- 1Herbarium, Department of Botany, Research Center for Plant Science, Ferdowsi University of Mashhad, Mashhad, Iran
- 2Herbarium, Research Institute of Forests and Rangelands, Tehran, Iran
- 3Department of Botany and Biodiversity Research, University of Vienna, Vienna, Austria
Although the mountains in South-West Asia are a global biodiversity hotspot, our understanding of their biodiversity, especially in the commonly remote alpine and subnival zones, is still limited. This is well exemplified here by Aethionema umbellatum (Brassicaceae), a species considered to have a wide yet disjoint distribution in the Zagros and Yazd-Kerman mountains of western and central Iran. Morphological and molecular phylogenetic data (based on plastid trnL-trnF and nuclear ITS sequences) show that A. umbellatum is restricted to a single mountain range in southwestern Iran (Dena Mts., southern Zagros), whereas populations from central Iran (Yazd-Kerman and central Zagros) and from western Iran (central Zagros) belong to species new to science, A. alpinum and A. zagricum, respectively. Both new species are phylogenetically and morphologically close to A. umbellatum, with which they share unilocular fruits and one-seeded locules. However, they are easily distinguishable by leaf shape, petal size, and fruit characters. This study confirms that the alpine flora of the Irano-Anatolian region is still poorly known. As the proportion of rare and local endemic species in alpine habitats is high, these habitats are of prime interest for conservation efforts.
1 Introduction
Mountains are known as biodiversity hotspots with a high proportion of endemic and narrowly distributed species (Hoorn et al., 2013; Vanderplank et al., 2014; Hoorn et al., 2018; Noroozi et al., 2019a; Noroozi et al., 2019b), with alpine habitats being the centers of endemism (Testolin et al., 2021). Tectonic uplift, geographic isolation, climatic oscillations, and strong microhabitat differentiation in high mountains are among the major reasons for allopatric speciation in high elevations (Körner, 1995; Agakbanjanz and Breckle, 2002; Dirnböck et al., 2011; Sandel et al., 2011; Parisod, 2022). A particularly high rate of endemic species is found in the mountains of South-West Asia, a global biodiversity hotspot (Mittermeier et al., 2011; Noroozi et al., 2019a; Noroozi et al., 2019b), where the rate of endemism of the subalpine and alpine flora exceeds 75% (Noroozi et al., 2021). These mountains are characterized by high elevational amplitude, topographic complexity, and strong habitat isolation (Noroozi et al., 2018; Noroozi et al., 2021), likely drivers of the high plant diversity in this region. However, the high elevations of these mountains are floristically poorly explored, and it is estimated that ca. 7% of the alpine flora of the Iranian mountains are still taxonomically undescribed (Noroozi et al., 2016), indicating that the region’s diversity, although already high, is still considerably underestimated. This may be particularly the case for the isolated high mountains in Zagros in western and southwestern Iran, where during the last years, many new species have been described (such as Moazzeni et al., 2014; Moazzeni et al., 2016; Balali et al., 2019; Doostmohammadi and Mirtadzadini, 2019; Pahlevani and Amini Rad, 2019; Noroozi et al., 2020; Alipour et al., 2021).
Aethionema Aiton (Brassicaceae) comprises approximately 70 species distributed from the western Mediterranean region via South-West Asia to Central Asia, with the majority of species being found in the Irano-Anatolian region in Turkey and Iran (Al-Shehbaz et al., 2006; Mohammadin et al., 2017; Moazzeni et al., 2018). In South-West Asia, Aethionema species occur from lowlands up to the high alpine and subnival zones, and approximately one-third of the species are found, often exclusively, in alpine habitats (Hedge, 1965; Hedge, 1968; Moazzeni et al., 2018). The genus comprises perennial herbaceous plants and small shrubs, rarely annuals, with linear or narrowly oblong to ovate or orbicular leaves, pink, lilac, or white flowers, and glabrous mainly angustiseptate dehiscent (rarely indehiscent) silicles with one or two locules (Moazzeni et al., 2018). Morphological characters diagnostic on the species level include features of the filaments (entire, winged, or with teeth), leaves (shape), and fruits (shape, wing characters, number of locules; Moazzeni et al., 2018). Molecular phylogenetic studies confirmed that Aethionema is sister group to the rest of Brassicaceae (= core Brassicaceae; Couvreur et al., 2010), established the monophyly of the genus (Lenser et al., 2016; Mohammadin et al., 2017) after transferal of five traditionally recognized Aethionema species to Noccaea (Al-Shehbaz, 2012), and provided hypotheses on phylogenetic structure within the genus (Lenser et al., 2016; Mohammadin et al., 2017). Being a sister group to all other Brassicaceae, Aethionema has become the object for studies on multiple aspects of plant biology, such as dimorphic seeds, seed germination, and seed coat differentiation (Bhattacharya et al., 2019; Mérai et al., 2019; Arshad et al., 2021), as well as phylogeny and taxonomy (Mohammadin et al., 2017; Moazzeni et al., 2018).
In this paper, we focus on Aethionema umbellatum (Boiss.) Bornm., which, although known for a long time only from its locus classicus on Dena Mts. in southern Zagros, is currently considered a widely, yet disjointly distributed species (Hedge, 1968; Moazzeni et al., 2018). However, initial morphological investigations of material from different regions including the type locality indicated taxonomically relevant differences in leaf and fruit shape, suggesting that A. umbellatum, as hitherto understood, contains more than one species. To test this hypothesis, we conducted molecular phylogenetic analyses using nuclear ITS and plastid trnL-trnF sequences (as for these markers broad data sets across Aethionema are already available; Lenser et al., 2016; Mohammadin et al., 2017) and analyzed morphological characters, focusing on those known to be diagnostic in this genus (such as leaf shape and number of locules per fruit; Hedge, 1965; Hedge, 1968; Mohammadin et al., 2017; Moazzeni et al., 2018). Based on the results of these analyses, we taxonomically revise this group, including the description of two new Aethionema species.
2 Material and methods
2.1 Plant material
We included nine specimens identified as A. umbellatum [two from Shirkuh; one each from Damaneh, Zardkuh, and Oshtorankuh Mts.; and four individuals from the type location of A. umbellatum; material from the Kerman province (documented by a voucher in Edinburgh herbarium: E0061167) and one from central Zagros (W 0215578) were not available to us for molecular study; see Figure 1].
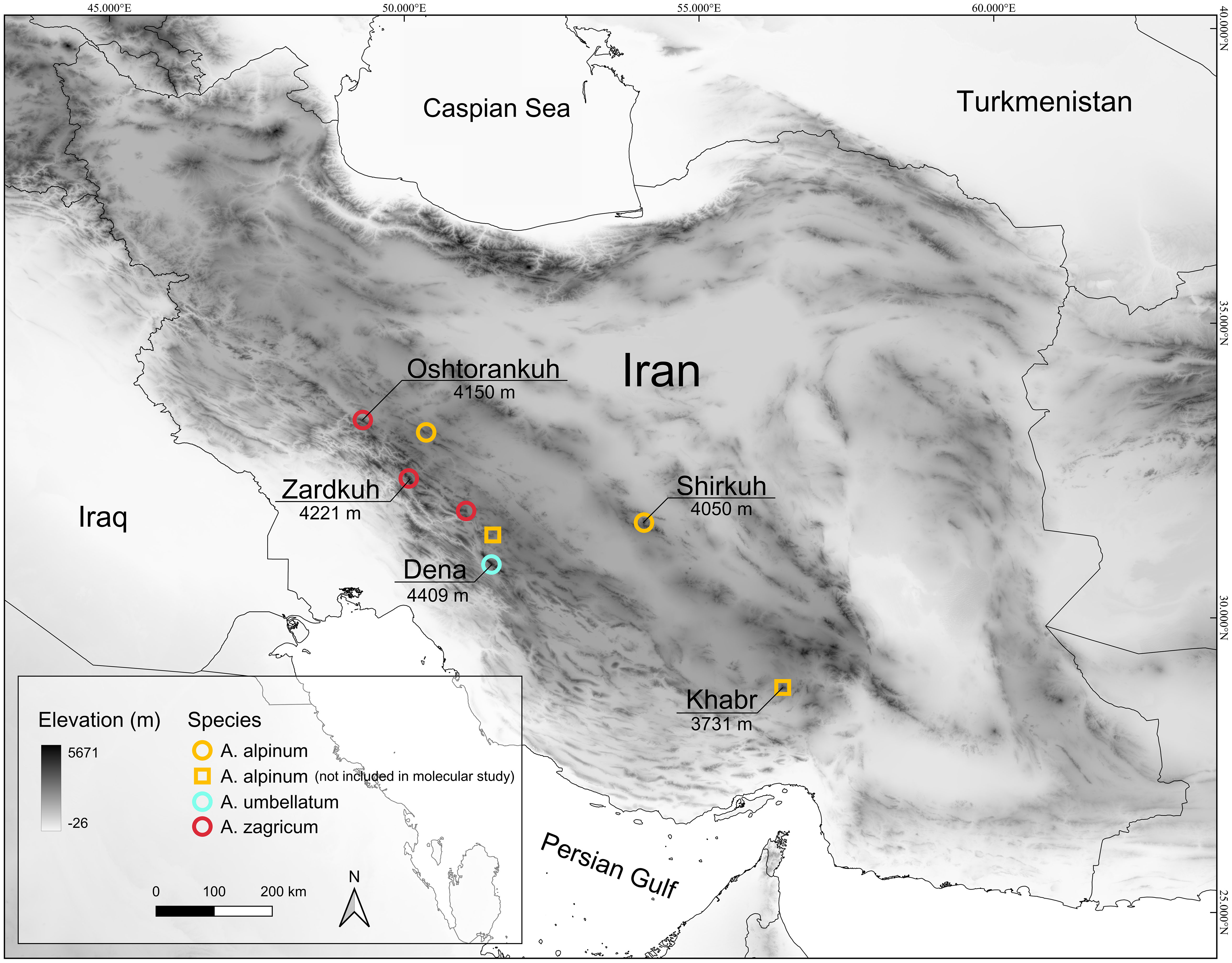
Figure 1 Distribution map of Aethionema alpinum, A. umbellatum, and A. zagricum in Iran. The highest mountain peaks of the Zagros, Yazd, and the southern Kerman mountains (where the species have been recorded) are presented on the map.
