- College of Life Science, Shanxi Normal University, Taiyuan, China
Salt Overly Sensitive 1 (SOS1) is one of the members of the Salt Overly Sensitive (SOS) signaling pathway and plays critical salt tolerance determinant in plants, while the characterization of the SOS1 family in potato (Solanum tuberosum) is lacking. In this study, 37 StSOS1s were identified and found to be unevenly distributed across 10 chromosomes, with most of them located on the plasma membrane. Promoter analysis revealed that the majority of these StSOS1 genes contain abundant cis-elements involved in various abiotic stress responses. Tissue specific expression showed that 21 of the 37 StSOS1s were widely expressed in various tissues or organs of the potato. Molecular interaction network analysis suggests that 25 StSOS1s may interact with other proteins involved in potassium ion transmembrane transport, response to salt stress, and cellular processes. In addition, collinearity analysis showed that 17, 8, 1 and 5 of orthologous StSOS1 genes were paired with those in tomato, pepper, tobacco, and Arabidopsis, respectively. Furthermore, RT-qPCR results revealed that the expression of StSOS1s were significant modulated by various abiotic stresses, in particular salt and abscisic acid stress. Furthermore, subcellular localization in Nicotiana benthamiana suggested that StSOS1-13 was located on the plasma membrane. These results extend the comprehensive overview of the StSOS1 gene family and set the stage for further analysis of the function of genes in SOS and hormone signaling pathways.
1 Introduction
High soil salinity is a major abiotic stress that significantly affects plant growth and ultimately reduces plant productivity by preventing the absorption of water and nutrients (Brindha et al., 2021; You et al., 2022). The Salt Overly Sensitive (SOS) signaling pathway plays an essential role in the response of plants to salt stress. It consists of three components: SOS1, SOS2, and SOS3 (Cheng et al., 2019). SOS1 is a Na+/H+ antitransporter that governs the efflux of Na+ into the root and loading into the xylem vessel for long-distance transport out of the root (Świeżawska et al., 2018). SOS2 exists as a form of protein kinase in the SOS signaling pathway, which in turn activates SOS1 to bring about sodium ion homeostasis and salt tolerance (Ali et al., 2021). SOS3, which encodes an EF-handed Ca2+ binding protein, can sense calcium signals elicited by salt stress, interact with SOS2, and activate SOS2 (Zhu et al., 2021).
SOS1 genes were firstly identified in Arabidopsis (Keisham et al., 2018) and designated as AtNHX1-AtNHX8. AtNHX7 (or AtSOS1) is a critical player in the SOS signaling pathway (Zhao C. et al., 2021). AtSOS1 locates in the plasma membrane (Shi et al., 2000). AtSOS1 is primarily expressed in epidermal cells at the root tip and in the parenchyma at the xylem-symplast boundary of root, stem, and leaf, hinting at the role of this transporter in the extrusion of Na+ into the growing medium and in controlling long-distance Na+ transport in plants (Gao et al., 2016). SOS1 behaves as a homodimer, with each monomer having 12 transmembrane domains at its N-terminal region and a long C-terminal region containing a cytosolic domain, a cyclic nucleotide binding domain, and an auto-inhibitory domain (Wu et al., 1996; Núñez-Ramírez et al., 2012). SOS proteins were involved in the regulation of plant tolerance to salinity (Zhu et al., 1998). Overexpression of SOS1 led to reduction of Na+ accumulation in the xylem and shoot (Shi et al., 2003).
In addition to Arabidopsis, the physiological roles of the associated SOS1 genes have been investigated in cash crop plants, such as soybean, maize, tomato, cotton (Chen et al., 2017; Wang Z. et al., 2021; Zhang M. et al., 2022; Zhou et al., 2022), and so on. In soybeans, significant accumulation of Na+ in the roots of GmSOS1 mutants resulted in an imbalance of Na+ and K+, suggesting that GmSOS1 played a critical role in soybean salt tolerance by maintaining Na+ homeostasis (Zhang et al., 2022). In maize, SOS pathway has a conserved salt tolerant effect, and its components (ZmSOS1 and ZmCBL8) have Na+ regulation and natural variations of salt tolerance, providing an important gene target for breeding salt-tolerant maize (Zhou et al., 2022). However, its role has not yet been investigated in potato (Solanum tuberosum).
Potato is an important crop in human food systems around the world (Dahal et al., 2019; Ceci et al., 2022) and their cultivation and production are often severely threatened by the various environmental stresses such as salinity and pathogens (Li et al., 2021; Yang et al., 2022). Identification and characterization of resistance genes to salt stress would therefore be helpful in improving potato production. Since the role of SOS1 in controlling ion homeostasis has been shown in several plants, this gene family is thought to also be valuable in the salt tolerance mechanism and quality improvement of potato. However, limited efforts have been made to identify gene families in the potato, and their expression patterns and regulatory mechanisms remain unclear.
In this study, we identified and analyzed the SOS1 gene family in potato. Extensive analysis including chromosomal localization, gene structure, and upstream promoter cis-acting elements of these gene family were conducted. The physicochemical properties, motifs, gene ontologies, and phylogenetic relationships between the encoded proteins were predicted using bioinformatics tools. Furthermore, the expression profiles of specific StSOS1s at salt stress were examined using RT-qPCR. In addition, their expression profiles in response to the exogenous phytohormone abscisic acid (ABA), methyl jasmonate (MeJA), gibberellin (GA) and salicylic acid (SA) were also investigated. The results indicate a diverse pattern of responses to abiotic stress via SOS and hormone signaling pathways. It may be beneficial to elucidate the resistance of the potato to abiotic stress, providing some theoretical basis for molecular breeding.
2 Materials and methods
2.1 Plant material and treatments
The potato (diploid cultivar Solanum phureja, DM1-3 516 R44) plants used in this study were obtained from Institute of Vegetable and Flowers, Chinese Academy of Agricultural Sciences (CAAS). The potato was grown in a growth chamber at 26 °C/18 °C (day/night) with a 16:8 light: dark cycle and 60-70% relative humidity according to (Ali et al., 2014). The roots of 7-8-leaves-old plantlets were watered with 200 mM NaCl solution (Ma et al., 2021). And the leaves were sprayed with 100 μM ABA, 50 μM MeJA, 350 μM GA and 50 μM SA, respectively. When spraying, moisten the positive and negative sides of all leaves with condensed water droplets without dropping. After the spraying, the plants were immediately wrapped in black plastic bags and treated only once (Yu et al., 2021). Then, the 1, 2, 3, 4 and 5 d (0 d as control) treated plant leaves were respectively quickly frozen in liquid nitrogen at -80 °C for later use (Li et al., 2021). And each treatment was repeated three times.
2.2 SOS1 genes identification in the potato
All protein sequences were obtained from potato genome data (SolTub_3.0)1. First, the HMM profile for the SOS1s domain (PF00999) was downloaded from the Pfam server2. Then, the HMMER program3 was used to identify the SOS1 proteins in the potato genome (Liang et al., 2017). Finally, the SOS1 (Na+/H+ exchanger, NHX) domain of all putative SOS1 proteins were determined through CDD4 and SMART databases5. A total of 37 putative SOS1 genes were identified.
2.3 Biophysical properties and chromosomal location analysis
Biophysical characteristics of SOS1 proteins were analyzed through ExPASy webserver6 (Wang T. et al., 2021) and NetPhos 3.17 (Naureen et al., 2023). The online prediction tool UniProt8 (Ilzhöfer et al., 2022) was applied to predict the tertiary structures of potato SOS1s. Subcellular location of protein was predicted using the Cell-PLoc 2.0 prediction tool9. The physical positions of the StSOS1s along each chromosome were identified from the potato genome database and the distribution of StSOS1s was plotted (Xiang et al., 2016).
2.4 StSOS1s cis-acting element analysis
The 2000 bp upstream region of the ATG start codon was submitted to PlantCARE10 (Koul et al., 2019) to identify the cis-acting elements and calculate the number of each element. These promoter sequences were represented as word clouds with the help of the WordArt tool11 (Sharma et al., 2021).
2.5 Conserved motifs and gene structure analysis
The conserved motifs in StSOS1s were identified to use the MEME website12 (Multiple Em for Motif Elicitation) (Zhang et al., 2021) with the maximum number of motifs was set to 10. Figures of phylogenetic tree along with gene conserved motifs and CDS/UTR structure of StSOS1s were drawn with TBtools (v1.098) (Chen et al., 2020) software. Gene Structure Display Server (GSDS)13 (Sun et al., 2022) and MEME webserver were employed for gene structure analysis.
2.6 StSOS1s tissue-specific expressions and GO enrichment
RNA-Seq data (fragments per kilobase of exon per million mapped, FPKM) (NCBI accession number ERP000527) in potato DM genotype (Wang J. et al., 2021) was used to analyze the expression level of StSOS1 genes. PlantRegMap14 (Li H. et al., 2020) was used to functionally re-annotate the proteome of up or down-regulated genes and to plot gene ontology (GO) annotations. Protein-protein interaction (PPI) enrichment was computed by STRING15 (Fayez et al., 2022) tool, in which Cytoscape software was used for reconstructing the PPI network, modules and to detect the relationship between overall targeted genes.
2.7 Evolutionary tree construction and collinearity analysis
The SOS1 protein sequences of Arabidopsis, tomato, pepper and tobacco were downloaded from the EnsemblPlants (Contreras-Moreira et al., 2022). Homologous sequences were fed into the MEGA7 software and the Clustalw program was used to perform multi-sequence alignment. The results of the output multi-sequence alignment were used to construct an evolutionary tree using the proximity method (He et al., 2022). The collinearity of the sequences of potato with other four species was extracted using TBtools (Zhang C. et al., 2022).
2.8 RNA isolation and RT-qPCR analysis
The leaves samples were ground into powder in liquid nitrogen, total RNA was extracted using TransZol Up Plus RNA kit (Trans, Beijing, China), following the manufacturer’s instruction. Then the extracted RNA was employed as a template with TransScript® One-Step gDNA Removal and cDNA Synthesis SuperMix for qPCR (Trans, Beijing, China) for the first strand cDNA synthesis. All primer sequences used in this study were designed by Primer Blast website16 of NCBI (Table S1). The RT-qPCR was performed on a QuantStudio-3 system (Thermo Fisher Scientific, Shanghai, China). The reaction system was 20 µL (cDNA 1 µL, SOS1-F 0.4µL, SOS1-R 0.4µL, SuperMix 10 µL, DyeII 0.4µL, Water 7.8 µL). The reaction system was 94 °C 30 s, (94 °C 5 s, 60 °C 30 s) ×40 Cycles. Three replications were performed and the expression values were calculated by using the 2−ΔΔCT method (Mo et al., 2022).
2.9 Subcellular localization of StSOS1-13
For the localization and expression of StSOS1-13 in potato, the CDS without the stop codon was cloned into pCAMBIA1300. Firstly, the complete coding region of StSOS1-13 (1 734bp) was amplified from the cDNA by PCR using a pair of primers with a homologous arm and inserted into the pCAMBIA1300 vector linearized by the restriction enzyme NcoI. Then, the obtained pStSOS1-13-GFP fusion plasmid was converted into Escherichia coli DH5α for verified by bacterial liquid PCR and company sequencing (Sangon, Shanghai, China), further inserted into individual Agrobacterium tumefaciens strain GV3101 cells and a single colony was selected for PCR positive identification. Finally, the expression vectors were injected into tobacco leaves for the transient expression experiments (Luo et al., 2022). GFP expression was analyzed using scanning confocal laser microscopy.
3 Results
3.1 Identification of SOS1 genes in the potato
To identify the SOS1s family members in potato, the similar protein sequences were searched in the HMMER program with the query sequence SOS1s motif (PF00999). The SMART tool was then used to confirm whether the candidates contained the Na+/H+ exchanger (NHX) domain. In total, 37 SOS1 genes were retrieved from the potato genome and renamed StSOS1-1 to StSOS1-37 based on their relative linear order on each chromosome, following the widely used nomenclature (Figure 1). Meantime, we found four pairs of tandem duplicated genes existed in 37 StSOS1 genes. The analysis showed that there was one pair of tandem duplicated genes (StSOS1-2 and StSOS1-3) on Chr1, one pair (StSOS1-7 and StSOS1-8) on Chr2, one pair (StSOS1-26 and StSOS1-27) on Chr6, and one pair (StSOS1-30 and StSOS1-31) on Chr9.
