVID-19. Digital Chin. Med. 5, 18–32. doi: 10.1186/s13020-022-00576-w
Yamada, Y., Yoshimoto, T., Yoshida, S. T., Sato, F. (2016). Characterization of the promoter region of biosynthetic enzyme genes involved in berberine biosynthesis in coptis japonica. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01352
Yan, H. Y., Feng, L., Zhao, Y. F., Feng, L., Zhu, C. P., Qu, Y. F., et al. (2020). Predicting the potential distribution of an invasive species, Erigeron canadensis L., in China with a maximum entropy model. Glob Ecol. Conserv. 21, e00822. doi: 10.1016/j.gecco.2019.e00822
Yang, Z. B., Bai, Y., Alatalo, J. M., Huang, Z. D., Yang, F., Pu, X. Y., et al. (2021). Spatio-temporal variation in potential habitats for rare and endangered plants and habitat conservation based on the maximum entropy model. Sci. Total Environ. 784. doi: 10.1016/j.scitotenv.2021.147080
Yang, Y., Peng, J., Li, F., Liu, X., Deng, M., Wu, H., et al. (2017). Determination of alkaloid contents in various tissues of coptis chinensis franch. by reversed phase-high performance liquid chromatography and ultraviolet spectrophotometry. J. Chromatogr Sci. 55, 556–563. doi: 10.1093/chromsci/bmx009
Yang, L., Wen, K. S., Ruan, X., Zhao, Y. X., Wei, F., Wang, Q., et al. (2018). Response of plant secondary metabolites to environmental factors. Molecules 23, 1–26. doi: 10.3390/molecules23040762
Ye, X. P., Yu, X. P., Yu, C. Q., Tayibazhaer, A., Xu, F. J., Skidmore, A. K., et al. (2018). Impacts of future climate and land cover changes on threatened mammals in the semi-arid Chinese Altai Mountains. Sci. Total Environ. 612, 775–787. doi: 10.1016/j.scitotenv.2017.08.191
Zhang, J., Chen, T., Wang, J., Chen, Q., Luo, Y., Zhang, Y., et al. (2016). Genetic diversity and population structure in cherry (Cerasus pseudocerasus (Lindl). G. Don) along Longmenshan Fault Zones in China with newly developed SSR markers. Sci. Hortic-Amsterdam 212, 11–19. doi: 10.1016/j.scienta.2016.09.033
Zhang, K. L., Zhang, Y., Tao, J. (2019). Predicting the Potential Distribution of Paeonia veitchii (Paeoniaceae) in China by Incorporating Climate Change into a Maxent Model. Forests 10, 190. doi: 10.3390/f10020190
Keywords: Coptis, genetic diversity, population structure, environment variables, distribution of suitable area
Citation: Chi Y, Liu C, Liu W, Tian X, Hu J, Wang B, Liu D and Liu Y (2024) Population genetic variation and geographic distribution of suitable areas of Coptis species in China. Front. Plant Sci. 15:1341996. doi: 10.3389/fpls.2024.1341996
Received: 21 November 2023; Accepted: 06 March 2024;
Published: 19 March 2024.
Edited by:
Robert Philipp Wagensommer, Free University of Bozen-Bolzano, ItalyReviewed by:
Enrico Vito Perrino, International Centre for Advanced Mediterranean Agronomic Studies, ItalyCarlos I. Arbizu, Universidad Nacional Toribio Rodríguez de Mendoza de Amazonas, Peru
Copyright © 2024 Chi, Liu, Liu, Tian, Hu, Wang, Liu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yifei Liu, liuyifei@hbtcm.edu.cn; Di Liu, liuditcm@126.com
 Yujie Chi1
Yujie Chi1 Xufang Tian
Xufang Tian Bo Wang
Bo Wang Yifei Liu
Yifei Liu