2.2 Molecular analyses
Total genomic DNA was extracted from silica-dried material or herbarium specimens using the CTAB protocol (Doyle and Doyle, 1987). Voucher information and GenBank accession numbers for all newly included specimens are listed in Table S1.
The internal transcribed spacer (ITS) and the trnL-trnF region were amplified and sequenced using primers 18F and 26S-82R (Mummenhoff et al., 1997) and c and f (Taberlet et al., 1991), respectively. The standard 25-μl PCR mixtures contained 1 μl of each primer (10 μm, BGI, Hong Kong, China), 12.5 μl of 2× Taq DNA Polymerase Master Mix RED (Amplicon, Denmark), 1 μl of unquantified template DNA, and 1 μl of dimethyl sulfoxide (DMSO, 78.13 g mol, Merck, Darmstadt, Germany); deionized water was added to achieve a final volume of 25 μl. Amplification was done with optimized annealing temperatures (ITS: 38°C; trnL-trnF: 51°C). Amplification products for both markers were purified using PEG (Joly et al., 2006). Sequencing was performed by BGI (Hong Kong, China) using the PCR primers.
The newly generated sequences (23 sequences) were trimmed and assembled using Geneious 6.1.2 (https://www.geneious.com). Sequences were added to the ITS and trnL-trnF data sets (including only Aethionema samples to avoid alignment issues with too distant outgroups) obtained from Mohammadin et al. (2017) and Lenser et al. (2016), respectively, thus including sequences from jointly 54 accessions (Table S1). Based on previous results (Mohammadin et al., 2017 and references therein), A. spinosum and A. lepidioides were used as outgroups. Both ITS and trnL-trnF data sets were aligned with MAFFT 1.3 as implemented in Geneious 6.1.2 under default settings. Alignments were manually corrected in Geneious 6.1.2. Using the AIC criterion as implemented in jModelTest 2.1.4 (Darriba et al., 2012), the GTR + G + I and GTR + G models were chosen as best-fit substitution models for the ITS and the trnL-trnF alignments, respectively. All aligned matrices are available in Data Sheet 2 of the Supplementary Material.
Phylogenetic analyses were performed using maximum likelihood (ML) and Bayesian inference (BI). The ML analysis was carried out on the IQ-TREE web server (http://iqtree.cibiv.univie.ac.at/; Minh et al., 2020) using the default settings, assessing branch support via 1,000 bootstrap replicates employing the ultrafast bootstrap approximation (UFBoot; Minh et al., 2013). BI was performed using MrBayes 3.2.7 (Ronquist et al., 2012) at the CIPRES portal (https://www.phylo.org/index.php; Miller et al., 2010) with default prior settings, running the four chains for 10 million MCMC generations each sampling every 1,000 generations. The quality of the analysis was checked by comparing likelihood values and parameter estimates from different runs in Tracer 1.6 (http://tree.bio.ed.ac.uk/software/tracer/) as well as checking the average standard deviation of split frequencies (to be <0.01) and ESS values (to be >200). After discarding the initial 25% of trees as burn-in, the remaining trees were summarized in a 50% majority-rule consensus tree.
After a preliminary phylogenetic analysis, samples with ambiguous phylogenetic positions were removed (Table S1). Due to the general morphological similarity among Aethionema species (Moazzeni et al., 2018) and/or considerable intraspecific morphological variation (Andersson et al., 1983), misidentification may be the main reason for the dubious phylogenetic placement of some Aethionema samples. As we did not have access to the vouchers of these samples and thus could not check their identification, we adopted the strategy to remove them.
2.3 Morphological analyses
The evolution of two morphological characters (leaf shape and locus number), which are important features to identify Aethionema species, was inferred using ML-based ancestral state reconstructions as implemented in Mesquite 3.70 (Maddison and Maddison, 2021) on the better-resolved phylogenetic hypothesis inferred from trnL-trnF sequences. To this end, leaf shape was coded as linear or narrowly oblong versus ovate, obovate, or spathulate, while locus number was coded as unilocular, bilocular, or both. ML reconstructions were performed under a single-rate Mk likelihood model for discrete morphological characters, described by Lewis (2001). The “trace character over trees” option was used to summarize the ancestral state reconstructions over the set of posterior trees.
3 Results
3.1 Molecular phylogeny
The newly obtained sequences are available in GenBank under accession numbers OQ695471–OQ695480 for ITS and OQ726488–OQ726500 for trnL-trnF. Tree topologies obtained via maximum likelihood and Bayesian Inference were essentially identical for both markers (Data Sheet 2 of the Supplementary Material). In both data sets, Aethionema split into three moderately to well-supported main clades [bootstrap support (BS)/posterior probability (PP) of 85–100/0.85–1.0; Figures 2, 3] in agreement with previous studies based on plastome and rRNA-cistron data (Lenser et al., 2016; Mohammadin et al., 2017). All samples presumably identified as A. umbellatum were placed within clade A, where they, as inferred from ITS data (Figure 2), formed a monophyletic group (BS/PP 92/0.85) or, as inferred from trnL-trnF data (Figure 3), were diphyletic (A. umbellatum from Dena Mts.: BS/PP 76/0.92; A. umbellatum from elsewhere: BS/PP 89/0.96) in a trichotomy (BS/PP 94/1) with a clade including, among others, Aethionema saxatile (BS/PP 75/0.89), the species inferred as a sister (BS/PP 99/0.88) to the umbellatum clade by ITS data (Figure 2). Whereas the accessions from Damaneh plus Shirkuh and from Dena Mts. formed clades (BS/PP 76–99/0.92–0.98), those from Zardkuh plus Oshtorankuh Mts. were inferred as clade only by the trnL-trnF data (BS/PP 99/1; Figure 3).