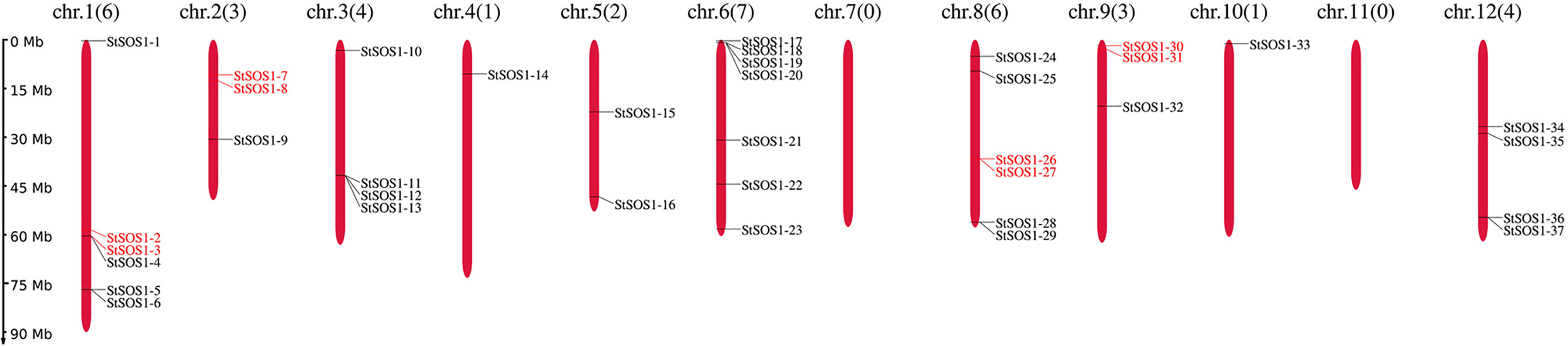
Figure 1 Distribution of the StSOS1 genes in the potato on 12 chromosomes. The nomenclature for StSOS1 members was based on the physical position from top to bottom on the chromosome, the names were displayed on the right-hand side of each chromosome, the number of chromosomes and the StSOS1 genes were indicated at the top of each chromosome, and the scale of the genome size was given on the left-hand side. All protein sequences were obtained from potato genome data (SolTub_3.0).
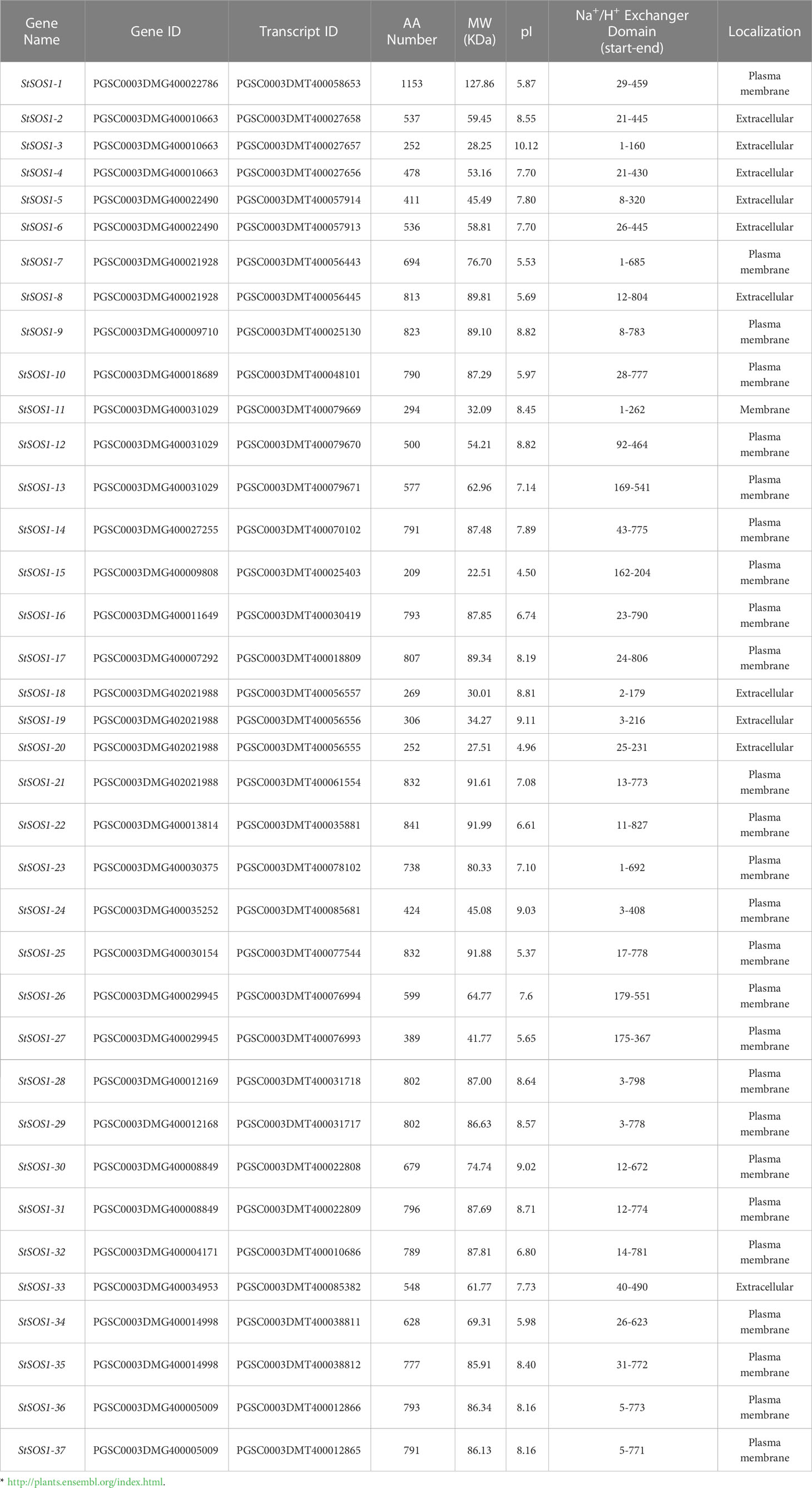
We further determined the biophysical properties of the potato SOS1 genes including the locus ID, protein length (aa), predicted protein molecular weight (MW), isoelectric points (pI), and NHX domain. The statistical results showed that the protein length ranged from 209 (StSOS1-15) to 1153 (StSOS1-1) amino acids, the average amino acids length and molecular weights ranged from 22.51 KDa (StSOS1-15) to 127.86 KDa (StSOS1-1). PI varying from 4.96 (StSOS1-20) to 10.12 (StSOS1-3). The subcellular localization of these StSOS1s predicted through Cell-PLoc 2.0 tool revealed that most of the StSOS1 proteins were localized in the plasma membrane (Table 1). The results of the NetPhos 3.1 server revealed that StSOS1 proteins were phosphorylated, and phosphorylated residues were Serine (Ser), threonine (Thr) and tyrosine (Tyr) (Table S2), among which serine prediction sites ranged from 11 (StSOS1-24) to 72 (StSOS1-1). The threonine prediction sites ranged from 5 (StSOS1-15) to 32 (StSOS1-1), and the tyrosine prediction sites ranged from 0 (StSOS1-15 and StSOS1-20) to 9 (StSOS1-21). Three-dimensional protein models were constructed by sequence similarity search using UniProt PDB database and the homology modeling was predicted by DS Visualizer (Figure S1). The structures of StSOS1-7, StSOS1-8, StSOS1-9, StSOS1-16, StSOS1-17, StSOS1-21, StSOS1-22, StSOS1-25, StSOS1-28, StSOS1-29, StSOS1-31, StSOS1-32, StSOS1-36, and StSOS1-37 are similar and suggest shared functionality, as do StSOS1-18 and StSOS1-19. These provide an initial basis for understanding the molecular function of the StSOS1 proteins.
3.2 Prediction of cis-elements in the promoter sequences of StSOS1 genes
To clarify which hormonal, environmental stress, or developmental-related signal elements are involved in these StSOS1s, we performed a promoter analysis using the PlantCARE server. A large number of basic components were discovered in the upstream sequence (2000 bp) regions, including WRE3, GATA-motif, CAT-box and G-Box, but also P-box, TCA-element, AuxRR-core, TGACG-motif, ABRE and ERE hormonal response-related elements; as-1, LTR, ARE, GC-motif, MBS environmental stress-related components and A-box development-related elements (Figures 2A, B). Hormonal response elements were detected in the promoters of 37 potato StSOS1 genes, including 15 SA, 19 MeJA, 26 ABA and 30 auxin response. The cis-elements involved in the GA response are present in all promoters of StSOS1s. The promoters of 10, 16, and 20 StSOS1 genes contained MYB binding sites involved in low-temperature response, defense and stress response cis-elements and drought-inducibility, respectively (Figure 2C). These results suggest that the StSOS1 genes may play a critical role not only in phytohormones, but also in biological and abiotic responses in the potato.
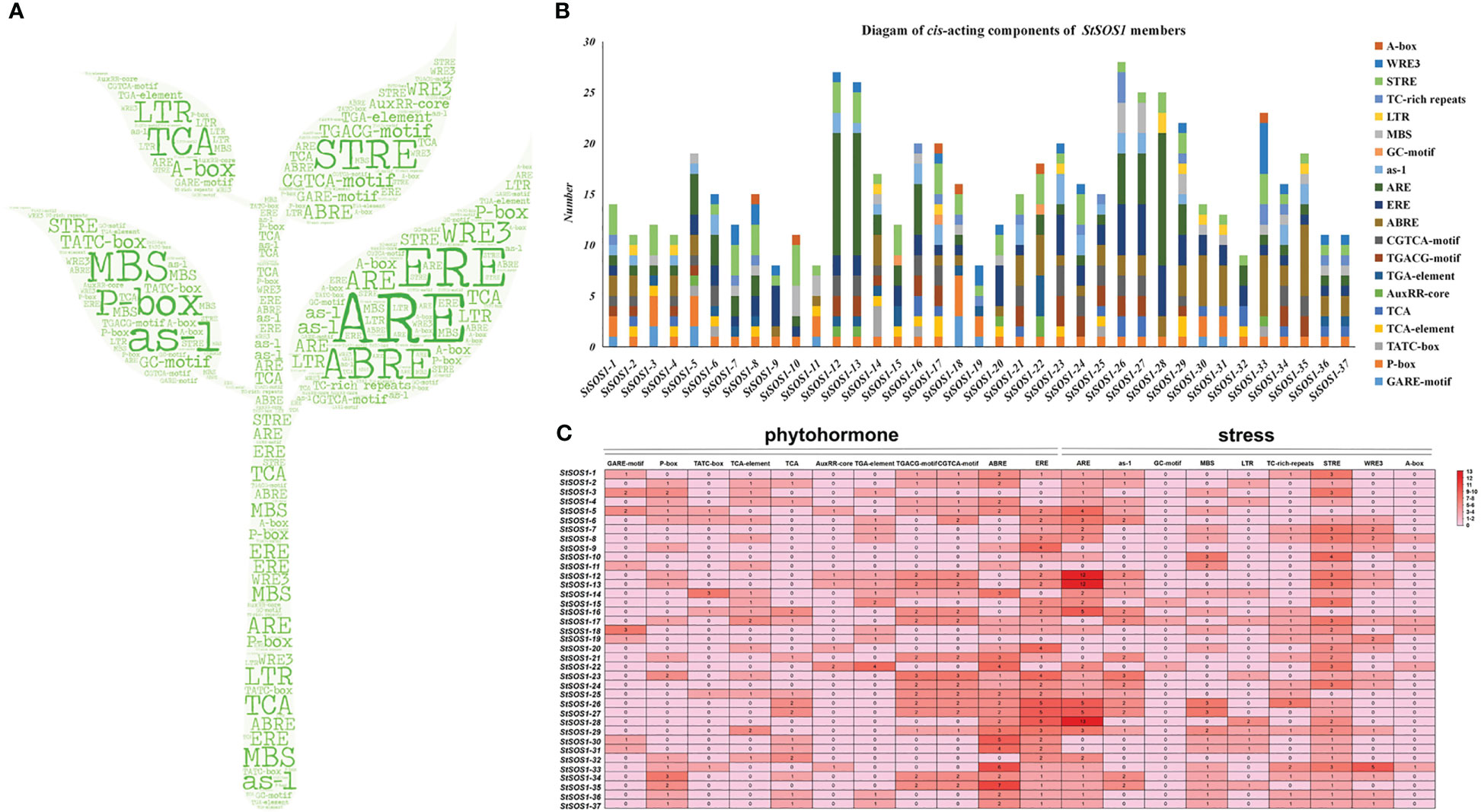
Figure 2 Analysis of the cis-acting elements. (A) Word clouds representing different cis-regulatory elements present at 2000 bp upstream of StSOS1 genes sequences. (B) Graphical representation of 37 StSOS1 genes with various roles in hormonal response, abiotic stress, plant development, defense, and stress response. (C) Cis-elements were denoted by different colors according to their number. The darker the color, the higher the occurrence frequency, and the number indicates the number of cis-elements.
3.3 Gene structure and conserved motifs of StSOS1s
In order to better understand the relationship between the structure and function of these StSOS1 proteins, gene structure and conserved motifs were analyzed to construct individual phylogenies. Depending on the different branches of the evolutionary tree, it has been found that the motif architectures remain consistent within the same evolutionary branch, and thus they may have a similar function (Figures 3A, B). The results showed that the number of intron in StSOS1 genes ranged from two (StSOS1-22, StSOS1-23, StSOS1-34) to 20 (StSOS1-13, StSOS1-26, StSOS1-27). Furthermore, closely related genes share a similar structural architectures with different introns lengths (Figure 3C). The shortest StSOS1 protein was just 209 aa in length (StSOS1-15), while the longest was StSOS1-1, with a length of 1153 aa (Table 1). The functional sites in the conserved motifs were analyzed using the Eukaryotic Linear Motif resource server (ELM) and the results showed that there was a great functional divergency among these sites and most of the functional sites are related to phosphorylation, kinase phosphorylation, binding and sorting signal responsible for the interaction (Table S3).
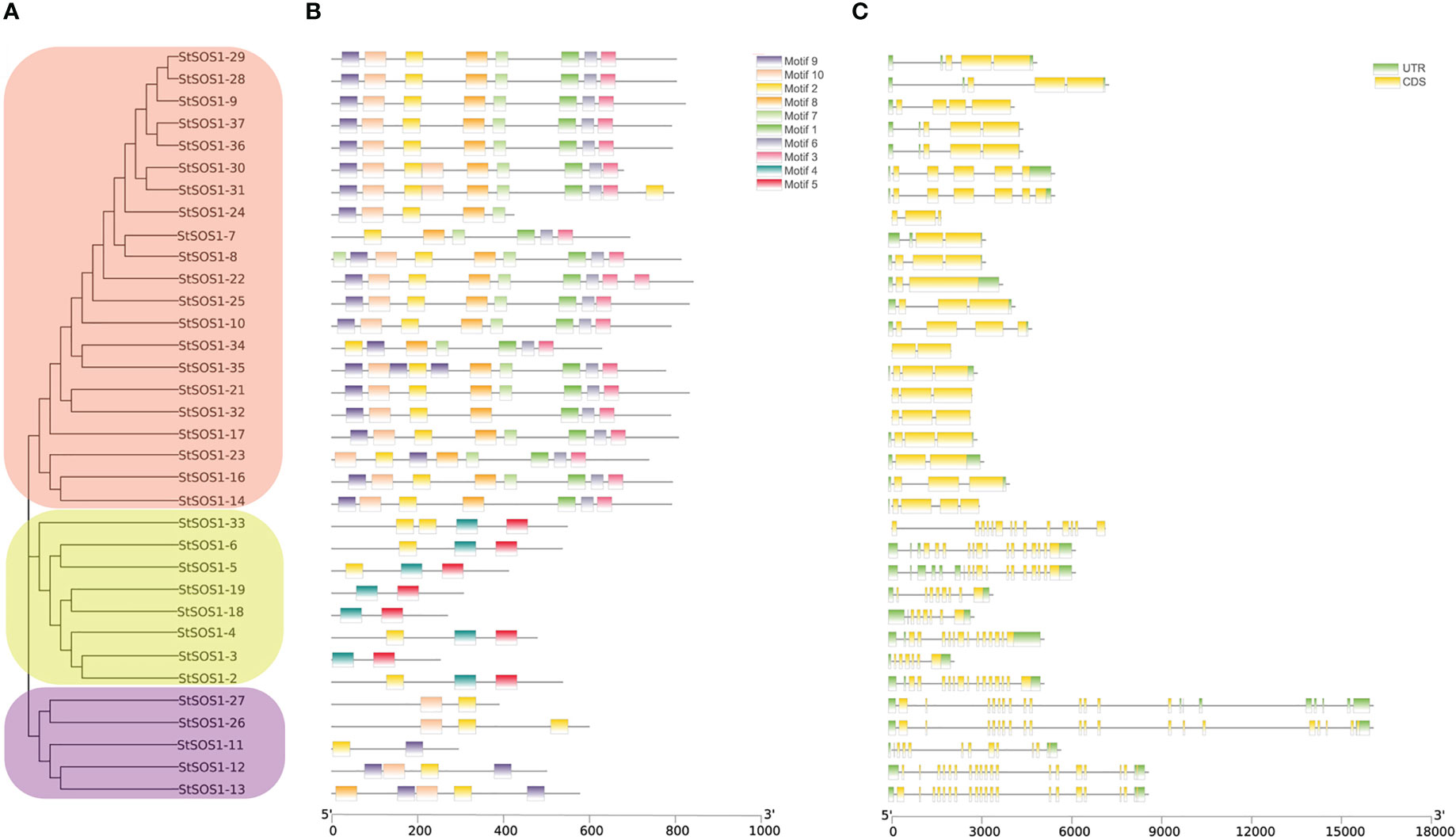
Figure 3 Phylogenetic relationships, structures, and motifs of members of the StSOS1s family [StSOS1-1 (1153 aa), StSOS1-15 (209 aa), and StSOS1-20 (252 aa) excepted]. (A) The phylogenetic tree of the StSOS1 proteins was constructed using the Maximum Likelihood method, which was based on conserved motifs and CDS/UTR structure. Different subgroups were represented by different background colors. (B) The conserved motifs of the StSOS1 proteins. Different patterns were represented by boxes of various colors, 5 ‘and 3’ represent the N and C ends. (C) Gene structures, exons and untranslated regions (UTR) are shown in green and yellow boxes, while black lines indicated introns. Phylogenetic trees, conserved motifs, and gene structures were predicted using TBtools, and their lengths were estimated using bottom ruler.
3.4 Expression characterization of StSOS1s
To investigate the biological function of StSOS1s in different tissues, expression profiles of all identified StSOS1 genes were analyzed in six different tissues, including roots, tubers, stolons, leaves, whole mature flowers, and mature whole fruit (Figure 4A). Of all the 21 StSOS1 genes, StSOS1-2 exhibits the highest levels of expression in almost all the tissues except the tubers. Some members of StSOS1 exhibit highly tissue-specific expression, such as the expression of StSOS1-10, StSOS1-16, StSOS1-17, StSOS1-19, StSOS1-22, StSOS1-23, StSOS1-32, and StSOS1-35 throughout the mature flower, suggesting that the StSOS1 genes exhibit differential tissue-specific expression patterns. Then we analyzed spatio-temporal expression patterns in stolon, tuber pith, tuber peel, tuber cortex, young tuber, mature tuber and tuber sprout using RNA-seq data (Figure 4B). It showed that two genes (StSOS1-14 and StSOS1-32) had a very low abundance in these tissues or organs. StSOS1-16, StSOS1-19, and StSOS1-31 were predominantly expressed in stolon; StSOS1-6 and StSOS1-13 were predominantly expressed in tuber sprout. StSOS1-1 was highly expressed in tuber pith, tuber peel, tuber cortex, young tuber and tuber sprouts. To have a better understand the function of StSOS1s under biotic stress, the expression pattern was observed responding to Phytophthora infestans, β-aminobutyric acid (BABA) and benzothiadiazole (BTH) treatment (Figure 4C). StSOS1-2 was the only member to exhibit down-regulation under all three biotic stress conditions. Some genes show up-regulation, in particular one type of stress treatment; StSOS1-6, StSOS1-13, StSOS1-28 and StSOS1-29 showed up-regulation only in response to BABA treatment. For the abiotic stresses and phytohormones responsiveness of StSOS1s, we analyzed their transcript profiling in response to three abiotic stress and four phytohormone conditions mannitol, water-stress, heat, IAA, GA3, BAP and ABA (Figure 4D). StSOS1-6 was found to be highly up-regulated in the three stress conditions of mannitol, water-stress and ABA. StSOS1-10, StSOS1-17, StSOS1-22, StSOS1-23, StSOS1-32, and StSOS1-35 showed low or no expression in the eight tissues.
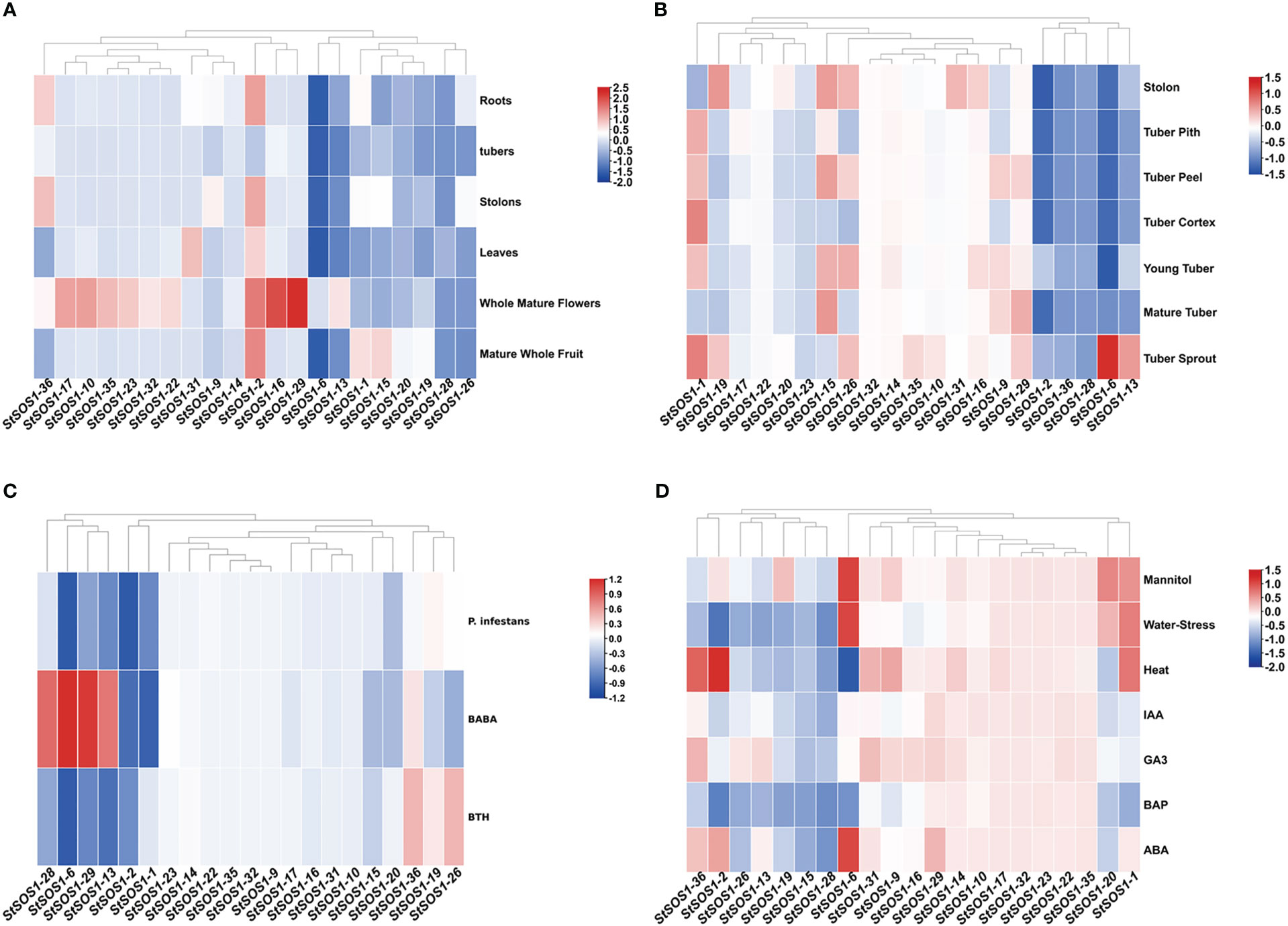
Figure 4 Expression levels of StSOS1 genes in biotic, abiotic stress, and in different tissues and developmental stages. (A) Expression profiles of StSOS1s in different tissues and developmental stages. (B) Expression profiles of StSOS1s in different tissues and developmental stages of the potato tuber. (C) Expression of StSOS1s transcripts was altered in response to biotic stress. (D) Expression profiles of StSOS1s at abiotic stress and phytohormones. In the heat map, red, blue and white represent up-regulated, down-regulated, and unchanged (log10 ratio), respectively. Heat map and hierarchical clustering were performed by average linage (default) method.