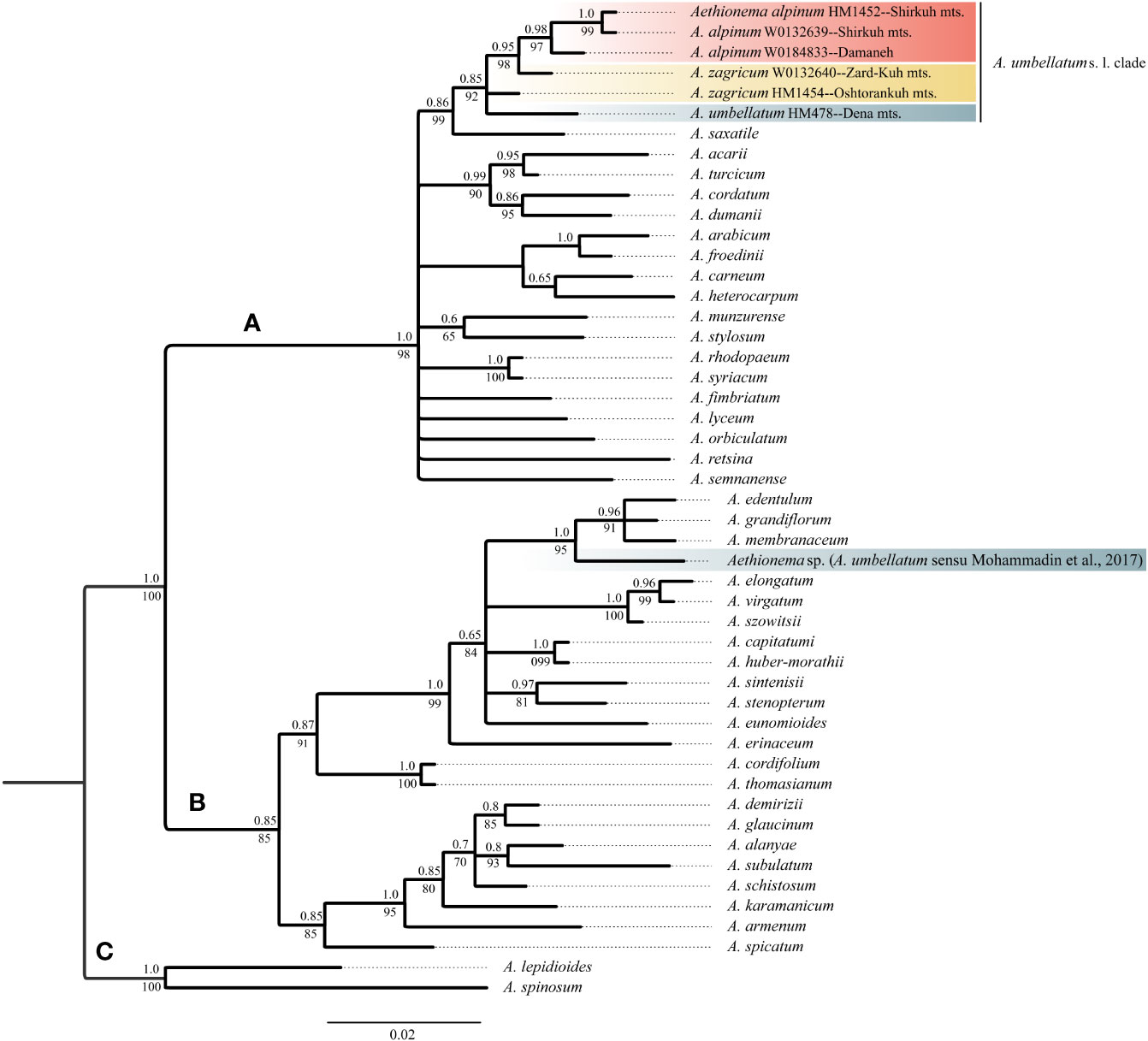
Figure 2 Majority rule consensus tree of Bayesian analysis of nuclear ITS sequences showing the phylogenetic position of Aethionema alpinum, A. zagricum, and A. umbellatums. str. Numbers above the branches indicate posterior probabilities, and those below the branches indicate maximum likelihood bootstrap values; capital letters indicate major clades.
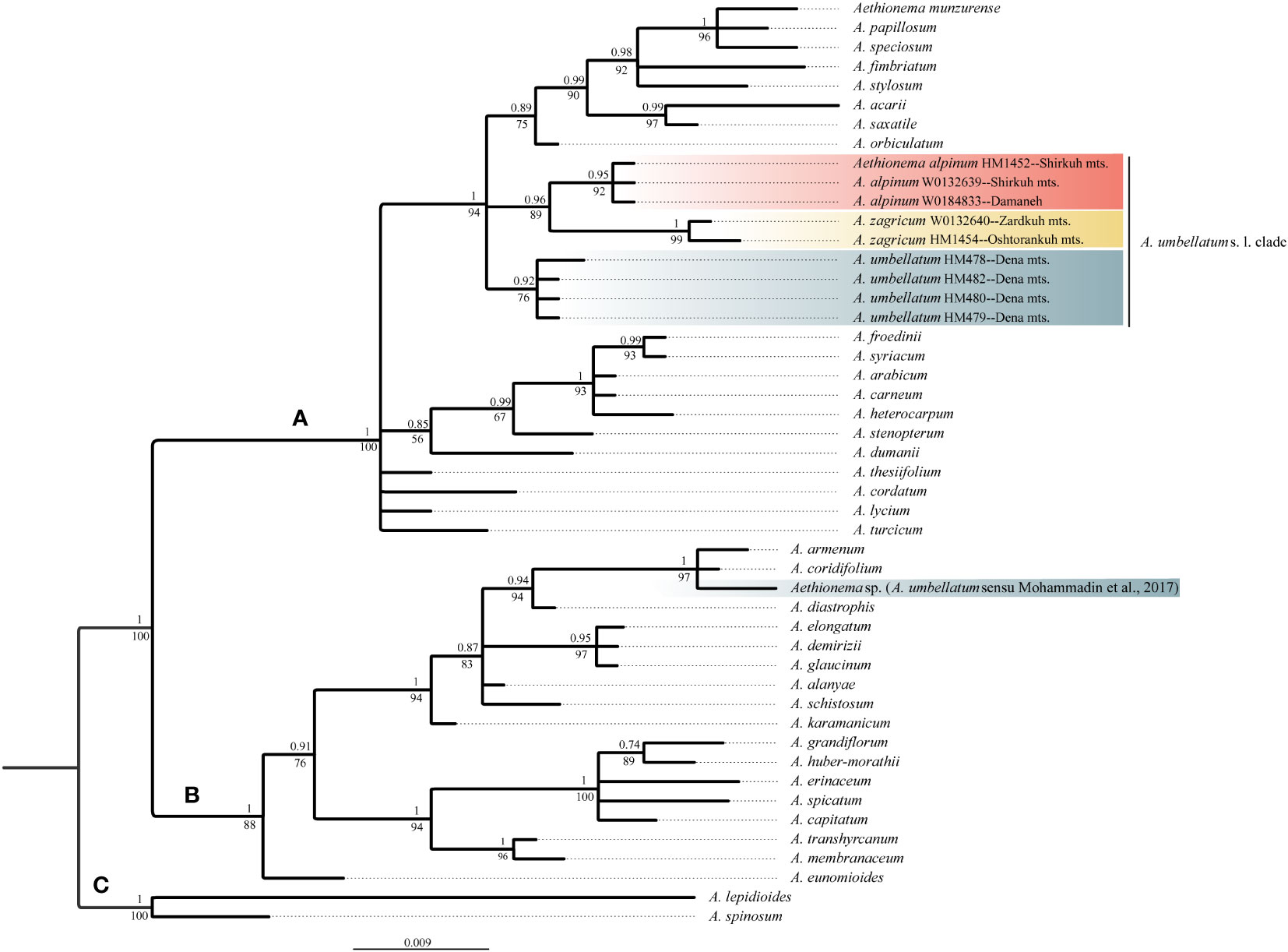
Figure 3 Majority rule consensus tree of Bayesian analysis of plastid trnL-trnF sequences showing the phylogenetic position of Aethionema alpinum, A. zagricum, and A. umbellatum s. str. Numbers above the branches indicate posterior probabilities, and those below the branches indicate maximum likelihood bootstrap values; capital letters indicate major clades.
3.2 Ancestral character state reconstruction
From the ancestral leaf shape, inferred to be linear or narrowly oblong, there were two changes to a broader leaf shape in single species in clade B. In clade A, ovate, obovate, or spathulate leaf shapes dominated, only a few species, including A. umbellatum from Dena Mts., retaining the ancestral state of linear to narrowly oblong leaves (Figure 4A). Starting from bilocular fruits, estimated to be the ancestral state in Aethionema, changes to unilocular fruits were inferred in the outgroup, several times independently in clade B, and on the branch leading to the ancestor of clade A (Figure 4B). Within clade A, several reversals to bilocular fruits were inferred, but A. umbellatum s. l. retained the ancestral condition of clade A, i.e., unilocular fruits.
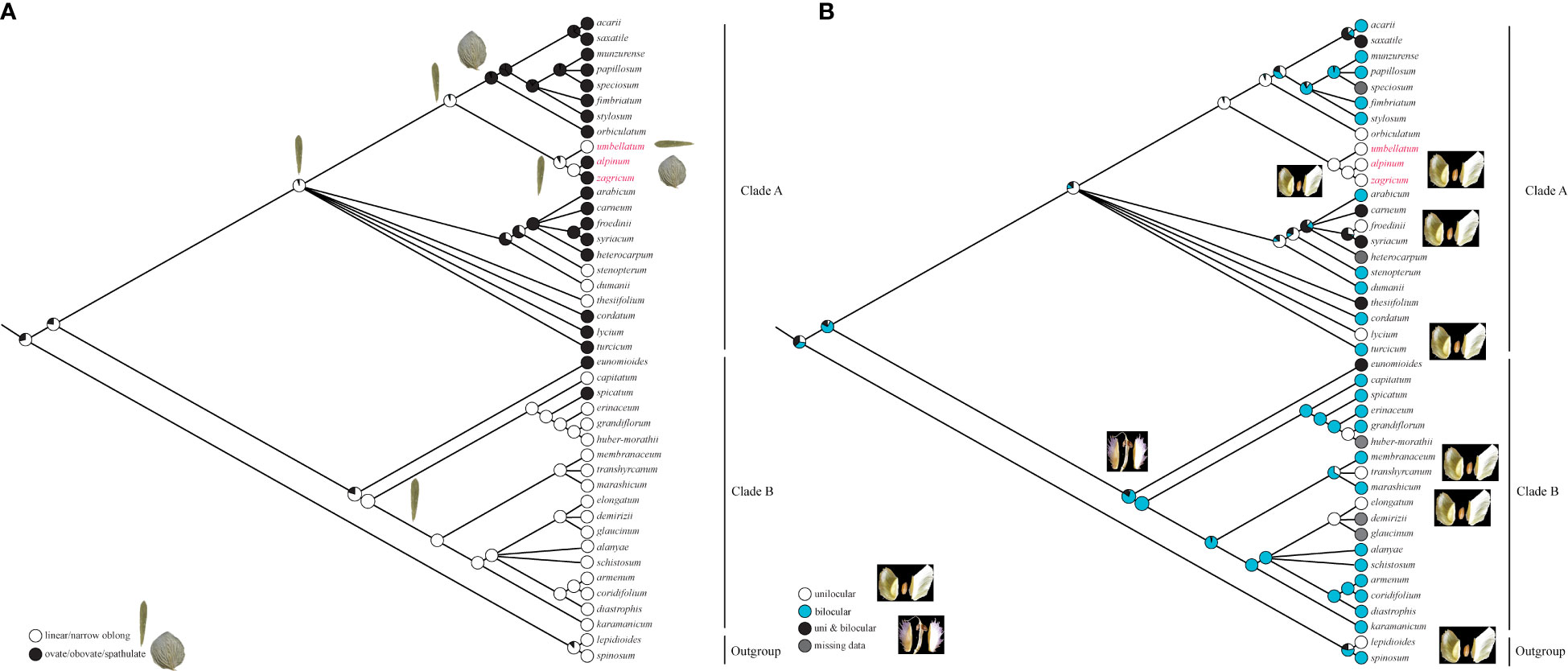
Figure 4 Evolution of (A) leaf shape and (B) the number of locules per fruit in Aethionema inferred using maximum likelihood on the Bayesian tree inferred from plastid trnL-trnF sequences.
3.3 Taxonomic treatment
Based on the phylogenetic results and inspection of 13 specimens of members of A. umbellatum s. l., we taxonomically revise this group to include three species, two of which are (based on five and three specimens, respectively) described herein as new to science.
Aethionema alpinum Moazzeni & Noroozi sp. nov. (Figure 5).
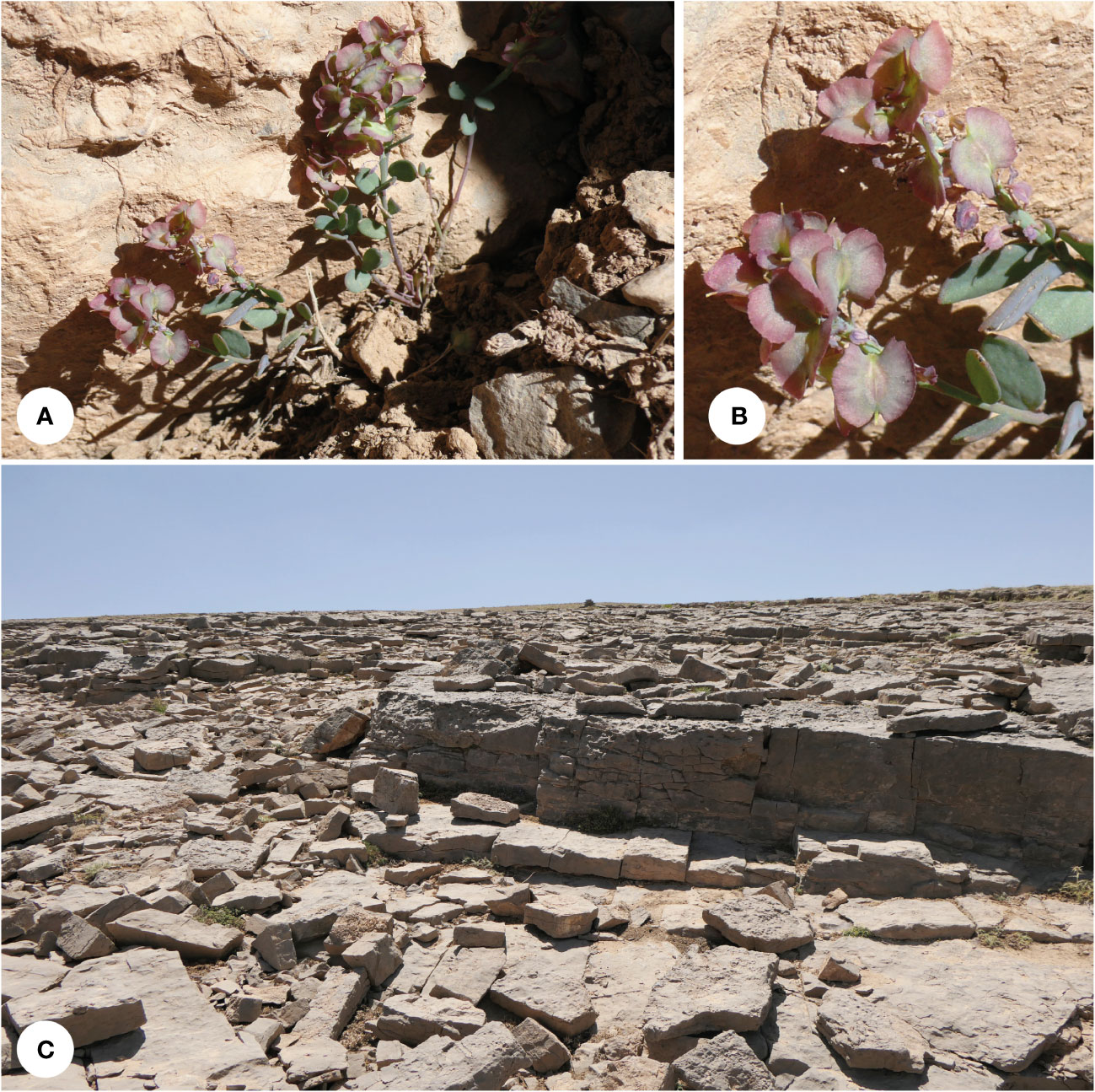
Figure 5 Aethionema alpinum: (A) plants in their natural habitat, (B) close-up of the fruits, and (C) habitat (Shirkuh Mt. 3,800 m a.s.l.; photographs by JN).
Type: Iran, Yazd, Shirkuh Mts., 31°37′N; 54°04′E, 3,800 m a.s.l., 2012 July 05, J. Noroozi & M. Mahmoodi 2843 (Holotype: W 0132639, Isotype: TARI [98638-TARI])
Diagnosis: — Differt ab A. umbellato foliis majoribus et latioribus, foliorum mediorum alternantibus, fructibus minoribus et angustioribus, fructibus alatis undulate.