3.5 Gene ontology analyses of StSOS1s
To identify functions of up and down-regulated genes, GO analysis was performed and genes belonging to different categories of Biological Processes (BP), Molecular Functions (MF) and Cellular Compartments (CC) were identified (Figure 5). The BP categorized results showed that the up-regulated genes were significantly enriched in transport and cellular process. For MF, these up-regulated states enriched in transport activity. Moreover, up-regulated genes in the CC category are significantly enriched in both membrane and membrane-like components. In addition, the most significantly enriched GO terms for down-regulated genes were detection of hydrogen transport (BP), and transporter activity and antiporter activity (MF). It is important to note that the membrane integral, the membrane intrinsic and membrane fraction are all present in both up- and down-regulated genes in CC. The difference is that the up-regulated genes have a late endosome while the down-regulated genes do not. In summary, most GO terms are involved in membrane transport and composition, suggesting that they are likely to play an important role in maintaining proper ion homeostasis in the cytoplasm.

Figure 5 Gene ontology analyse of StSOS1s. Functional classification of up- and down-regulated genes by GO analysis into categories of Biological Process, Molecular Function and Cellular Component. The number in each pie represents the number of times the function (BP, CC, MF) was expressed in the data (abundance). GO-based classifications of up-regulated genes were shown in red, while those of down-regulated genes were shown in blue.
The verification of PPI is a defining aspect of molecular biology. PPI analysis was conducted to analyze the interactions among the SOS1s (Figure S2). The biological pathways and cellular compartments (retrieved from the GO) associated with these proteins were similar. Here, the interaction network between 96 SOS1-related genes was also mapped using the STRING database and Cytoscape software for function analyse, seven clusters were identified, including the pathways of biological regulation, membrane, ion transmembrane transporter activity, calcium ion binding, potassium ion transmembrane transport, response to salt stress and cellular process (Figure S3). Only 25 StSOS1s interact with other genes, and the most PPI was observed between proteins involved in potassium ion transmembrane transport, response to salt stress and cellular processes. These studies inform the biochemical mechanism of StSOS1 and provide a new reference for the interplay between ion homeostasis and transmembrane transport during plant salt tolerance.
3.6 Phylogenetic and collinearity analyses of StSOS1s
For the evolutionary relationship of SOS1s among Arabidopsis, tomato, pepper, potato and tobacco, we extracted and compared the protein sequences of SOS1s in these species, and constructed the phylogenetic tree of neighbor junction (NJ) (Figure 6A). Potato SOS1s are named based on their position relative to orthologs from four other species on the tree. 134 SOS1 candidates of five species were grouped into four distinct classes (I-IV) based on sequence conservation. Among them, the subgroup I had 13 members (11.19%), subgroup II 27 (20.14%) and subgroup III 37 (27.61%), respectively. The subgroup IV contained 57 genes and had the most members (42.54%). The phylogenetic relationships indicate that the SOS1 proteins in the potato are more strongly homologous to pepper and tomato than to Arabidopsis and tobacco. Gene duplication has always played a key role in the expansion of genes and the occurrence of novel functions of genes. To explore the evolution of SOS1 genes, we studied the replication patterns of the five species and performed genetic correlation analysis (Figure 6B). The results showed that there were 17, 8, 5, and 1 SOS1 members participating in the potato-tomato, potato-pepper, potato-Arabidopsis and potato-tobacco synteny relations, respectively. Among the above collinear gene pairs, StSOS1-11 with SlSOS1-23, CaSOS1-10, AtSOS1-58, respectively; StSOS1-28 with SlSOS1-26, CaSOS1-1, AtSOS1-47, respectively; and StSOS1-37 with SlSOS1-26, CaSOS1-37, AtSOS1-55, respectively, had simultaneously collinear relations.
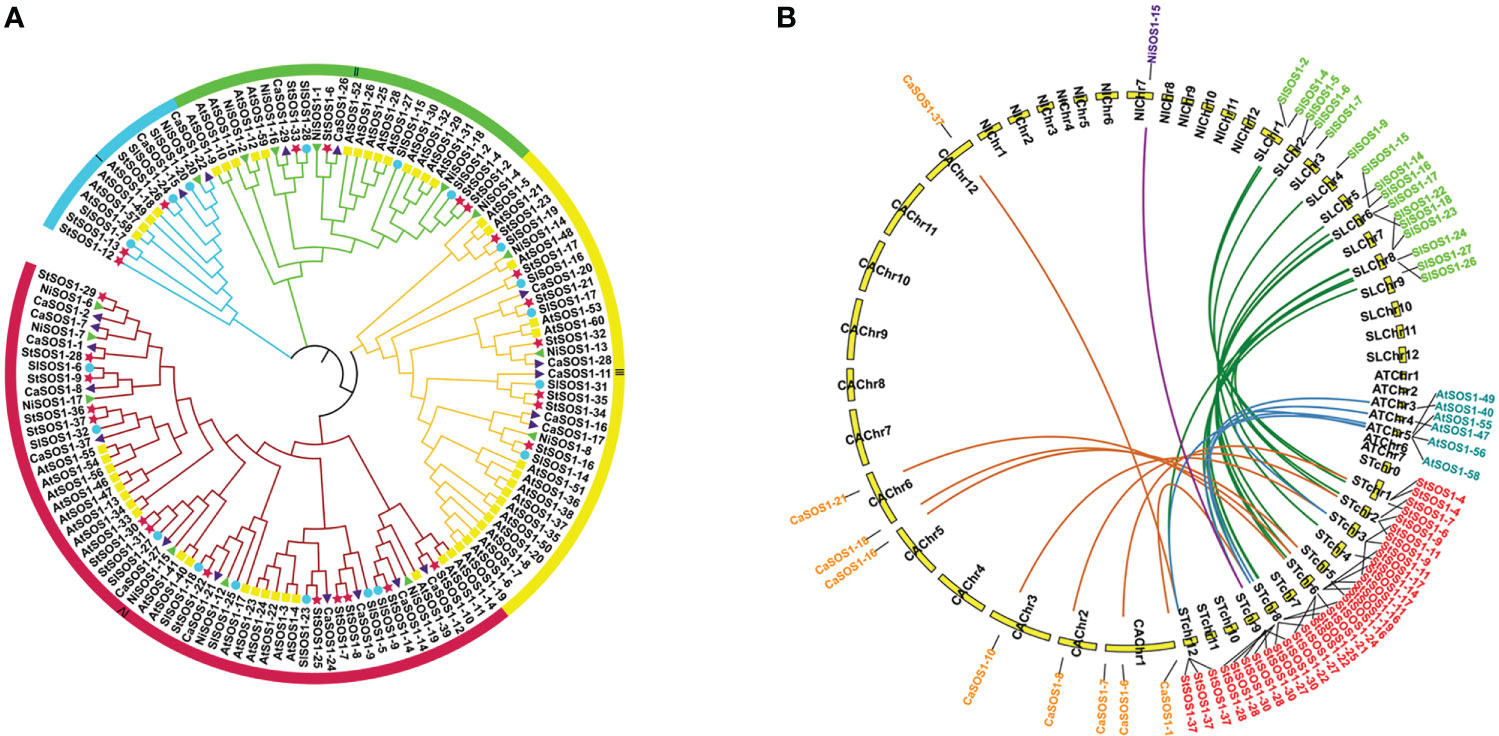
Figure 6 A Phylogenetic analysis of SOS1 proteins. (A) phylogenetic tree of SOS1 proteins was constructed with neighbor-junction (NJ) phylogenetic tree. The four subgroups were shown in different colors. The red stars represent potato SOS1s (StSOS1s), the green triangles represent tobacco SOS1s (NiSOSs), the purple triangles represent pepper SOS1s (CaSOSs), the yellow boxes represent Arabidopsis SOS1s (AtSOSs) and the blue circles represent tomato SOS1s (SlSOSs). (B) Collinearity analysis of SOS1s in potato and other plants. The green, orange, blue and purple lines in the background correspond to collinear gene pairs in potato and tomato, potato and pepper, and potato and Arabidopsis, potato and tobacco, respectively.
3.7 Expression analysis of StSOS1 genes under different abiotic stresses
The SOS pathway plays an important role in maintaining proper ion homeostasis in the cytoplasm and in regulating plant tolerance to salinity. However, there is limited information on SOS1’s response to potato salt stress. In order to investigate the potato response to salt stress, the StSOS1 genes were analyzed using the transcriptomic data of potato exposed to NaCl treatment. Only 21 StSOS1 genes showed differential gene expression pattern and were identified and visualized in a heat map (Figure 7A). Furthermore, six StSOS1 genes in potato leaves of different grow stages under salt stress were randomly selected and quantitative analyzed by RT-qPCR (Figures 7B–G). These results suggested that these six genes were significantly differentially up-regulated under salt stress, which may positively regulate salt tolerance in the potato, this is not consistent with the heat map, which may be related with different levels of expression under different levels of salt stress treatment. StSOS1-2, StSOS1-6 and StSOS1-28 occurred two up-regulated expressions phenomenon under salt stress, this could be related to the response period of the SOS1 signaling pathway. Notably, the expression of StSOS1-13 was 14-fold higher at 3 d after salt treatment compared to expression levels before salt stress, and then reached 32-fold higher at 4 d, suggesting that StSOS1-13 may be an important candidate gene involved in the salt stress response.