Description: Low-growing perennial herbs, (4–)6.5–7 cm tall. Lower leaves opposite, middle and upper ones alternate, ovate to rarely elliptic, cuneate at the base, rounded at the apex, 4–8 × (1.2–)1.5–2(2.5) mm, entire. Flower characters not known (no flowering material available). Infructescence 10–17 mm long; fruiting pedicels 2–3(–4) mm long, erect. Fruit suborbicular, unilocular, one-seeded; wing undulate, 1.5–2 mm wide, developed over the entire circumference, apical notch ca. 1–1.5 mm deep; locule 3–3.5 × 3–3.5 mm; style 0.6–1 mm long. Seeds ca. 1.5 × 1 mm.
Phenology: Flowering probably in June and July (we have not seen flowers) and fruiting in June and July.
Distribution and ecology: This plant is known from several collections in high alpine habitats, growing on calcareous bare grounds dominated by rocks and big stones (Figure 5C). This species is limited to the high mountains of central Iran (Yazd-Kerman mountains) and the eastern side of Zagros. These mountains have a more continental climate and receive less precipitation compared with the western sides and the central parts of Zagros.
Etymology: The epithet “alpinum” refers to the alpine habitats the species is restricted to.
Conservation status: Based on the current data, A. alpinum is restricted to an area of 6.4 ha [extent of occurrence (EOO) = 63,892 km2, area of occurrence (AOO) = 16 km2]; thus, following the IUCN Red List criteria (IUCN, 2022), it is categorized as Least Concern (LC).
Additional specimens: Iran, Esfahan, in montibus prope Damaneh 35 km SE Daran, 115 km NW Esfahan, substr. calc. 1974.06.10, J. Renz 47646 (W 0184833); Iran, Kerman, Kuh-e Khabr, 1977.06.08, Assadi, Edmonson & Miller, 1760 (E00061167); Persia: W: Qashqai: Pashma Kuh, a Semirom 5 km occidentem versus, substr. calc., 2,700–3,000 m; 1974 June 06, K.H. Rechinger 47450 (W 0215578).
Aethionema zagricum Moazzeni & Mahmoodi sp. nov. (Figure 6).

Figure 6 Aethionema zagricum: (A) plant in its natural habitat, (B) close-up of the flowers and fruits, and (C) habitat (Oshtorankuh Mt., 3,700 m a.s.l.; photographs by MM).
Type: Iran, Lorestan, Azna, Oshtorankuh, between Bidesraneh village and Sanboran summit, 3,700 m a.s.l., 2012.07.23, Mahmoodi and Hoseini (Holotype: TARI-98504)
Diagnosis: — Differt ab A. umbellato et A. alpino inflorescentiis majoribus, stylis longioribus et alis fructuum angustioribus, et a speciebus Turcicis foliis et petalis brevioribus et angustioribus.
Description: Low-growing perennial herbs, 7–13 cm tall. Lower leaves opposite, upper and middle alternate, elliptic to oblong to rarely obovate, cuneate at the base, rounded at the apex, 4–7 × 1.5–2 mm, entire. Racemes with up to 15 flowers. Sepals 2–3 mm long; petals spathulate, pink, 3–3.5 × 1–1.3 mm, emarginate at the apex; median filaments 3 mm long, lateral ones 2.5 mm long, entire. Infructescence 10–17 mm long; fruiting pedicels 2–3(–4) mm long, erect. Fruit orbicular to rarely suborbicular, unilocular, one-seeded; wing more or less crenulate, 0.5–1.3 mm wide, developed over the entire circumference, apical notch ca. 0.2 mm deep; locule 3.5–5.5 × 3.5–5.5 mm; style 1–1.5 mm long. Seeds not seen.
Phenology: Flowering and fruiting in July and August.
Distribution and ecology: Endemic to the alpine and subnival zones of central Zagros, growing on limestone screes above 3,700 m a.s.l. The mountains in this region (Zardkuh and Oshtorankuh) receive a high amount of precipitation (more than 1,500 mm per year), mostly as snow during the wintertime, but the growing season is usually dry (Mediterranean precipitation regime).
Etymology: The epithet “zagricum” refers to the Zagros Mountain range, where the new species is restricted to.
Conservation status: The new species is known so far only from the highest elevations of Mts. Oshtorankuh and Zardkuh above 3,700 m. Since the highest summits of these mountains are 4,150 and 4,221 m, respectively, the species is strongly restricted elevationally and geographically. As A. zagricum is restricted to a small area of 2.8 ha (EOO = 2,857 km2, AOO = 12 km2), following the IUCN Red List criteria (IUCN 2022), it is categorized as Endangered (EN).
Additional specimens: Iran, Chahār Mahāl va Bakhtīārī, In lapide cacum Kellal, 1868.09, Haussknecht, s.n. (JE 00034617); Zardeh-Kuh, above Kurrang Valley: N.E. facing slope in stony clay, immediately below extensive snow fields. 4,265 m, 1966.08.05 Archibald, 2968 (W0132640).
Aethionema umbellatum (Boiss.) Bornmüller (1911c: 535) (Figure 7)

Figure 7 Aethionema umbellatum: (A, B) plant in its natural habitat, (C) habit, and (D) close-up of the fruits (Dena Mt., 4,100 m a.s.l.; photographs by JN).
≡ Moriera umbellata Boissier (1846: 16) ≡ Crenularia umbellata (Boiss.) Boissier (1867: 338). — Holotype: IRAN. Persia, in glareosis reg[ionum] summarum alpis Kuh-Daëna [Dena], Kotschy 780 (G-BOIS [G00154010]; isotypes: B [B100241892], BM [BM001254112], E [E00061169, E00061170], FI [FI005705], G [G00002123, G00002124], K [K000075747], KW [KW000127991], LE [LE00012860], MO [MO2001061, MO2001062], P [P00835122, P00835123, P00835124, P00835126, P00835127], W [W0005554, W18890020458], WAG [WAG0003966]).
Low-growing perennial herbs, (4–)5–13 cm tall, with a pleiocorm, stems glabrous. Leaves sessile, linear-elliptic, cuneate at the base, rounded at the apex, 4–6 × c. 1 mm, entire, lower and middle opposite, upper alternate. Racemes with up to 30 flowers. Sepals 2–3 mm long; petals spathulate, purple, 2.5–3 × 1.5–2 mm, emarginate at the apex; median filaments 1.5 mm long, lateral ones 1 mm long, entire. Infructescence 8–15 mm long; fruiting pedicels 1–3(–4) mm long, erect. Fruit suborbicular to elliptic, unilocular, one-seeded; wing entire, 1 mm wide, developed over the entire circumference, apical notch ca. 0.5 mm deep; locule 2.5–3.5 × 2–3 mm; style 1 mm long. Seeds ca. 1.5 × 1 mm.
Additional specimens: Iran: Kohgiluyeh and Boyer-Ahmad Province, Sisakht, Dena Mts., near the summit of Hoz-Dal in southern slopes. 30.9058°N; 51.4787°E, 4,000–4,300 m a.s.l., 2019.07.19, Jalil Noroozi 4059 (W! duplicate FUMH)!.
Phenology: Flowering and fruiting in July and August.
Distribution and habitat: It is a local endemic of Dena Mts., southern Zagros, adapted to limestone screes of the subnival zone (Figures 7A, B). The region receives a high amount of annual precipitation as in central Zagros. The accompanying plants of A. umbellatum are Astragalus melanodon Boiss. (Fabaceae), Bromus frigidus Boiss. & Hausskn. (Poaceae), Chenopodium foliosum Asch. (Amaranthaceae), Elymus longearistatus (Boiss.) Tzvelev (Poaceae), Euphorbia aucheri Boiss. (Euphorbiaceae), Galium pseudokurdicum (Ehrend.) Schönb.-Tem. (Rubiaceae), Nepeta lasiocephala Bwnth. (Lamiaceae), Physoptychis gnaphalodes (DC.) Boiss. (Brassicaceae), Potentilla flaccida Th.Wolf ex Bornm. (Rosaceae), Pseudocamelina aphragmodes (Boiss.) N. Busch (Brassicaceae), Silene daenensis Melzh. (Caryophallaceae), and Zerdana anchonioides Boiss. (Brassicaceae).
Conservation status: Aethionema umbellatum is known only from the highest elevations of Dena Mts. above 4,000 m. We estimated the population size to be 20 to 50 individuals. As A. umbellatum is restricted to a small area of 4 km2 (EOO = incomputable, AOO = 4 km2), following the IUCN Red List criteria (IUCN 2022), it is categorized as Critically Endangered (CR).
4 Discussion
4.1 Underestimated diversity in Aethionema umbellatum sensu lato
Our study complements recent studies that the alpine flora in SW Asia, although widely recognized as highly diverse and rich in endemics (Noroozi et al., 2016; Noroozi et al., 2021), is still poorly explored. This is well exemplified here in A. umbellatum s. l., where morphological and molecular data uncovered cryptic species diversity allowing three instead of a single species to be recognized.
Whereas molecular data do not reject current taxonomy (i.e., a broadly defined A. umbellatum), the geographic structure of phylogenetic lineages as well as morphological data suggest the presence of several distinct species within A. umbellatum s. l. (Table 1). The combination of ovate leaves (common in species of clade A, but rare in clade B: Figure 4A; Mohammadin et al., 2017) and unilocular fruits (relatively common in clade A, but rare in clade B: Figure 4B) separates A. alpinum and A. zagricum, which differ from each other in fruit size and leaf number (Table 1), from A. umbellatum s. str. (Figure 4). Such a discrepancy between diagnostically relevant morphological data (here leaf shape and the number of locules per fruit) and molecular data is seen in many cruciferous genera (Meyer, 1973; Mummenhoff et al., 1997; Moazzeni et al., 2007; Moazzeni et al., 2010), and confirms that morphological characters in Brassicaceae often are homoplaseous, although they can be useful features for species identification (Franzke et al., 2011; Al-Shehbaz, 2012; Huang et al., 2016; Walden et al., 2020).
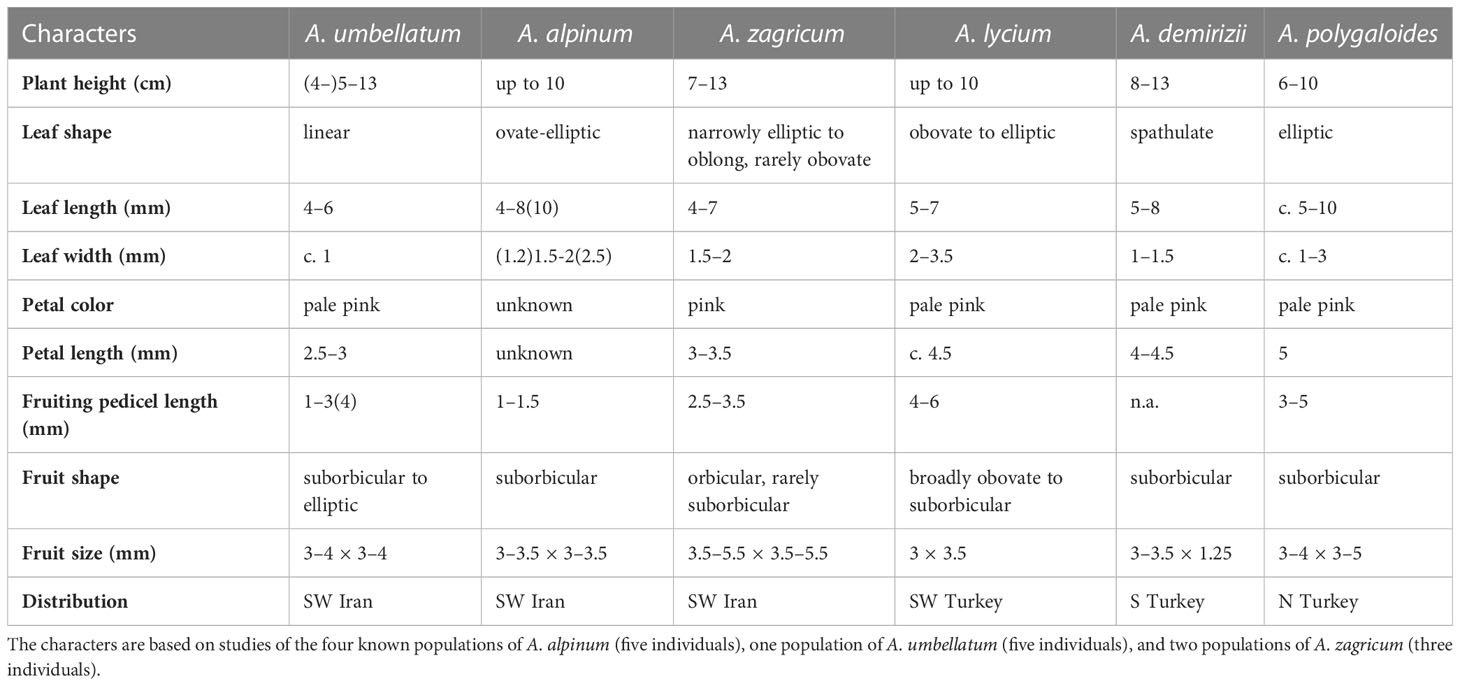
Table 1 Comparison of the morphological characteristics of Aethionema alpinum, A. umbellatum, A. zagricum, and their most similar relatives.
With respect to habit, fruit size, and shape as well as the number of locules per fruit and seeds per locule, A. alpinum and A. zagricum resemble, apart from A. umbellatum s. str., some species from Turkey, such as A. demirizii, A. lycium, and A. polygaloides. Aethionema alpinum can be readily distinguished from A. umbellatum s. str. by larger and broader leaves, by alternate phyllotaxis of the middle leaves, and by smaller and narrower fruits that have deeper notches and from the Turkish species by their shorter height and broader fruits (Table 1). Aethionema zagricum can be distinguished from A. umbellatum s. str. and from A. alpinum by larger inflorescences, longer styles, and narrower fruit wings and from the Turkish species by having shorter and narrower leaves and petals (Table 1).
The accession of A. umbellatum used by Mohammadin et al. (2017) not only did not group with the other accessions of this species collected at its type locality, but actually fell in a different major clade (clade B instead of clade A; Figures 2, 3). The plants used by Mohammadin et al. (2017) were collected from Chalus road in northern Iran (Lenser et al., 2016), but a voucher specimen of A. umbellatum is missing (although intended to be stored at the herbarium of Wageningen University: Lenser et al., 2016). Extensive field studies in the Chalus Road area as well as investigation of herbarium specimens located in main Iranian herbaria (such as TARI, TUH, and IRAN) have never shown the presence of A. umbellatum s. l. in this region, suggesting that the plant used by Mohammadin et al. (2017) has been misidentified and actually belongs to a different species, whose identity cannot be ascertained without the voucher specimen.
4.2 Determination key
Key to the low-growing (up to 30 cm tall) perennial species of Aethionema with unilocular fruits and one seed per locus (species marked with an asterisk are endemic to Iran, the remaining ones are endemic to Turkey):
1. Filaments with a conspicuous tooth in the upper third; fruit cymbiform ...............................................................A. sabzevaricum*
- Filaments entire; fruit not cymbiform .......................................2
2. Leaves < 10, fruit broad, 4–5.5 mm wide ............A. alpinum*
- Leaves at least 10, fruit 3–3.5 mm wide ....................................3
3. Leaves linear, petals 2–3.5 mm long .........................................4
- Leaves ovate to spathulate, petals 4.5–5.5 mm long ...............5
4. Leaves c. 1 mm wide, raceme with up to 30 flowers, petals pale pink, 2.5–3 mm long, style ≤1 mm long, fruit wing entire ...................................................................................A. umbellatum*
- Leaves 1.5–2 mm wide, raceme with up to 15 flowers, petals lilac, 3–3.5 mm long, style 1–1.5 mm long, fruit wing more or less crenulate ...................................................................A. zagricum*
5. Leaves spathulate, fruit 3 mm wide.........................A. demirizii
- Leaves ovate to obovate, fruit 1.5 mm wide ..............................6
6. Lower leaves alternate, ovate; fruit suborbicular .....................................................................................A. polygaloides
- Lower leaves opposite, elliptic to obovate to elliptic, fruit broadly obovate ....................................................................A. lycium
5 Conclusion
Our explorations over the last years of high elevations of the mountains of Iran have resulted in the description of several species new to science (Noroozi et al., 2010; Mahmoodi et al., 2013; Noroozi and Ajani, 2013; Noroozi et al., 2020), confirming that the alpine and subnival flora of the Irano-Anatolian biodiversity hotspot is still insufficiently known. The proportion of rare and local endemic species at high elevations is very high (Noroozi et al., 2021), rendering these habitats very important for conservation biology. This is exacerbated considering that the region is under a high pressure by human activities mainly overgrazing and ongoing climate change, but at the same time suffers from poor conservation management (Jowkar et al., 2016). The three species studied in this manuscript and many other rare and local endemic species restricted to the upper limit of vascular plants of these mountains have no alternative habitats to ascend to under the impact of climate change. Therefore, high attention to the conservation of the biodiversity of these habitats is strongly recommended.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
HM and JN developed the idea. HM conducted the final morphological and molecular analyses. HM and JN wrote the draft of the manuscript and all authors contributed to the editing process. All authors contributed to the article and approved the submitted version.