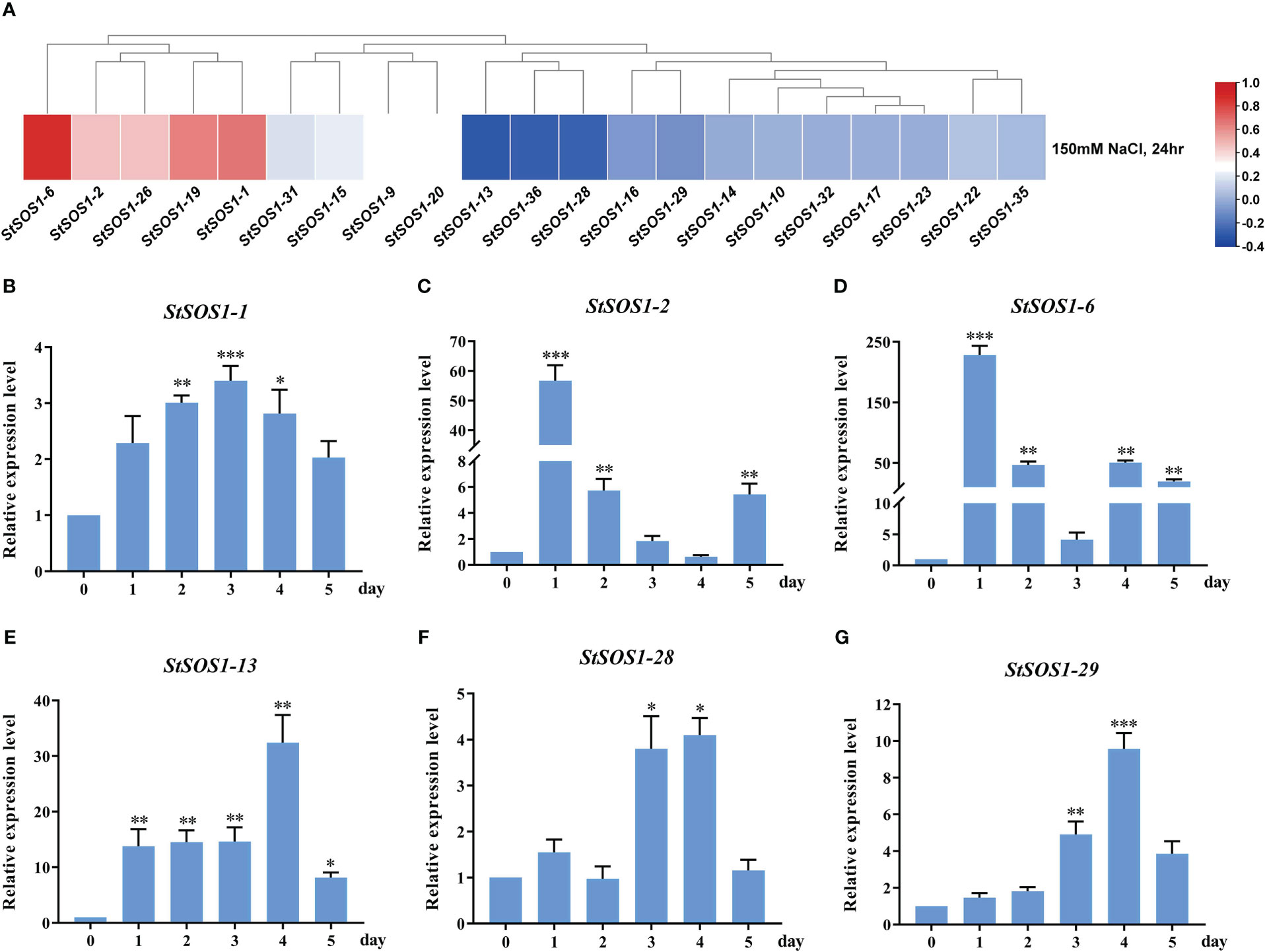
Figure 7 The expression pattern of StSOS1s under salt stress. (A) Expression profiles of StSOS1s at NaCl stress based on RNA seq-date. (B-G) RT-qPCR profiles of StSOS1 genes under salt stress. The expression level of StSOS1s on control was normalized as “1”. The vertical bars indicate the standard error of the mean. Asterisks indicate significant differences based T test (*, p < 0.05, **, p < 0.01, ***, p < 0.001, ****, p < 0.0001).
To further understand potential function changes in StSOS1-13 gene in response to abiotic stress, RT-qPCR was used to analyze the expression patterns of the selected StSOS1-13 gene in phytohormone treatment (Figure 8). It was observed that the StSOS1-13 was up-regulated on exposure to ABA, GA, and SA treatment, and the magnitude of up-regulation was higher in ABA treatment as compared to GA, SA treatment. Conversely, for the MeJA treatment, expression in the leaves decreased after 0-2 d and then increased continuously, with the highest levels of expression in the leaves at 5 d. Overall, these results indicated that StSOS1-13 may play a critical regulatory role in response to abiotic stress.
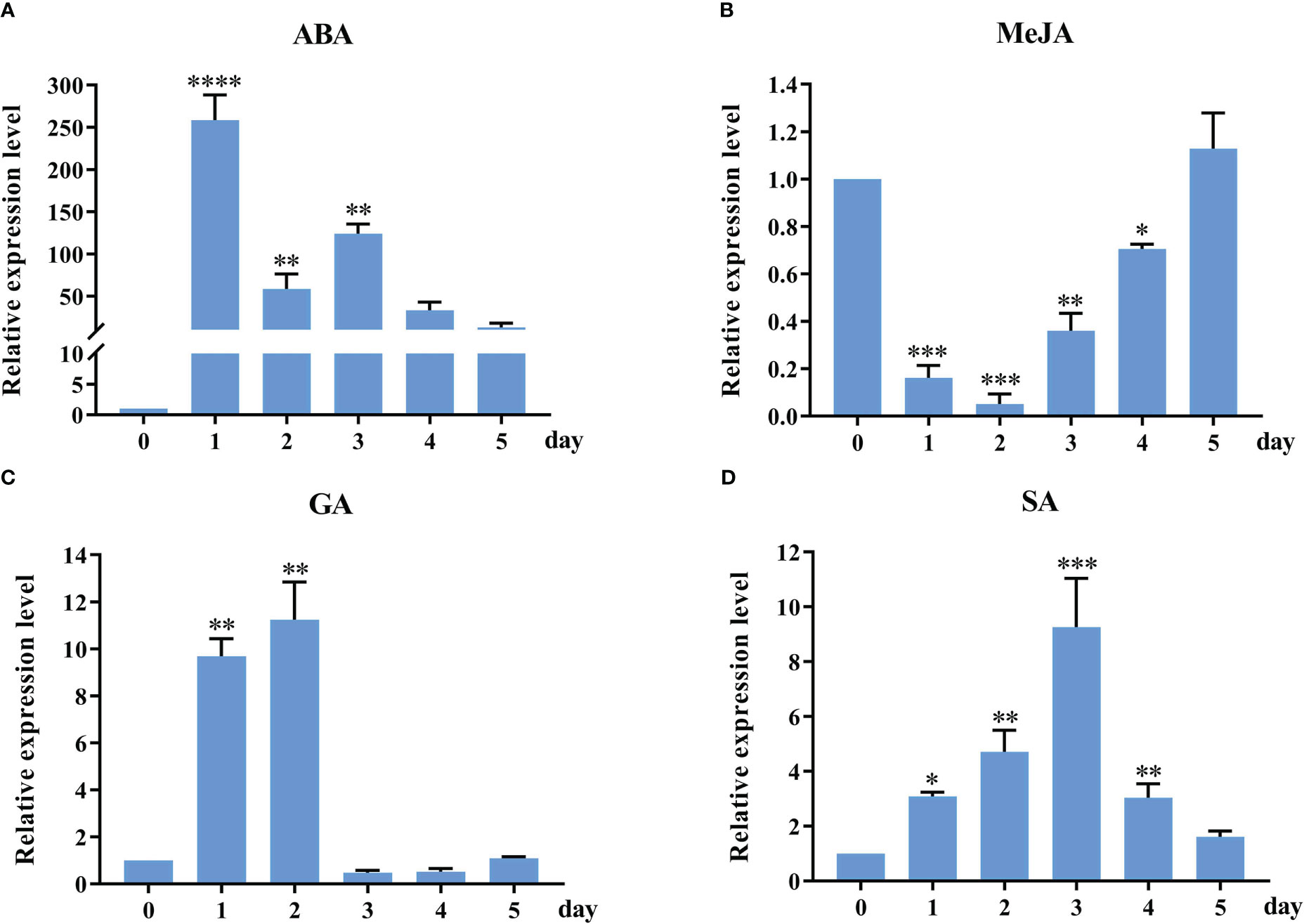
Figure 8 RT-qPCR profiles of StSOS1-13 gene under phytohormone treatment. The StSOS1-13 expression level of control was normalized as “1”. The vertical bars indicate the standard error of the mean. Asterisks indicate significant differences based on T test (*, p < 0.05, **, p < 0.01, ***, p < 0.001, ****, p < 0.0001).
3.8 Subcellular localization of StSOS1-13
Detecting the subcellular localization of StSOS1-13 is essential to elucidate their function. The subcellular localization of StSOS1-13 predicted by the Cell-PLoc 2.0 tool revealed that the StSOS1-13 protein was localized in the plasma membrane. To further verify the location of StSOS1-13 protein, the full-length coding sequence of StSOS1-13 deleted stop codon was fused with green fluorescence protein (GFP) and the transient expression was performed under the control of 35S promoter in tobacco. The results showed that the StSOS1-13 protein is localized in the plasma membrane (Figure 9), this is consistent with the result of bioinformatics analysis.
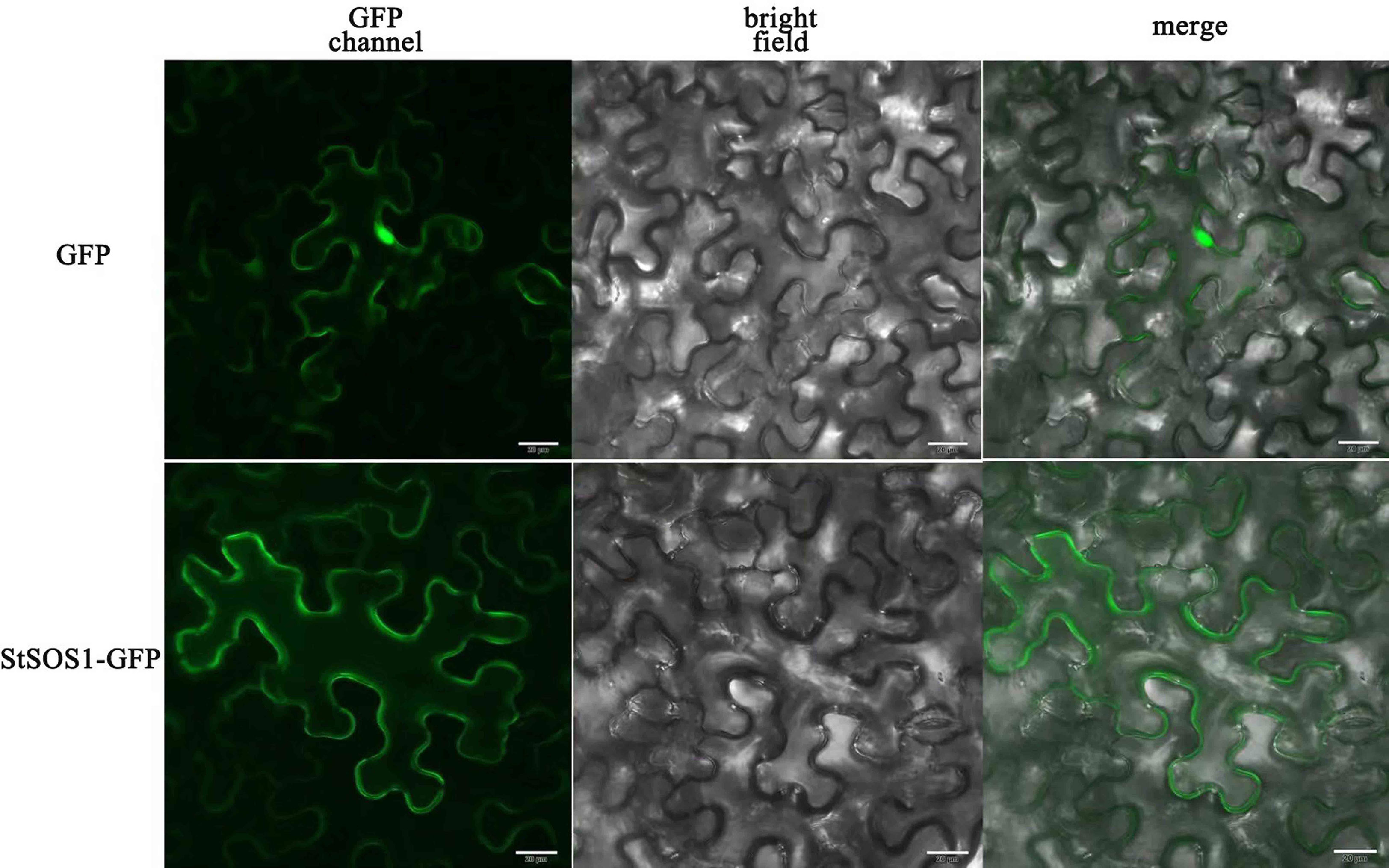
Figure 9 Subcellular localization of StSOS1-13 in Nicotiana benthamiana. The leaves were injected with a strain of Agrobacterium tumefaciens containing 35S::StSOS1-13-GFP, and the empty vector 35S::GFP were used as a control. After 48 h of injection, pStSOS1-13-GFP fusion protein and GFP alone transiently expressed separately in leaves, the dark field was green fluorescence and the white field was cell morphology, with Confocal combined detection. GFP, GFP fluorescence (green). Bright, bright fields. Merge, superimpose GFP and bright-field images. The experiment repeated three times with similar results. Scale bar, 20 µm.
4 Discussion
Soil salinity is one of the most significant abiotic stresses faced by crop plants in agricultural fields worldwide (Świeżawska et al., 2018), reducing crop yield and production (Rolly et al., 2020). Plants have evolved the SOS pathway to achieve salt tolerance (Cha et al., 2022), the SOS pathway comprising SOS1, SOS2 and SOS3 has been proposed to regulate cellular signaling during salt stress to mediate ion homeostasis (Luo et al., 2022). SOS1 is a critical salt tolerance determinant in plants (Świeżawska et al., 2018). SOS1 genes have been reported to improve the tolerance to salt stresses in plants such as Arabidopsis (Wu et al., 1996), soybean (Zhang et al., 2022), and maize (Zhou et al., 2022). Potato is one of the most crucial crops in the world due to its nutritional quality (Takeuchi et al., 2022). The crop can also be used as a commercial health food because it is high in antioxidants, minerals, and dietary fibers (Kumar et al., 2021). In addition, potato plants are often subjected to various types of abiotic stress during growth and development (Yang et al., 2020; Kumar et al., 2021). It was reported that soil salinization negatively affected the growth and yield of potato crops, especially in arid and semi-arid climates (Li et al., 2022), which caused osmotic and oxidative stress, ion imbalance, mineral deficiency, and ion toxicity problems (Hamooh et al., 2021). Therefore, the selection and breeding of salt-tolerant genes has become a promising approach for improving the yield and adaptability of potato (Zhu et al., 2022). Previous studies have shown that a gene encoding SOS2 (PGSC0003DMG400006384) is up-regulated, indicating that this gene plays an active regulatory role in salt stress response. However, the complete SOS pathway for salt stress response in potato has not been established, and only a few genes of this pathway have been reported (Li Q. et al., 2020). The aim of this study is to screen for key StSOS1 genes that are more sensitive to abiotic stress and to lay the groundwork for further unraveling the regulatory mechanisms of SOS1 genes in potato.
In this study, a total of 37 SOS1 family members were identified in potato (Table 1) and they locate in 10 of 12 chromosomes (Figure 1) which were significantly lower than Arabidopsis (60 SOS1s in Figure 6A). Gene duplication may explain the difference in the number of SOS1 family members between the potato and Arabidopsis. A possible explanation for this is that SOS1 genes in the potato may have a higher rate of gene loss than in Arabidopsis, and frequent gene loss has been reported in various plant species during genome duplication events (Li et al., 2020), indicating a key role of gene duplication over the course of evolution in various species (Zhang et al., 2021). Some of the duplicated genes may be retained in its descendants, which could provide the original genetic resource for the adaptive evolution of plants (Flagel and Wendel, 2009). The number of SOS1 genes in the potato was similar to that in the pepper. Phylogenetic analysis demonstrated that the Solanaceae SOS1 genes were generally classified into four clades (Figure 6A). Interestingly, four subfamilies were present in all five plant species, suggesting that genetic expansion occurred prior to the divergence of these plant species. By comparing the syntenic analysis of SOS1 genes in potato and four other plants (tomato, pepper, Arabidopsis and tobacco), we found that the sequence similarity between the SOS1 gene pairs within potato was much higher than that between the tomato and the pepper (Figure 6B), which is consistent with the phenomenon in chrysanthemum (Gao et al., 2016), indicating the similarity of evolutionary relationship among different species in the same group. The conserved motif analysis of SOS1s family revealed the occurrence of 10 conserved motifs (Figure 3) might be related to specific functions shared among SOS1 family members. In addition, StSOS1s within the same subfamily share a high degree of similarity in exon-intron structures and conserved motifs. The loss and gain of introns may reflect evolutionary trends in genes with similar functions (Rogozin et al., 2003), which had been demonstrated in Brassica juncea (Cheng et al., 2019).
In salt-acclimated tobacco, the compartmentalization of Na+ in vacuoles may be mediated by vesicle transport (Garcia de la Garma et al., 2015), which represents an over-sensitive mechanism of the Na+/H+ antitransporter SOS1 to accommodate salt stress (Hamaji et al., 2009; Zhao S. et al., 2021). When the SOS signaling pathway is activated, the Na+/H+ antiport activity of SOS1 is enhanced and the accumulated Na+ is transported out of the cell (Xie et al., 2022). For further functional analysis, we use GO annotation enrichment analysis to functionally annotate different StSOS1s. Gene ontology is a fundamental analysis that predicts the contribution of putative functions across living organisms. In the present study, GO analysis revealed the significant role of StSOS1s with cellular process, transport and component of membrane (Figure 5). To support this argument, we have constructed an additional PPI network with StSOS1 proteins as the core (Figure S2). Among the numerous functional modulated by the SOS1 network, there are the regulation pathways of biological regulation, membrane, ion transmembrane transporter activity, calcium ion binding, potassium ion transmembrane transport, response to salt stress and cellular process (Figure S3). Most of the StSOS1 genes are involved in cellular transport process (Figure S2), suggesting that they probably play a vital role in maintaining appropriate ion homeostasis in the cytoplasm.
The cis-elements and functional characteristics of SOS genes promoters have been identified in many species, such as Brassica juncea var. Tumida (Cheng et al., 2019), B. juncea (Kaur et al., 2015), and Arabidopsis (Feki et al., 2015). To further explore the possible function of SOS1s in potato, we performed an analysis of cis-acting regulatory elements in the promoter region in this study. Cis-regulatory elements were found to include phytohormone (SA, MeJA, ABA, auxin, GA) and abiotic stresses (cold, defense and stress response, drought) (Figure 2), which is consistent with the report about the previous studies in other species. More importantly, the cis-elements involved in the GA response are present in the promoters of all StSOS1s, and more than half of the promoters of StSOS1s have MYB elements involved in drought-inducibility. Interestingly, in the heat map (Figure 4D), StSOS1s could be induced by both auxin and GA, two important plant hormones in regulation. Most of the StSOS1s notably up-regulate under both mannitol and NaCl stress conditions. Overall, the results presented above revealed that StSOS1s may play a significant role in the response to phytohormone and abiotic stresses.
In wheat, most TaSOS1 genes expressed in different tissues, including shoots, leaves, spikes, and grains (Jiang et al., 2021). In Arabidopsis, AtSOS1 promoter-driven GUS expressed primarily in the roots, inflorescences and leaves (Yang et al., 2009). Our results revealed StSOS1-2 and StSOS1-31 were specifically expressed in leaves whereas StSOS1-10, StSOS1-16, StSOS1-17, StSOS1-19, StSOS1-22, StSOS1-23, StSOS1-32 and StSOS1-35 had clear expression preference in whole mature flowers (Figure 4A). These suggested that the StSOS1s played a significant role in the growth and development of different potato organs. In addition, the StSOS1s had the similar tissue-specific expression patterns with the AtSOS1s, this suggested that the SOS1 gene family played a conserved function in both Arabidopsis and potato.
Under salt stress, SOS1 gene expression levels of Populus euphratica and Chrysanthemum crassum were up-regulated (Wu et al., 2007; Song et al., 2012). There were differences in SOS1 gene expression in cotton at different time intervals (Akram et al., 2020). In this study, compared with the control, the expression level of StSOS1s in leaves was immediately up-regulated under salt stress, and the results of RT-qPCR of StSOS1-1, StSOS1-2 and StSOS1-6 were highly consistent with the results of heat map (Figures 7B–D). In addition, in wheat, the expression of SOS1 in leaves under salt stress was consistent with mRNA abundance (Xu et al., 2008). However, the RT-qPCR results of StSOS1-13, StSOS1-28 and StSOS1-29 were contrary to the down-regulated results of heat map within 24 h (Figures 7E–G). In purslane (Sesuvium portulacastrum), the RT-qPCR results also differ from the heat map results. That is, the quantitative expression level of SpSOS1 in roots increased sharply within 3-6 h and then decreased to the basic level, while the transcription abundance of SOS1 in leaves did not change significantly within 48 h of NaCl treatment (Zhou et al., 2015). In addition, the expressions of StSOS1-2, StSOS1-6 and StSOS1-28 in leaves were up-regulated twice (Figures 7C–D, F). Similarly, the expression level of GhSOS1 under salt stress also showed this phenomenon (Chen et al., 2017). In conclusion, the mechanism of SOS1 in potato salt stress resistance is relatively complex and more studies are needed to determine the function of SOS1s in potato in the future.
Exogenous ABA, MeJA, SA treatment can improve the yield of potato (Pérez-Alonso et al., 2021). Under ABA stress, the expression of BjSOS genes increased with increasing stress duration in both contrasting genotypes (Nutan et al., 2018). Several reports have suggested co-expression of many stress-responsive genes at both salinity and ABA (Takahashi et al., 2004). Our results of RT-qPCR analysis indicated that the StSOS1-13 was expressed under four phytohormone treatment (Figure 8). StSOS1-13 was significantly up-regulated about 3, 9, and 250 times at 1 d in leaves under SA, GA, and ABA treatment, respectively, while StSOS1-13, was down-regulated under MeJA treatment. The promoter biological function is further corroborated by the expression analysis of StSOS1-13 in response to hormonal stress. StSOS1s may regulate the expression of genes involved in the transduction of hormone signals, and thus participate in plant growth and development.
Studies have reported that the excessive Na+ ions in soil can cause imbalance in vivo, moisture deficiency and ion toxicity (Tester and Davenport, 2003), so some plants formed a Na+ efflux and Na+ segment processing. As a result, some plants have developed Na+ efflux and Na+ segment treatments to maintain low intracellular Na+ concentrations to accommodate the effects of salt stress on plant growth and development. The SOS pathway studied previously is a more classical salt signaling pathway (Chinnusamy et al., 2004). Arabidopsis salt-tolerant site SOS1 encodes Na+/H+ antiporter. Confocal imaging of a green fluorescent protein fusion protein of SOS1 in a transgenic Arabidopsis plant revealed that SOS1 is localized in the plasma membrane (Shi et al., 2002). SOS3 and SOS2, which are located in the cytoplasm, regulate SOS1 on the cytoplasmic membrane, which will therefore achieve an intracellular balance of Na+ (Hill et al., 2013). Protein subcellular location is key in determining the function and accumulation patterns of plant proteins (Hooper et al., 2020). In Chrysanthemum crassum, CcSOS1 was expressed close to the plasma membrane in transiently transformed onion epidermal cells (Song et al., 2012). Like the A. thaliana homologue AtSOS1 (Shi et al., 2002), CcSOS1 is regulated by salinity, especially in the roots after stress, and could play an important role in salt tolerance in C. crassum. In rice (Gupta et al., 2021) and cotton (Guo et al., 2020), SOS1 genes were also predicted to express in plasma membrane. To investigate the subcellular localization of StSOS1-13, the cassette encoding StSOS1-13-Green Fluorescent protein (GFP) fusion protein driven by the CaMV 35S promoter (35S::StSOS1-13-GFP) was transformed into Nicotiana benthamiana leaves, and the fluorescence was observed using the confocal microscope. Fluorescence localization verified that the selected StSOS1-13 was expressed in the plasma membrane (Figure 9), demonstrating the reliability and accuracy of the predicted results.
5 Conclusions
This study provides a genome-wide analysis of the StSOS1 genes, with 37 StSOS1s in the potato identified and divided into three subfamilies. We found that segmental and tandem duplication contribute to the expansion of StSOS1 gene family. These StSOS1s phylogenetically cluster with SlSOS1s and CaSOS1s. The exon-intron structures and motifs of StSOS1s further suggest that the potato SOS1 proteins were highly conserved within the subfamilies. In addition, subcellular localization in Nicotiana benthamiana suggested that StSOS1-13 was located on the plasma membrane. The RT-qPCR results suggested the crucial role of the StSOS1s in response to salt and homologous stress, and suggested that some specific up-regulated genes such as StSOS1-1, StSOS1-13, and StSOS1-29 would be potential candidates for potato salt-tolerant seeding. The results presented in this study will provide essential clues in elucidating the role of the StSOS1s in abiotic stress and the mechanisms underlying the tolerance to salt stress in potato mediated by the StSOS1 proteins.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the corresponding author GG (ggsxnu@126.com), without undue reservation. The potato RNA-Seq data in this article can be download in NCBI with accession number ERP000527.