Funding
This study was funded through the Austrian Science Fund (FWF P31898 to JN).
Acknowledgments
We would like to thank Dominik Metschina and Michael HJ Barfuss for DNA extraction and sequencing some of the materials.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1182073/full#supplementary-material
References
Agakbanjanz, O., Breckle, S.-W. (2002). “Plant diversity and endemism in high mountains of central Asia, the Caucasus and Siberia,” in Mountain biodiversity, Eds. Körner, C., Spehn, E. M. (London, UK: Routledge), pp. 117–127.
Alipour, S., Mehregan, I., Lidén, M. (2021). Dionysia splendens (Primulaceae), a new species from the Fars province of Iran. Edinb. J. Bot. 78, 1–5. doi: 10.24823/EJB.2021.397
Al-Shehbaz, I. A. (2012). A generic and tribal synopsis of the Brassicaceae (Cruciferae). Taxon 61, 931–954. doi: 10.1002/tax.615002
Al-Shehbaz, I. A., Beilstein, M. A., Kellogg, E. A. (2006). Systematics and phylogeny of the Brassicaceae (Cruciferae): an overview. Plant Syst. Evol. 259, 89–120. doi: 10.1007/s00606-006-0415-z
Andersson, I., Carlström, A., Franzén, R., Karlen, T. D., Nybom, H. (1983). A revision of the Aethionema saxatile complex (Brassicaceae). Willdenowia 13, 3–42.
Arshad, W., Lenser, T., Wilhelmsson, P. K. I., Chandler, J. O., Steinbrecher, T., Marone, F., et al. (2021). A tale of two morphs: developmental patterns and mechanisms of seed coat differentiation in the dimorphic diaspore model Aethionema arabicum (Brassicaceae). Plant J. 107, 166–181. doi: 10.1111/tpj.15283
Balali, G., Asri, Y., Afsharzadeh, S., Amoli, S. (2019). Nepeta azadkouhensis (Lamiaceae), a new alpine-subnival species from the central Alborz of Iran. Phytotaxa 395, 129–133. doi: 10.11646/395.2.6
Bhattacharya, S., Sperber, K., Özüdoğru, B., Leubner-Metzger, G., Mummenhoff, K. (2019). Naturally-primed life strategy plasticity of dimorphic Aethionema arabicum facilitates optimal habitat colonization. Sci. Rep. 9, 16108. doi: 10.1038/s41598-019-52520-y
Couvreur, T. L. P., Franzke, A., Al-Shehbaz, I. A., Bakker, F. T., Koch, M. A., Mummenhoff, K. (2010). Molecular phylogenetics, temporal diversification, and principles of evolution in the mustard family (Brassicaceae). Mol. Biol. Evol. 27, 55–71. doi: 10.1093/molbev/msp202
Darriba, D., Taboada, G. L., Doallo, R., Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9, 772–772. doi: 10.1038/nmeth.2109
Dirnböck, T., Essl, F., Rabitsch, W. (2011). Disproportional risk for habitat loss of high-altitude endemic species under climate change. Global Change Biol. 17, 990–996. doi: 10.1111/j.1365-2486.2010.02266.x
Doostmohammadi, M., Mirtadzadini, M. (2019). Paracaryum lalezarense (Boraginaceae), a new species from high alpine elevations of Lalezar mountains, in Iran. Phytotaxa 400:291–297. doi: 10.11646/phytotaxa.400.5.5
Doyle, J. J., Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15.
Franzke, A., Lysak, M. A., Al-Shehbaz, I. A., Koch, M. A., Mummenhoff, K. (2011). Cabbage family affairs: the evolutionary history of Brassicaceae. Trends Plant Sci. 16, 108–116. doi: 10.1016/j.tplants.2010.11.005
Hedge, I. C. (1965). “Aethionema W.T. Aiton” in Flora of Turkey and the East Aegean islands. Ed. Davis, P. H. (Edinburgh: Edinburgh University Press), 314–332.
Hedge, I. C. (1968). “Aethionema R. Br. & Moriera Boiss.” in Flora iranica. Eds. Hedge, I. C., Rechinger, K. H. (Graz: Academische Drucku. Verlagsanstalt), 100–110.
Hoorn, C., Mosbrugger, V., Mulch, A., Antonelli, A. (2013). Biodiversity from mountain building. Nat. Geosci. 6, 154–154. doi: 10.1038/ngeo1742
Hoorn, C., Perrigo, A., Antonelli, A. (2018). Mountains, climate and biodiversity (Oxford, UK: Wiley-Blackwell).
Huang, C. H., Sun, R., Hu, Y., Zeng, L., Zhang, N., Cai, L., et al. (2016). Resolution of Brassicaceae phylogeny using nuclear genes uncovers nested radiations and supports convergent morphological evolution. Mol. Biol. Evol. 33, 394–412. doi: 10.1093/molbev/msv226
IUCN. (2022). Guidelines for using the IUCN Red List categories and criteria version 15.1. Available at: https://nc.iucnredlist.org/redlist/content/attachment_files/RedListGuidelines.pdf [Accessed March 1, 2023].
Joly, S., Starr, J. R., Lewis, W. H., Bruneau, A. (2006). Polyploid and hybrid evolution in roses east of the Rocky Mountains. Am. J. Bot. 93, 412–425. doi: 10.3732/ajb.93.3.412
Jowkar, H., Ostrowski, S., Tahbaz, M., Zahler, P. (2016). The conservation of biodiversity in Iran: threats, challenges and hopes. Iranian Stud. 49, 1065–1077. doi: 10.1080/00210862.2016.1241602
Körner, C. (1995). “Alpine plant diversity: a global survey and functional interpretations,” in Arctic And alpine biodiversity: patterns, causes and ecosystem consequences. Eds. Chapin, F. S., Körner, C. (Berlin, Heidelberg: Springer), 45–62.
Lenser, T., Graeber, K., Cevik Ö, S., Adigüzel, N., Dönmez, A. A., Grosche, C., et al. (2016). Developmental control and plasticity of fruit and seed dimorphism in Aethionema arabicum. Plant Physiol. 172, 1691–1707. doi: 10.1104/pp.16.00838
Lewis, P. O. (2001). A likelihood approach to estimating phylogeny from discrete morphological character data. Syst. Biol. 50, 913–925. doi: 10.1080/106351501753462876
Maddison, W. P., Maddison, D. R. (2021) Mesquite: a modular system for evolutionary analysis. Available at: http://www.mesquiteproject.org.
Mahmoodi, M., Maassoumi, A. A., Noroozi, J. (2013). A new alpine species and a new record of Astragalus sect. Stereothrix (Fabaceae) from Iran, with comments on the phytogeography of the section. Willdenowia 43, 263–270. doi: 10.3372/wi.43.43205
Mérai, Z., Graeber, K., Wilhelmsson, P., Ullrich, K. K., Arshad, W., Grosche, C., et al. (2019). Aethionema arabicum: a novel model plant to study the light control of seed germination. J. Exp. Bot. 70, 3313–3328. doi: 10.1093/jxb/erz146
Meyer, F. K. (1973). Conspectus der “Thlaspi” Arten Europas, Afrikas und Vorderasiens. Feddes Repert 84, 449–469. doi: 10.1002/fedr.19730840503
Miller, M. A., Pfeiffer, W., Schwartz, T. (2010). “Creating the CIPRES science gateway for inference of large phylogenetic trees,” in IEEE Proceedings 2010 Gateway Computing Environments Workshop (GCE). 1–8. doi: 10.1109/GCE.2010.5676129
Minh, B. Q., Nguyen, M. A. T., Von Haeseler, A. (2013). Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30, 1188–1195. doi: 10.1093/molbev/mst024
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., Von Haeseler, A., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534. doi: 10.1093/molbev/msaa015
Mittermeier, R. A., Turner, W. R., Larsen, F. W., Brooks, T. M., Gascon, C. (2011). “Global biodiversity conservation: the critical role of hotspots,” in Biodiversity hotspots: distribution and protection of conservation priority areas. Eds. Zachos, F. E., Habel, J. C. (Berlin, Heidelberg: Springer), 3–22.
Moazzeni, H., Al-Shehbaz, I., German, D., Assadi, M., Müller, J., Joharchi, M., et al. (2018). A taxonomic revision of the genus Aethionema s.l. (Brassicaceae) in Iran. Phytotaxa 356, 241–266. doi: 10.11646/phytotaxa.356.4.1
Moazzeni, H., Assadi, M., Zare, G., Mirtadzadini, M., Al-Shehbaz, I. (2016). Taxonomic novelties in Erysimum for the flora of Iran: E. polatschekii, a new alpine endemic and E. scabrum, a new record. Phytotaxa 269, 47. doi: 10.11646/phytotaxa.269.1.6
Moazzeni, H., Zarre, S., Al-Shehbaz, I. A., Mummenhoff, K. (2007). Seed-coat microsculpturing and its systematic application in Isatis (Brassicaceae) and allied genera in Iran. Flora 202, 447–454. doi: 10.1016/j.flora.2006.10.004
Moazzeni, H., Zarre, S., Al-Shehbaz, I. A., Mummenhoff, K. (2010). Phylogeny of Isatis (Brassicaceae) and allied genera based on ITS sequences of nuclear ribosomal DNA and morphological characters. Flora 205, 337–343. doi: 10.1016/j.flora.2009.12.028
Moazzeni, H., Zarre, S., Assadi, M., Joharchi, M. R., German, D. (2014). Erysimum hezarense, a new species and Rhammatophyllum gaudanense, a new record of Brassicaceae from Iran. Phytotaxa 175, 241–248. doi: 10.11646/phytotaxa.175.5.1
Mohammadin, S., Peterse, K., Van De Kerke, S. J., Chatrou, L. W., Dönmez, A. A., Mummenhoff, K., et al. (2017). Anatolian origins and diversification of Aethionema, the sister lineage of the core Brassicaceae. Am. J. Bot. 104, 1042–1054. doi: 10.3732/ajb.1700091
Mummenhoff, K., Franzke, A., Koch, M. (1997). Molecular phylogenetics of Thlaspi s.l. (Brassicaceae) based on chloroplast DNA restriction site variation and sequences of the internal transcribed spacers of nuclear ribosomal DNA. Can. J. Bot. 75, 469–482. doi: 10.1139/b97-051
Noroozi, J., Ajani, Y. (2013). A new alpine species of Nepeta sect. Capituliferae (Labiatae) from northwestern Iran. Novon 22, 297–303. doi: 10.3417/2012022
Noroozi, J., Ajani, Y., Nordenstam, B. (2010). A new annual species of Senecio (Compositae-Senecioneae) from subnival zone of southern Iran with comments on phytogeographical aspects of the area. Compositae Newslett. 48, 43–62.
Noroozi, J., Khalvati, S., Nafisi, H., Kaveh, A., Nazari, B., Zare, G., et al. (2021). Endemics determine bioregionalization in the alpine zone of the irano-Anatolian biodiversity hotspot (South-West Asia). Alp. Bot. 131, 177–186. doi: 10.1007/s00035-021-00266-7
Noroozi, J., Moser, D., Essl, F. (2016). Diversity, distribution, ecology and description rates of alpine endemic plant species from Iranian mountains. Alp. Bot. 126, 1–9. doi: 10.1007/s00035-015-0160-4
Noroozi, J., Talebi, A., Doostmohammadi, M., Rumpf, S. B., Linder, H .P., Schneeweiss, G. M. (2018). Hotspots within a global biodiversity hotspot - areas of endemism are associated with high mountain ranges. Sci. Rep. 8, 10345. doi: 10.1038/s41598-018-28504-9
Noroozi, J., Pirani, A., Moazzeni, H., Mahmoodi, M., Zare, G., Noormohammadi, A., et al. (2020). The new locally endemic genus Yazdana (Caryophyllaceae) and patterns of endemism highlight the high conservation priority of the poorly studied Shirkuh Mountains (central Iran). J. Syst. Evol. 58, 339–353. doi: 10.1111/jse.12575
Noroozi, J., Talebi, A., Doostmohammadi, M., Manafzadeh, S., Asgarpour, Z., Schneeweiss, G. M. (2019a). Endemic diversity and distribution of the Iranian vascular flora across phytogeographical regions, biodiversity hotspots and areas of endemism. Sci. Rep. 9, 12991. doi: 10.1038/s41598-019-49417-1
Noroozi, J., Zare, G., Sherafati, M., Mahmoodi, M., Moser, D., Asgarpour, Z., et al. (2019b). Patterns of endemism in Turkey, the meeting point of three global biodiversity hotspots, based on three diverse families of vascular plants. Front. Ecol. Evol. 7. doi: 10.3389/fevo.2019.00159
Pahlevani, A. H., Amini Rad, M. (2019). Two new alpine species of Euphorbia (Euphorbiaceae) from Iran. Kew Bull. 74, 49. doi: 10.1007/s12225-019-9830-5
Parisod, C. (2022). Plant speciation in the face of recurrent climate changes in the Alps. Alp. Bot. 132, 21–28. doi: 10.1007/s00035-021-00259-6
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Sandel, B., Arge, L., Dalsgaard, B., Davies, R., Gaston, K., Sutherland, W., et al. (2011). The influence of late Quaternary climate-change velocity on species endemism. Science 334, 660–664. doi: 10.1126/science.1210173
Taberlet, P., Gielly, L., Pautou, G., Bouvet, J. (1991). Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol. 17, 1105–1109. doi: 10.1007/BF00037152
Testolin, R., Attorre, F., Borchardt, P., Brand, R. F., Bruelheide, H., Chytrý, M., et al. (2021). Global patterns and drivers of alpine plant species richness. Global Ecol. Biogeogr. 30, 1218–1231. doi: 10.1111/geb.13297
Vanderplank, S. E., Moreira-Muñoz, A., Hobohm, C., Pils, G., Noroozi, J., Clark, V. R., et al. (2014). “Endemism in mainland regions – case studies,” in Endemism in vascular plants. Ed. Hobohm, C. (Dordrecht: Springer), 205–308.
Keywords: Brassicaceae, conservation, endemism, phylogeny, species nova, taxonomy
Citation: Moazzeni H, Mahmoodi M, Jafari M, Schneeweiss GM and Noroozi J (2023) Underestimated diversity in high elevations of a global biodiversity hotspot: two new endemic species of Aethionema (Brassicaceae) from the alpine zone of Iran. Front. Plant Sci. 14:1182073. doi: 10.3389/fpls.2023.1182073
Received: 08 March 2023; Accepted: 13 April 2023;
Published: 26 May 2023.
Edited by:
Michael Eric Schranz, Wageningen University and Research, NetherlandsReviewed by:
Nora Walden, Wageningen University and Research, NetherlandsJudita Zozomova-Lihova, Institute of Botany (SAS), Slovakia
Copyright © 2023 Moazzeni, Mahmoodi, Jafari, Schneeweiss and Noroozi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hamid Moazzeni, hmoazzeni@um.ac.ir; Jalil Noroozi, jalil.noroozi@univie.ac.at
†ORCID: Hamid Moazzeni, orcid.org/0000-0002-2406-2666
Mohammad Mahmoodi, orcid.org/0000-0003-3942-2086
Mohammad Jafari, orcid.org/0000-0002-9555-4756
Gerald M. Schneeweiss, orcid.org/0000-0003-2811-3317
Jalil Noroozi, orcid.org/0000-0003-4124-2359
 Hamid Moazzeni
Hamid Moazzeni Mohammad Mahmoodi
Mohammad Mahmoodi Mohammad Jafari
Mohammad Jafari Gerald M. Schneeweiss
Gerald M. Schneeweiss Jalil Noroozi
Jalil Noroozi