Author contributions
LL conceived and designed the study. LG analyzed and mapped the bioinformatics content, designed and performed the experimental work, interpreted and analyzed the data, and wrote the manuscript. YZ and ZH carried out the experimental work. WW, XZ, YW, XL, and SG helped to supplement the bioinformatics content and beautify the images. GG and WL supervised the project and critically revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (31771858), Natural Science Foundation of Shanxi Province (202203021211249, 202203021211259), Postgraduate Innovation Project of Shanxi Normal University (2021XSYO27, 2022XSY004), Innovation and Entrepreneurship Training Program for College Students of Shanxi Province (20220303).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1201730/full#supplementary-material
Footnotes
- ^ http://plants.ensembl.org/index.html
- ^ http://pfam.xfam.org
- ^ http://hmmer.janelia.org/
- ^ http://www.ncbi.nlm.nih.gov/cdd
- ^ http://smart.emblheidelberg.de/
- ^ http://www.expasy.org/
- ^ https://services.healthtech.dtu.dk/services/NetPhos-3.1/
- ^ https://www.uniprot.org/
- ^ http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2/
- ^ http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
- ^ https://wordart.com
- ^ http://memesuite.org/
- ^ http://gsds.cbi.pku.edu.cn/
- ^ http://plantregmap.gao-lab.org/go.php
- ^ https://cn.string-db.org/
- ^ https://www.ncbi.nlm.nih.gov/tools/primer-blast/
References
Świeżawska, B., Duszyn, M., Jaworski, K., Szmidt-Jaworska, A. (2018). Downstream targets of cyclic nucleotides in plants. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01428
Akram, U., Song, Y., Liang, C., Abid, M. A., Askari, M., Myat, A. A., et al. (2020). Genome-wide characterization and expression analysis of NHX gene family under salinity stress in Gossypium barbadense and its comparison with Gossypium hirsutum. Genes (Basel) 11, 803. doi: 10.3390/genes11070803
Ali, A., Alexandersson, E., Sandin, M., Resjö, S., Lenman, M., Hedley, P., et al. (2014). Quantitative proteomics and transcriptomics of potato in response to Phytophthora infestans in compatible and incompatible interactions. BMC Genomics 15, 497. doi: 10.1186/1471-2164-15-497
Ali, A., Raddatz, N., Pardo, J. M., Yun, D. J. (2021). HKT sodium and potassium transporters in Arabidopsis thaliana and related halophyte species. Physiol. Plant 171, 546–558. doi: 10.1111/ppl.13166
Brindha, C., Vasantha, S., Raja, A. K., Tayade, A. S. (2021). Characterization of the salt overly sensitive pathway genes in sugarcane under salinity stress. Physiol. Plant 171, 677–687. doi: 10.1111/ppl.13245
Ceci, A. T., Franceschi, P., Serni, E., Perenzoni, D., Oberhuber, M., Robatscher, P., et al. (2022). Metabolomic characterization of pigmented and non-pigmented potato cultivars using a joint and individual variation explained (JIVE). Foods 11, 1708. doi: 10.3390/foods11121708
Cha, J. Y., Kim, J., Jeong, S. Y., Shin, G. I., Ji, M. G., Hwang, J. W., et al. (2022). The Na(+)/H(+) antiporter SALT OVERLY SENSITIVE 1 regulates salt compensation of circadian rhythms by stabilizing GIGANTEA in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 119, e2207275119. doi: 10.1073/pnas.2207275119
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp
Chen, X., Lu, X., Shu, N., Wang, D., Wang, S., Wang, J., et al. (2017). GhSOS1, a plasma membrane Na+/H+ antiporter gene from upland cotton, enhances salt tolerance in transgenic Arabidopsis thaliana. PloS One 12, e0181450. doi: 10.1371/journal.pone.0181450
Cheng, C., Zhong, Y., Wang, Q., Cai, Z., Wang, D., Li, C. (2019). Genome-wide identification and gene expression analysis of SOS family genes in tuber mustard (Brassica juncea var. tumida). PloS One 14, e0224672. doi: 10.1371/journal.pone
Chinnusamy, V., Schumaker, K., Zhu, J. K. (2004). Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. J. Exp. Bot. 55, 225–236. doi: 10.1093/jxb/erh005
Contreras-Moreira, B., Naamati, G., Rosello, M., Allen, J. E., Hunt, S. E., Muffato, M., et al. (2022). Scripting analyses of genomes in ensembl plants. Methods Mol. Biol. 2443, 27–55. doi: 10.1007/978-1-0716-2067-0_2
Dahal, K., Li, X. Q., Tai, H., Creelman, A., Bizimungu, B. (2019). Improving potato stress tolerance and tuber yield under a climate change scenario - a current overview. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00563
Fayez, A. G., Esmaiel, N. N., Salem, S. M., Ashaat, E. A., El-Saiedi, S. A., El Ruby, M. O. (2022). miR-454-3p and miR-194-5p targeting cardiac sarcolemma ion exchange transcripts are potential noninvasive diagnostic biomarkers for childhood dilated cardiomyopathy in Egyptian patients. Egypt Heart J. 74, 65. doi: 10.1186/s43044-022-00300-x
Feki, K., Brini, F., Ben Amar, S., Saibi, W., Masmoudi, K. (2015). Comparative functional analysis of two wheat Na(+)/H (+) antiporter SOS1 promoters in Arabidopsis thaliana under various stress conditions. J. Appl. Genet. 56, 15–26. doi: 10.1007/s13353-014-0228-7
Flagel, L. E., Wendel, J. F. (2009). Gene duplication and evolutionary novelty in plants. New Phytol. 183, 557–564. doi: 10.1111/j.1469-8137
Gao, J., Sun, J., Cao, P., Ren, L., Liu, C., Chen, S., et al. (2016). Variation in tissue na(+) content and the activity of SOS1 genes among two species and two related genera of chrysanthemum. BMC Plant Biol. 16, 98. doi: 10.1186/s12870-016-0781-9
Garcia de la Garma, J., Fernandez-Garcia, N., Bardisi, E., Pallol, B., Asensio-Rubio, J. S., Bru, R., et al. (2015). New insights into plant salt acclimation: the roles of vesicle trafficking and reactive oxygen species signalling in mitochondria and the endomembrane system. New Phytol. 205, 216–239. doi: 10.1111/nph.12997
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40 (Database issue), D1178–D1186. doi: 10.1093/nar/gkr944
Guo, W., Li, G., Wang, N., Yang, C., Zhao, Y., Peng, H., et al. (2020). A Na(+)/H(+) antiporter, K2-NhaD, improves salt and drought tolerance in cotton (Gossypium hirsutum l.). Plant Mol. Biol. 102, 553–567. doi: 10.1007/s11103-020-00969-1
Gupta, B. K., Sahoo, K. K., Anwar, K., Nongpiur, R. C., Deshmukh, R., Pareek, A., et al. (2021). Silicon nutrition stimulates salt-overly sensitive (SOS) pathway to enhance salinity stress tolerance and yield in rice. Plant Physiol. Biochem. 166, 593–604. doi: 10.1016/j.plaphy.2021.06.010
Hamaji, K., Nagira, M., Yoshida, K., Ohnishi, M., Oda, Y., Uemura, T., et al. (2009). Dynamic aspects of ion accumulation by vesicle traffic under salt stress in arabidopsis. Plant Cell Physiol. 50, 2023–2033. doi: 10.1093/pcp/pcp143
Hamooh, B. T., Sattar, F. A., Wellman, G., Mousa, M. A. A. (2021). Metabolomic and biochemical analysis of two potato (Solanum tuberosum l.) cultivars exposed to in vitro osmotic and salt stresses. Plants (Basel) 10, 98. doi: 10.3390/plants10010098
He, F., Shi, Y. J., Chen, Q., Li, J. L., Niu, M. X., Feng, C. H., et al. (2022). Genome-wide investigation of the PtrCHLP family reveals that PtrCHLP3 actively mediates poplar growth and development by regulating photosynthesis. Front. Plant Sci. 13. doi: 10.3389/fpls
Hill, C. B., Jha, D., Bacic, A., Tester, M., Roessner, U. (2013). Characterization of ion contents and metabolic responses to salt stress of different arabidopsis AtHKT1;1 genotypes and their parental strains. Mol. Plant 6, 350–368. doi: 10.1093/mp/sss125
Hooper, C. M., Castleden, I. R., Aryamanesh, N., Black, K., Grasso, S. V., Millar, A. H. (2020). CropPAL for discovering divergence in protein subcellular location in crops to support strategies for molecular crop breeding. Plant J. 104, 812–827. doi: 10.1111/tpj.14961
Ilzhöfer, D., Heinzinger, M., Rost, B. (2022). SETH predicts nuances of residue disorder from protein embeddings. Front. Bioinform. 2. doi: 10.3389/fbinf
Jiang, W., Pan, R., Buitrago, S., Wu, C., Abou-Elwafa, S. F., Xu, Y., et al. (2021). Conservation and divergence of the TaSOS1 gene family in salt stress response in wheat (Triticum aestivum l.). Physiol. Mol. Biol. Plants. 27, 1245–1260. doi: 10.1007/s12298-021-01009-y
Kaur, C., Kumar, G., Kaur, S., Ansari, M. W., Pareek, A., Sopory, S. K., et al. (2015). Molecular cloning and characterization of salt overly sensitive gene promoter from Brassica juncea (BjSOS2). Mol. Biol. Rep. 42, 1139–1148. doi: 10.1007/s11033-015-3851-4
Keisham, M., Mukherjee, S., Bhatla, S. C. (2018). Mechanisms of sodium transport in plants-progresses and challenges. Int. J. Mol. Sci. 19, 647. doi: 10.3390/ijms19030647
Koul, A., Sharma, D., Kaul, S., Dhar, M. K. (2019). Identification and in silico characterization of cis-acting elements of genes involved in carotenoid biosynthesis in tomato. 3 Biotech. 9, 287. doi: 10.1007/s13205-019-1798-1
Kumar, P., Kumar, P., Sharma, D., Verma, S. K., Halterman, D., Kumar, A. (2021). Genome-wide identification and expression profiling of basic leucine zipper transcription factors following abiotic stresses in potato (Solanum tuberosum l.). PloS One 16, e0247864. doi: 10.1371/journal.pone.0247864
Li, H., Guan, H., Zhuo, Q., Wang, Z., Li, S., Si, J., et al. (2020). Genome-wide characterization of the abscisic acid-, stress- and ripening-induced (ASR) gene family in wheat (Triticum aestivum l.). Biol. Res. 53, 23. doi: 10.1186/s40659-020-00291-6
Li, J., Li, X., Han, P., Liu, H., Gong, J., Zhou, W., et al. (2021). Genome-wide investigation of bHLH genes and expression analysis under different biotic and abiotic stresses in Helianthus annuus l. Int. J. Biol. Macromol. 189, 72–83. doi: 10.1016/j.ijbiomac
Li, Q., Qin, Y., Hu, X., Jin, L., Li, G., Gong, Z., et al. (2022). Physiology and gene expression analysis of potato (Solanum tuberosum l.) in salt stress. Plants (Basel) 11, 1565. doi: 10.3390/plants11121565
Li, Q., Qin, Y., Hu, X., Li, G., Ding, H., Xiong, X., et al. (2020). Transcriptome analysis uncovers the gene expression profile of salt-stressed potato (Solanum tuberosum l.). Sci. Rep. 10, 5411. doi: 10.1038/s41598-020-62057-0
Liang, Y., Wan, N., Cheng, Z., Mo, Y., Liu, B., Liu, H., et al. (2017). Whole-genome identification and expression pattern of the vicinal oxygen chelate family in rapeseed (Brassica napus l.). Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00745
Luo, B., Guang, M., Yun, W., Ding, S., Ren, S., Gao, H. (2022). Camellia sinensis chloroplast fluoride efflux gene CsABCB9 is involved in the fluoride tolerance mechanism. Int. J. Mol. Sci. 23, 7756. doi: 10.3390/ijms23147756
Ma, R., Liu, W., Li, S., Zhu, X., Yang, J., Zhang, N., et al. (2021). Genome-wide identification, characterization and expression analysis of the CIPK gene family in potato (Solanum tuberosum l.) and the role of StCIPK10 in response to drought and osmotic stress. Int. J. Mol. Sci. 22, 13535. doi: 10.3390/ijms222413535
Mo, F., Li, L., Zhang, C., Yang, C., Chen, G., Niu, Y., et al. (2022). Genome-wide analysis and expression profiling of the Phenylalanine ammonia-lyase gene family in Solanum tuberosum. Int. J. Mol. Sci. 23, 6833. doi: 10.3390/ijms23126833
Naureen, U., Khosa, A. N., Mukhtar, M. A., Nabi, F., Ahmed, N., Saleem, M. (2023). Genetic biodiversity and posttranslational modifications of protease serine endopeptidase in different strains of Sordaria fimicola. BioMed. Res. Int. 2023, 2088988. doi: 10.1155/2023/2088988
Núñez-Ramírez, R., Sánchez-Barrena, M. J., Villalta, I., Vega, J. F., Pardo, J. M., Quintero, F. J., et al. (2012). Structural insights on the plant salt-overly-sensitive 1 (SOS1) Na(+)/H(+) antiporter. J. Mol. Biol. 424, 283–294. doi: 10.1016/j.jmb.2012.09.015
Nutan, K. K., Kumar, G., Singla-Pareek, S. L., Pareek, A. (2018). A salt overly sensitive pathway member from Brassica juncea BjSOS3 can functionally complement ΔAtsos3 in arabidopsis. Curr. Genomics 19, 60–69. doi: 10.2174/1389202918666170228133621
Pérez-Alonso, M. M., Ortiz-García, P., Moya-Cuevas, J., Pollmann, S. (2021). Mass spectrometric monitoring of plant hormone cross talk during biotic stress responses in potato (Solanum tuberosum l.). Methods Mol. Biol. 2354, 143–154. doi: 10.1007/978-1-0716-1609-3_7
Rogozin, I. B., Wolf, Y. I., Sorokin, A. V., Mirkin, B. G., Koonin, E. V. (2003). Remarkable interkingdom conservation of intron positions and massive, lineage-specific intron loss and gain in eukaryotic evolution. Curr. Biol. 13, 1512–1517. doi: 10.1016/s0960-9822(03)00558-x
Rolly, N. K., Imran, Q. M., Lee, I. J., Yun, B. W. (2020). Salinity stress-mediated suppression of expression of salt overly sensitive signaling pathway genes suggests negative regulation by AtbZIP62 transcription factor in Arabidopsis thaliana. Int. J. Mol. Sci. 21, 1726. doi: 10.3390/ijms21051726
Sharma, B., Saxena, H., Negi, H. (2021). Genome-wide analysis of HECT E3 ubiquitin ligase gene family in Solanum lycopersicum. Sci. Rep. 11, 15891. doi: 10.1038/s41598-021-95436-2
Shi, H., Ishitani, M., Kim, C., Zhu, J. K. (2000). The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. U. S. A. 97, 6896–6901. doi: 10.1073/pnas.120170197
Shi, H., Lee, B. H., Wu, S. J., Zhu, J. K. (2003). Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat. Biotechnol. 21, 81–85. doi: 10.1038/nbt766
Shi, H., Quintero, F. J., Pardo, J. M., Zhu, J. K. (2002). The putative plasma membrane Na(+)/H(+) antiporter SOS1 controls long-distance na(+) transport in plants. Plant Cell. 14, 465–477. doi: 10.1105/tpc.010371
Song, A., Lu, J., Jiang, J., Chen, S., Guan, Z., Fang, W., et al. (2012). Isolation and characterisation of Chrysanthemum crassum SOS1, encoding a putative plasma membrane Na(+)/H(+) antiporter. Plant Biol. (Stuttg). 14, 706–713. doi: 10.1111/j.1438-8677
Sun, H., Ren, M., Zhang, J. (2022). Genome-wide identification and expression analysis of fibrillin (FBN) gene family in tomato (Solanum lycopersicum l.). PeerJ 10, e13414. doi: 10.7717/peerj.13414
Takahashi, S., Seki, M., Ishida, J., Satou, M., Sakurai, T., Narusaka, M., et al. (2004). Monitoring the expression profiles of genes induced by hyperosmotic, high salinity, and oxidative stress and abscisic acid treatment in arabidopsis cell culture using a full-length cDNA microarray. Plant Mol. Biol. 56, 29–55. doi: 10.1007/s11103-004-2200-0
Takeuchi, A., Akatsu, Y., Asahi, T., Okubo, Y., Ohnuma, M., Teramura, H., et al. (2022). Procedure for the efficient acquisition of progeny seeds from crossed potato plants grafted onto tomato. Plant Biotechnol. (Tokyo). 39, 195–197. doi: 10.5511/plantbiotechnology.21.1119a
Tester, M., Davenport, R. (2003). Na+ tolerance and na+ transport in higher plants. Ann. Bot. 91, 503–527. doi: 10.1093/aob/mcg058
Wang, T., Gao, X., Chen, S., Li, D., Chen, S., Xie, M., et al. (2021). Genome-wide identification and expression analysis of ethylene responsive factor family transcription factors in Juglans regia. PeerJ 9, e12429. doi: 10.7717/peerj.12429
Wang, Z., Hong, Y., Li, Y., Shi, H., Yao, J., Liu, X., et al. (2021). Natural variations in SlSOS1 contribute to the loss of salt tolerance during tomato domestication. Plant Biotechnol. J. 19, 20–22. doi: 10.1111/pbi.13443
Wang, J., Zhang, Y., Xu, N., Zhang, H., Fan, Y., Rui, C., et al. (2021). Genome-wide identification of CK gene family suggests functional expression pattern against Cd(2+) stress in Gossypium hirsutum l. Int. J. Biol. Macromol. 188, 272–282. doi: 10.1016/j.ijbiomac
Wu, Y., Ding, N., Zhao, X., Zhao, M., Chang, Z., Liu, J., et al. (2007). Molecular characterization of PeSOS1: the putative Na(+)/H (+) antiporter of Populus euphratica. Plant Mol. Biol. 65, 1–11. doi: 10.1007/s11103-007-9170-y
Wu, S. J., Ding, L., Zhu, J. K. (1996). SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell. 8, 617–627. doi: 10.1105/tpc.8.4.617
Xiang, X. H., Wu, X. R., Chao, J. T., Yang, M. L., Yang, F., Chen, G., et al. (2016). Genome-wide identification and expression analysis of the WRKY gene family in common tobacco (Nicotiana tabacum l.). Yi Chuan. 38, 840–856. doi: 10.16288/j.yczz
Xie, Q., Zhou, Y., Jiang, X. (2022). Structure, function, and regulation of the plasma membrane Na(+)/H(+) antiporter salt overly sensitive 1 in plants. Front. Plant Sci. 13. doi: 10.3389/fpls
Xu, H., Jiang, X., Zhan, K., Cheng, X., Chen, X., Pardo, J. M., et al. (2008). Functional characterization of a wheat plasma membrane Na+/H+ antiporter in yeast. Arch. Biochem. Biophys. 473, 8–15. doi: 10.1016/j.abb
Yang, Q., Chen, Z. Z., Zhou, X. F., Yin, H. B., Li, X., Xin, X. F., et al. (2009). Overexpression of SOS (Salt overly sensitive) genes increases salt tolerance in transgenic Arabidopsis. Mol. Plant 2, 22–31. doi: 10.1093/mp/ssn058
Yang, G., Xu, C., Varjani, S., Zhou, Y., Wc Wong, J., Duan, G. (2022). Metagenomic insights into improving mechanisms of Fe(0) nanoparticles on volatile fatty acids production from potato peel waste anaerobic fermentation. Bioresour. Technol. 361, 127703. doi: 10.1016/j.biortech.2022.127703
Yang, X., Yuan, J., Luo, W., Qin, M., Yang, J., Wu, W., et al. (2020). Genome-wide identification and expression analysis of the class III peroxidase gene family in potato (Solanum tuberosum l.). Front. Genet. 11. doi: 10.3389/fgene.2020.593577
You, X., Nasrullah, Wang, D., Mei, Y., Bi, J., Liu, S., et al. (2022). N(7) -SSPP fusion gene improves salt stress tolerance in transgenic arabidopsis and soybean through ROS scavenging. Plant Cell Environ. 45, 2794–2809. doi: 10.1111/pce.14392
Yu, R. M., Suo, Y. Y., Yang, R., Chang, Y. N., Tian, T., Song, Y. J., et al. (2021). StMBF1c positively regulates disease resistance to Ralstonia solanacearum via it’s primary and secondary upregulation combining expression of StTPS5 and resistance marker genes in potato. Plant Sci. 307, 110877. doi: 10.1016/j.plantsci
Zhang, M., Cao, J., Zhang, T., Xu, T., Yang, L., Li, X., et al. (2022). A putative plasma membrane Na(+)/H(+) antiporter GmSOS1 is critical for salt stress tolerance in Glycine max. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.870695
Zhang, Q., Hou, S., Sun, Z., Chen, J., Meng, J., Liang, D., et al. (2021). Genome-wide identification and analysis of the MADS-box gene family in Theobroma cacao. Genes (Basel) 12. doi: 10.3389/fpls.2022.870695
Zhang, C., Liu, S., Liu, D., Guo, F., Yang, Y., Dong, T., et al. (2022). Genome-wide survey and expression analysis of GRAS transcription factor family in sweetpotato provides insights into their potential roles in stress response. BMC Plant Biol. 22, 232. doi: 10.1186/s12870-022-03618-5
Zhao, C., William, D., Sandhu, D. (2021). Isolation and characterization of salt overly sensitive family genes in spinach. Physiol. Plant 171, 520–532. doi: 10.1111/ppl.13125
Zhao, S., Zhang, Q., Liu, M., Zhou, H., Ma, C., Wang, X. X. X. P. (2021). Regulation of plant responses to salt stress. Int. J. Mol. Sci. 22, 4609. doi: 10.3390/ijms22094609
Zhou, X., Li, J., Wang, Y., Liang, X., Zhang, M., Lu, M., et al. (2022). The classical SOS pathway confers natural variation of salt tolerance in maize. New Phytol. 236, 479–494. doi: 10.1111/nph.18278
Zhou, Y., Yin, X., Duan, R., Hao, G., Guo, J., Jiang, X. (2015). SpAHA1 and SpSOS1 coordinate in transgenic yeast to improve salt tolerance. PloS One 10, e0137447. doi: 10.1371/journal.pone.0137447
Zhu, L., Li, M., Huo, J., Lian, Z., Liu, Y., Lu, L., et al. (2021). Overexpression of NtSOS2 from halophyte plant N. tangutorum enhances tolerance to salt stress in arabidopsis. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.716855
Zhu, J. K., Liu, J., Xiong, L. (1998). Genetic analysis of salt tolerance in arabidopsis. evidence for a critical role of potassium nutrition. Plant Cell. 10, 1181–1191. doi: 10.1105/tpc.10.7.1181
Keywords: Solanum tuberosum L., SOS1, expression profiles, abiotic stress, genome-wide
Citation: Liang L, Guo L, Zhai Y, Hou Z, Wu W, Zhang X, Wu Y, Liu X, Guo S, Gao G and Liu W (2023) Genome-wide characterization of SOS1 gene family in potato (Solanum tuberosum) and expression analyses under salt and hormone stress. Front. Plant Sci. 14:1201730. doi: 10.3389/fpls.2023.1201730
Received: 07 April 2023; Accepted: 14 June 2023;
Published: 30 June 2023.
Edited by:
Yan Zhao, Shandong Agricultural University, ChinaReviewed by:
Wenjiao Zhu, Nanjing Agricultural University, ChinaGennady L. Burygin, Institute of Biochemistry and Physiology of Plants and Microorganisms (RAS), Russia
Copyright © 2023 Liang, Guo, Zhai, Hou, Wu, Zhang, Wu, Liu, Guo, Gao and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gang Gao, ggsxnu@126.com; Weizhong Liu, liuwzh@sxnu.edu.cn
†These authors have contributed equally to this work and share first authorship
 Liqin Liang†
Liqin Liang† Liuyan Guo
Liuyan Guo Yifan Zhai
Yifan Zhai Gang Gao
Gang Gao Weizhong Liu
Weizhong Liu